Abstract
Automated microscopy was introduced two decades ago and has become an integral part of the discovery process as a high-content screening platform with noticeable challenges in executing cell-based assays. It would be of interest to use it to screen for reversers of a transformed cell phenotype. In this report, we present data obtained from an optimized assay that identifies compounds that reverse a transformed phenotype induced in NIH-3T3 cells by expressing a novel oncogene, KP, resulting from fusion between platelet derived growth factor receptor alpha (PDGFRα) and kinase insert domain receptor (KDR), that was identified in human glioblastoma. Initial image acquisitions using multiple tiles per well were found to be insufficient as to accurately image and quantify the clusters; whole-well imaging, performed on the IN Cell Analyzer 2000, while still two-dimensional imaging, was found to accurately image and quantify clusters, due largely to the inherent variability of their size and well location. The resulting assay exhibited a Z′ value of 0.79 and a signal-to-noise ratio of 15, and it was validated against known effectors and shown to identify only PDGFRα inhibitors, and then tested in a pilot screen against a library of 58 known inhibitors identifying mostly PDGFRα inhibitors as reversers of the KP induced transformed phenotype. In conclusion, our optimized and validated assay using whole-well imaging is robust and sensitive in identifying compounds that reverse the transformed phenotype induced by KP with a broader applicability to other cell-based assays that are challenging in HTS against chemical and RNAi libraries.
Introduction
Cancer is a deadly disease characterized by an uncontrolled cellular growth, invasion, and metastasis believed to be associated with activation of oncogenes.1–5 Oncogenes have been shown to induce transformed phenotypes in cultured cells and form foci, clusters, cocoons, or as often referred to as spheroids, and shown to grow to sizes ranging from 500 to over 1,000 μm in diameter. The resulting clusters have been imaged either by simple phase-contrast or confocal microscopy stained for several markers.6–8 The consequence of oncogene activation has provided a rational approach toward molecular targeted therapies with very few drugs that reverse the transformed phenotype as the majority of the approved drugs in the clinic are cytotoxic.4
Attempts to screen for inhibitors of the transformed phenotype have not been successful, even for those oncogenes where a function could be assayed and screened using high-throughput methodologies.5 It has also been argued that the lack of success could be associated with the fact that performing cell-based assays for screening using classical two-dimensional (2D) cell monolayer models are prone to artifacts as they are highly artificial and do not closely reflect or mimic physiological conditions and hence the lack of novel small molecule therapeutics for cancer.8–12 Nonetheless, several research groups have reported on novel assay technologies to screen three-dimensional (3D) cells in microtiter plate formats.8–10 Friedrich and co-workers reported a colorimetric method based on the quantification of the intracellular activity of alkaline phosphatase where p-nitrophenyl phosphate is hydrolyzed to p-nitrophenol and detected at 405 nm.8,10 Sittampalam and co-workers, on the other hand, also used a colorimetric method based on the reduction of tetrazolium salt to a formazan by-product by live cells only and measured at 490 nm.9 Both approaches successfully screen for cytotoxic compounds against the 3D spheroids or to study differential resistance of cancer cells upon formation of 3D spheroids to known cytotoxic agents, as their low-content approach would not allow for the identification of reversers of the transformed phenotype as these would score as inactive in these assay formats. Therefore, it is of great interest to develop assays that would detect and quantify reversal of a transformed phenotype achieved by compounds with minimal cytotoxic effects on cellular viability.
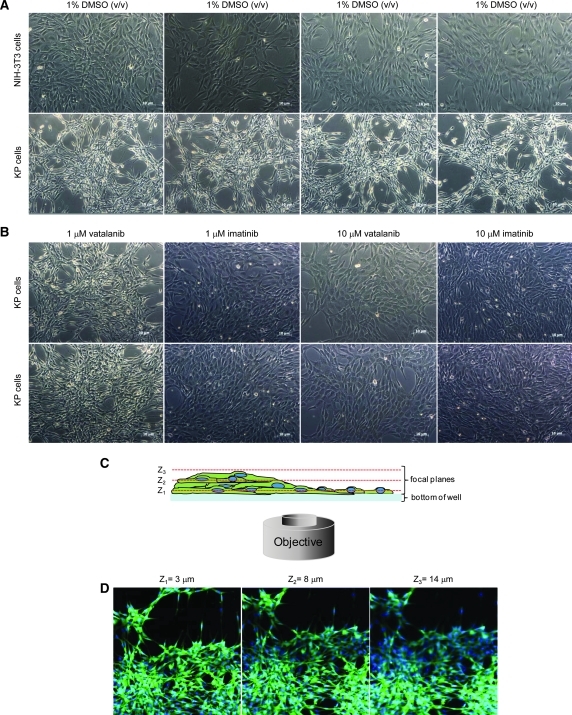
We have recently identified a novel oncogene in human glioblastoma through sequencing and refer to it as KP, a fusion protein between PDGFRα and KDR, which behaves as a constitutively active form of tyrosine kinase in vitro, propagating enhanced PDGFRα signaling through a combination of the MAPK and PI3K pathways.13 When introduced into NIH-3T3 cells, the resulting KP cells have a morphologically transformed phenotype characterized by the formation of clusters as imaged by brightfield microscopy (Fig. 1A). Interestingly, the KP-transformed phenotype can be reversed upon addition of the PDGFRα kinase inhibitors vatalanib or imatinib (Fig. 1B) and provides a controlled system in which to develop and establish assay methodologies to detect those specific compounds that reverse the KP-transformed phenotype. The assay can in principle be used to screen for compounds that reverse transformed phenotype induced by other oncogenes and may identify downstream signaling effectors in RNAi screening.
Fig. 1.
The transformed KP cells form clusters. (A) Brightfield images of NIH-3T3 and KP cells treated with 1% DMSO (v/v); resulting images show the transformed phenotype as compared with parental cell line that lacks any clusters. (B) Brightfield images of KP cells treated with either vatalanib or imatinib at 1 and 10 μM concentrations. Images clearly show reversal of KP-transformed phenotype by reducing clusters especially at 1 μM imatinib treatment as compared with the control untransformed NIH-3T3 cells. Scale bar: 10 μm (C) Schematic representation of KP cell captures in three different Z focal planes demonstrating the clustering nature of the KP cells. (D) Images of KP cells acquired by the IN Cell Analyzer 3000 in GFP (green) and Hoechst (blue) channels at three different focal planes; recapitulating the nature of the KP cells as per drawn schematics. DMSO, dimethyl sulfoxide; GFP, green fluorescent protein. Color images available online at www.liebertonline.com/adt.
In this article, we develop and validate a cell-based assay using a high-content screening (HCS) approach measuring cellular green fluorescent protein (GFP) expression in the green channel and nuclei stained with Hoechst in the blue channel, and as combined means to quantify and assess reversal of transformed phenotype in the KP-expressing NIH-3T3 cells. We compared image acquisition from the three imagers available in our facility and found that the IN Cell Analyzer 2000 (INCA2000) allows high-resolution whole-well imaging and characterized the geographical growth location of the clusters in the well. We developed and validated an analysis method allowing the quantification of the reversal of a transformed phenotype by measuring nuclei enrichment in any given cluster. The assay was further validated in a pilot screen against a focused library of known compounds and only identified those affecting the PDGFRα function. To our knowledge, this is the first report on successful development of a cell-based assay using HCS to identify compounds that reverse oncogenic transformed phenotype in viable cells.
Materials and Methods
Reagents
Dulbecco's modified Eagle's medium was purchased from American Tissue Culture Collection (ATCC). Penicillin, streptomycin, phosphate-buffered saline (PBS) without Mg2+, Ca2+, 0.25% Trypsin/EDTA, and Hoechst 33342 were purchased from Invitrogen Life Sciences. Heat-inactivated calf serum was purchased from Colorado Serum Co. About 32% paraformaldehyde (w/v) (PFA) was purchased from Electron Microscopy Science. Triton X-100 was purchased from Sigma-Aldrich.
Cell Culture
NIH-3T3-KP cells expressing GFP (KP cells) from the parental cell line NIH-3T3 (ATCC: CRL-1658) were prepared as described previously.13 KP cells and the parental NIH-3T3 cells were cultured under a humidified atmosphere at 37°C/5% CO2–95% air in complete Dulbecco's modified Eagle's medium (ATCC: 30-2002) containing 10% heat-inactivated calf serum (Colorado Serum Co.: CS1334), 100 units/mL penicillin, and 100 μg/mL streptomycin.
Image Acquisition
Images were acquired using the following three IN Cell Analyzer platforms (GE Healthcare): the IN Cell Analyzer 1000 (INCA1000), the INCA2000, and the IN Cell Analyzer 3000 (INCA3000).
The INCA1000 is an automated epifluorescence microscope modeled after the Nikon TE2000. The 10 × magnifying objective with flat field and apochromatic corrections (Plan Apo) and 0.45 numerical aperture (NA) was used. Images of GFP expressed in the cytoplasm were acquired in the FITC channel using 475/20 nm excitation (475 nm excitation peak; ± 20 nm bandpass), 535/50 nm emission, and Q505LP dichroic at an exposure time of 200 ms. Images of nuclei stained with Hoechst were acquired in the 4′,6-diamino-2-phenylindole (DAPI) channel using 360/40 nm excitation, 535/50 nm emission, and Q505LP dichroic at an exposure time of 200 ms. Four tiles were imaged per well covering 33% of the well, with an acquisition time of 16 s per well, and were collected with a Roper/Princeton Instruments CoolSnap HQ charged-coupled device (CCD) camera (12-bit, 6.45 μm pixels) using no binning. In total, acquisition time was 103 min per 384-well microtiter plate.
The INCA2000 is a wide-field automated epifluorescence microscope equipped with a large-chip CCD camera allowing for whole-well imaging. The 4 × magnifying objective with Plan Apo, chromatic aberration-free infinity (CF160) and 0.20NA was used. Images were acquired using a custom-made polychroic. Images of GFP were acquired in the FITC channel using 490/20 nm excitation, 525/36 nm emission, and utilizing the newly available 2D deconvolution feature. GFP was imaged using an exposure time of 500 ms. Images of Hoechst-stained nuclei in the DAPI channel were acquired using 350/50 nm excitation and 455/58 nm emission at an exposure time of 100 ms. One tile was imaged per well covering 100% of the well, with an acquisition time of 4 s per well and total of 24 min per 384-well microtiter plate.
The INCA3000 is a line-scanning confocal automated microscope that allows for high image resolution. This laser-scanning confocal imager comprises two laser light sources, three excitation lines, and three highly sensitive 12-bit CCD cameras allowing simultaneous imaging of 3 fluorophores with continuous laser-based autofocus. The 40 × magnifying objective with CFI60 and 0.6NA was used. GFP images and nuclear images were acquired using 488 nm excitation/535 nm emission and 364 nm excitation/450 nm emission, respectively, at an exposure time of 1.5 ms for both channels. Nine tiles were imaged per well covering 88% of well, with an acquisition time of 31 s per well, and were collected with two separate Roper/Princeton Instruments CCD cameras (12-bit, 6.45 μm pixels) using 2 × 2 binning. In total, acquisition time was 196 min per 384-well microtiter plate.
Image Analysis
Images acquired by the three IN Cell Analyzers were analyzed using the Developer Toolbox 1.7 software (GE Healthcare). For INCA1000 and INCA3000 images, object and nuclear segmentations were performed on the channel corresponding to nuclei staining and intensity segmentation was performed on the channel corresponding to the cytoplasm staining. Postprocessing, including sieving, dilation, and erosion, was used to obtain optimal outline of desired objects and intensities. Clusters were identified using an exclusion parameter based on perimeter and area measurements. INCA2000 images were processed using object-based segmentation. After postprocessing, clusters were identified using an exclusion parameter based on the cluster area on the cytoplasm channel and were defined as having an area >3,000 μm2. An overlap of both channels allowed us to extract parameters such as the number of clusters, the cluster area, the number of nuclei in clusters, the total number of nuclei, and the percentage nuclei in clusters. Using such parameters, we were able to calculate the nuclear enrichment factor (NEF). Specifically, the NEF is calculated by using a complex algorithm that determines the total number of nuclei within clusters. The number is then compared with the total number of nuclei to give a percentage of total nuclei. Finally, the percentage is multiplied by 100 to give a distinct separation between controls.
Assessment of Resolution by 2D Deconvolution
During acquisition with INCA2000, the FITC channel for GFP and the DAPI channel for nuclei were captured with or without 2D deconvolution using 4 × magnifying objective. The line scan analysis was performed with imageJ (http://rsb.info.nih.gov/ij/) by drawing a one-pixel line across several Hoechst-stained nuclei present in the DAPI channel. Plot profiles representing gray levels at each point across the line were measured.
Cell-Based Assay Development and Validation
To optimize cell seeding densities, KP cells were trypsinized and seeded in 384-well microtiter plates (Corning, #3985) at final cell densities of 1,000, 2,000, 3,000, or 5,000 cells per well in 45 μL of complete media using an automated Multidrop384 dispenser (Thermo Scientific). The plates were then incubated at 37°C and 5% CO2 in a Steri-cult incubator (Thermo Scientific) for 48, 72, or 96 h. At each time point, the medium was aspirated using an automated plate washer ELx405 (Biotek Instruments) and 50 μL of 4% PFA (w/v) in PBS was added using the Multidrop384. After incubating for 20 min, PFA was removed and the cells were washed twice with PBS and resuspended in 50 μL PBS. The nucleus of the cells was not stained. After the plates were sealed, image acquisition for GFP was performed on the INCA1000 using a 10 × magnifying objective.
After selecting a cell density of 2,000 cells per well as the optimal seeding density for the assay, KP cells were tested against varying concentrations of dimethyl sulfoxide (DMSO) to assess compound carrier effect on cluster formation. Cells were seeded in 45 μL of media using the Multidrop384. After 24 h of incubation at 37°C and 5% CO2 in the Steri-cult incubator, KP cells were treated with 5 μL of 1%, 5%, or 10% DMSO (v/v) to reach a final concentration of 0.1%, 0.5%, or 1% (v/v). Afterward, cells were further incubated for 48 h in Steri-cult. After fixation with PFA, cells were stained with 1 μM Hoechst in PBS containing 0.05% Triton X-100 (v/v) for 1 h in the dark. Hoechst solution was removed and the cells were washed twice with PBS and resuspended in 50 μL PBS. After the plates were sealed, image acquisition for GFP and nuclei was conducted as described above (data not shown).
After the establishment of assay conditions, a sensitivity assessment was performed as a dose–response run against the following five known PDGFRα inhibitors and two epidermal growth factor receptor (EGFR) inhibitors: imatinib,14 vatalanib,15 PD166326,16 SKI212221,16 SKI217520,16 gefitinib, and erlotinib. The dose–response was assessed using 12-point doubling dilutions with 10 μM compound concentration as the upper limit. Serial doubling dilutions of compounds were preplated in an intermediate 384-well poly-propylene plate (Thermo Scientific), and 5 μL of each dilution was transferred to the assay plates (Corning, #3985) using a PP-384-MM Personal Pipettor with a custom 384 head (Apricot Designs). Controls consisted of 1% DMSO (v/v) (high control) and 10 μM imatinib in 1% DMSO (v/v) (low control). The Z′ factor was used to assess assay performance.17 After the plates were sealed, image acquisition for GFP and nuclei was conducted on the INCA 2000 as described above. NEF was calculated for each compound as described above. Dose–response curves were fitted using a logistic 4-parameter equation of SigmaPlot (Systat Software Inc.) and IC50 values were determined. The optimized assay workflow is summarized in Table 1.
Table 1.
Workflow of the Optimized KP Assay
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Library compounds | 5 μL | 12 doubling dilutions from 10 or 1 μM in 1% DMSO (v/v) |
| 2 | Positive control | 5 μL | 10 μM imatinib in 1% DMSO (v/v) |
| 3 | Negative control | 5 μL | 1% DMSO (v/v) |
| 4 | Cell plating | 45 μL | 2,000 NIH3T3-KP cells in tissue culture media |
| 5 | Incubation time | 48 h | 37°C, 5% CO2 |
| 6 | Fix | 50 μL | 4% paraformaldehyde (w/v) |
| 7 | Incubation time | 20 min | At room temperature |
| 8 | Nuclear staining | 50 μL | 1 μM Hoechst, 0.05% Triton X-100 (v/v) |
| 9 | Incubation time | 1 h | At room temperature, in the dark |
| 10 | Wash | 50 μL | Twice with 1 × PBS |
| 11 | Assay readout | 350/455 nm and 490/525 nm (ex/em) | INCA2000 automated microscope |
| 12 | Image Analysis | – | Multiparametric analysis using Developer Toolbox 1.7 |
1. Dispensing on the PP-384-M Personal Pipettor using a custom 384 head
2. Dispensing on the PP-384-M Personal Pipettor using a custom 384 head
3. Dispensing on the PP-384-M Personal Pipettor using a custom 384 head
4. Dispensing with Multidrop 384 bulk liquid dispenser
5. 384-well assay plates stored in the Stericult; an automated temperature controlled incubator
6 & 10. Aspirating on the Biotek EL × 405 washer
6, 8, and 10. Dispensing with Multidrop 384 bulk liquid dispenser
11. Readout performed on automated platform; 4 s per well with total imaging time of 24 min per 384-well microtiter plate
12. Analysis of GFP and Hoechst channel, 8 min per 384-well microtiter plate
DMSO, dimethyl sulfoxide; GFP, green fluorescent protein; INCA2000, IN Cell Analyzer 2000; PBS, phosphate-buffered saline.
Focused Chemical Library
The focused chemical library used in this study was assembled by purchasing 58 compounds targeting various signaling pathways commercially available from LC laboratories, Sigma-Aldrich, Cayman Chemical, A.G. Scientific, and Biaffin GmbH, and is referred to in this article as the focused library (Supplementary Table S1, Supplementary Data are available online at www.liebertonline.com/adt).
Pilot Screen for Compounds that Reverse the KP Oncogenic Phenotype
A pilot screen was conducted with the 58 compounds from the focused library in duplicate, using 12-point doubling dilutions with 10 μM compound concentrations in 1% DMSO (v/v) as the upper limit. Serial doubling dilutions of compounds were preplated in an intermediate plate, and 5 μL of each dilution was transferred to the assay plates using the PP-384-MM Personal Pipettor. Controls consisted of 1% DMSO (v/v) (high control) and 10 μM imatinib in 1% DMSO (v/v) (low control). The assay was performed as described in Table 1. Dose–response curves for each data set were fitted separately using a logistic 4-parameter equation of SigmaPlot. IC50 values were averaged using replicate values and represented as IC50 plus or minus standard error.
Results
Imaging Cellular Clusters in 384-Well Microtiter Plates
Although brightfield microscopy is arguably the most common and simplest technique used in studying 3D cellular growth in culture (Fig. 1A), it lacks the resolution required to perform and quantify 3D cellular clusters for HCS. Therefore, to better observe the cytoplasm of cells, we used fluorescent GFP protein that was encoded by the plasmid used to express the oncogene KP into NIH-3T3 cells (KP cells).13 The resulting images taken at three different focal planes (3, 8, and 14 μm), using the INCA3000, clearly show that the KP-transformed cells form transformed clusters with cells pilling up on top of each other (Fig. 1C, D). As expected, the PDGFRα inhibitor vatalanib and the BCR-ABL inhibitor imatinib (which also inhibit PDGFRα16) fully revert the KP-transformed phenotype back to the parental morphology of the untreated NIH-3T3 cells observed using brightfield microscopy (Fig 1B).
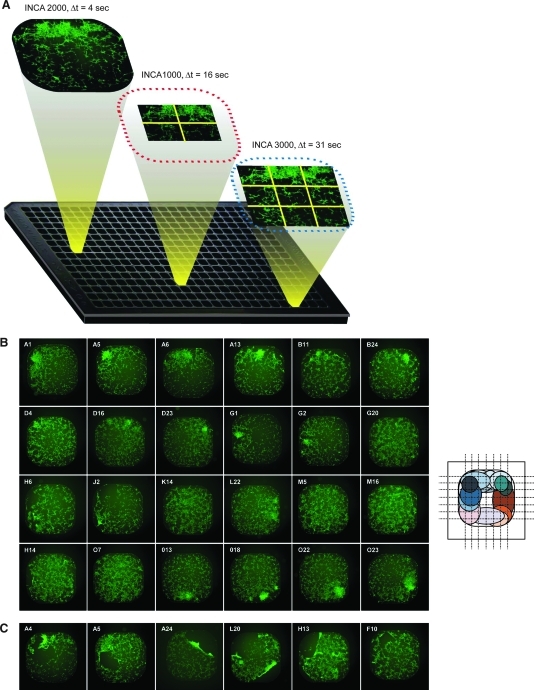
We initially optimized the number of cells plated per well and the concentration of DMSO used in the screen. In comparison to control NIH-3T3 cells, KP cells demonstrated the ability to grow and form distinct clusters at 2,000 cells per well while not forming large clusters as seen in the 3,000 and 5,000 cell densities (Supplementary Fig. S1). In comparison, lack of cell growth or distinct cluster formation was observed at the cell density of 1,000 cells per well. As such, the optimal cell seeding density was determined to be 2,000 cells per well. KP cells were then seeded in 384-well microtiter plates, grown for 2 days in the presence of 1% DMSO (v/v), and then fixed and stained with Hoechst. The plates were then imaged using three automated microscopes: the INCA1000, INCA3000, and the INCA2000 (Fig. 2A). The specifications of each of the three microscopes are summarized in Table 2. The INCA1000, a wide-field microscope equipped with a 10 × magnifying objective, allowed four tiles to be imaged at a fixed location within the well resulting in a well coverage of up to 33%. The INCA3000, a laser scanning confocal microscope equipped with a 10 × magnifying objective, allowed nine tiles to be imaged also at fixed locations, resulting in coverage of 88%, almost three-fold higher than the INCA1000. The INCA2000, a widefield microscope with a much larger camera size and a 4 × magnifying objective, allow imaging of the same assay plates and coverage of the whole well at 100% (Fig. 2A). The availability of an imaging platform such as the INCA2000 has allowed us for the first time to investigate cell cluster growth patterns and positional preferences in 384-well microtiter plates. Close examination of the obtained whole well images demonstrated that a dominating overall pattern of growth location with an outer well preference for growth with random geographical positions; very few wells showed a spread of growth across the well. Figure 2B summarizes an ensemble of obtained images. In some instances, clear detachment, loss of clusters, and cluster folding on each other were observed leaving some regions of the well devoid of cell structures. These events are most likely to be due to a combination of progressive transformation induced loss of adherence with the mechanical manipulation of cells during fixing and staining (Fig. 2C). These observations support the concept of whole-well imaging for cell-based assays, since their absence in some areas of the well partially reports the residual events, which in turn could be misinterpreted as reversal of transformed phenotype, especially since the resulting images from both the INCA1000 and 3000 were truncated.
Fig. 2.
Comparison of the three imaging platforms: advantages of whole-well imaging. (A) Imaging of KP cell clusters in 384-well microtiter plates using the three automated microscope platforms: the IN Cell Analyzer 3000 with nine tiles per well imaged in 31 s capturing 88% of well, the IN Cell Analyzer 1000 with four tiles per well imaged in 16 s capturing 33% of the well, and the INCA2000 with one tile per well imaged in 4 s capturing 100% of the well. Acquisition times described pertain to the capturing of both GFP (green) and Hoechst (not shown) channels. In addition, the 2D deconvolution feature was used to capture the GFP channel on the INCA2000. (B) Random and peripheral growth of KP cell clusters in 384-well microtiter plates: Evaluation of the random cluster distribution by whole-well imaging. Representative GFP images of cluster location, acquired by the INCA2000 at 4 × magnifying objective were retrieved for 24-wells from five different 384-well microtiter plates. The schematic view summarizes the dominating peripheral cluster locations in the wells with complete absence of centralized growth. (C) Representative images showing detached or missing clusters as a result of fixing and staining protocols. INCA2000, IN Cell Analyzer 2000; 2D, two-dimensional. Color images available online at www.liebertonline.com/adt.
Table 2.
Technical Specifications and Performance of the Three Imaging Platforms: the IN Cell Analyzer 1000, the IN Cell Analyzer 2000, and the IN Cell Analyzer 3000
| IN Cell Analyzer 1000 | IN Cell Analyzer 2000 | IN Cell Analyzer 3000 | |
|---|---|---|---|
| Illumination | Widefield | Widefield | Linescan laser confocal |
| Light source | Xenon | Enhanced metal arc | Argon ion and HeNe laser |
| Light power | 100 W | 200 W | 100 mW |
| Image size (pixels) | 1,392 × 1,040 | 2,048 × 2,048 | 1,280 × 1,280 |
| Field of view (mm) | 0.90 × 0.68 | 3.79 × 3.79 | 0.75 × 0.75 |
| Magnification | 10× | 4× | 40× |
| Depth of field (μm) | 3 | 18 | 1.6 (in addition to the 8 of confocal) |
| Exposure time for GFP channel (ms) | 200 | 500 | 1.5 |
| Exposure time for Hoescht channel (ms) | 200 | 100 | 1.5 |
| Fields acquired | 4 | 1 | 9 |
| Coverage per well | 33 | 100 | 88 |
| Mean nuclei count for fields acquired per well | 755 | 2,960 | 1,945 |
| Acquisition time (min, per 384-well microtiter plate) | 103 | 24 | 196 |
| Analysis time (min) | 20 | 8 | 20 |
| Data storage (GB) | 12 | 6 | 8 |
Acquisition time shown for the INCA2000 is based on imaging of the GFP channel using two-dimensional deconvolution.
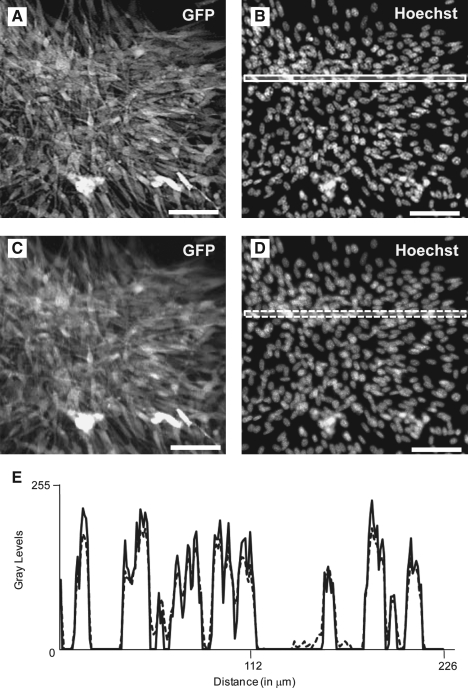
It is conceivable that whole-well imaging at low magnification would be inadequate for high-content assays that require high-resolution images. Therefore, we evaluated the image sharpness obtained with the INCA2000. We compared images acquired using the INCA2000 with (Fig. 3A, B) or without (Fig. 3C, D) the 2D deconvolution feature, which corrects blurring caused by the magnifying objective lens. We assessed the gray levels histogram for the entire image (data not shown) and found that the gray levels were more spread when 2D deconvolution was performed, suggesting better contrast. We further evaluated the resolution achieved with the INCA2000 by line scan analysis (Fig. 3E). The representative line scan plot for the Hoechst signal, for both deconvolved and nondeconvolved images, shows that maximal values for 2D deconvolution were often higher than without deconvolution. Moreover, the peaks of intensity were sharper when 2D deconvolution was used. However, the image analysis algorithm was not affected by including or excluding the 2D deconvolution feature on the Hoechst images (data not shown). Thus, 2D deconvolution feature was not used for the DAPI channel during acquisition. Overall, this shows that the signal-to-noise ratio (S/N) was enhanced with 2D deconvolution, resulting in a sharper image that would allow a more accurate measurement for subcellular features.
Fig. 3.
Assessment of resolution using 2D deconvolution during image acquisition with the INCA2000 at a 4 × magnifying objective. Magnified images (monochrome overlaid) of KP cells acquired by the INCA2000 at 4 × magnifying objective. Scale bar: 100 μm. (A, B) GFP and Hoechst channels captured with 2D deconvolution. (C, D) GFP and Hoechst channels captured without 2D deconvolution. (E) Representative line scan analysis of images acquired with (white solid line) (B) or without (white dashed line) (D) 2D deconvolution corresponding to gray levels across the lines and with minimal resolution differences of the obtained lines.
Other advantages of whole-well imaging and typical bottlenecks in HCS are acquisition times and file sizes. We compared the performance of the three platforms for these two critical parameters (Table 2). The exposure times used for each platform were selected on the basis of optimal S/N. Based on the exposure times and settings such as the number of fields imaged per well, we estimated the acquisition time per 384-well microtiter plate for each imaging platform. Acquisition time averaged 196 min per 384 microtiter plate for the INCA3000 due mainly to the highest number of fields imaged per well, whereas acquisition times on the INCA1000 and the INCA2000 were estimated to be 103 and 24 min, respectively. As for the file sizes, the most expensive platform was the INCA1000 with 12 GB per plate, followed by the INCA3000 with 8 GB per plate and lastly the INCA2000 with 6 GB per plate. Therefore, there is a clear advantage in using the INCA2000 whole-well imaging platform for both an acquisition time of 24 min per plate and translating into ∼18 days of imaging 1,074 plates the current size of our chemical library, and ∼7 TB of data generated for the same screen size.
Analyzing Images of Cellular Clusters and Quantifying Reversal of Transformed Phenotype
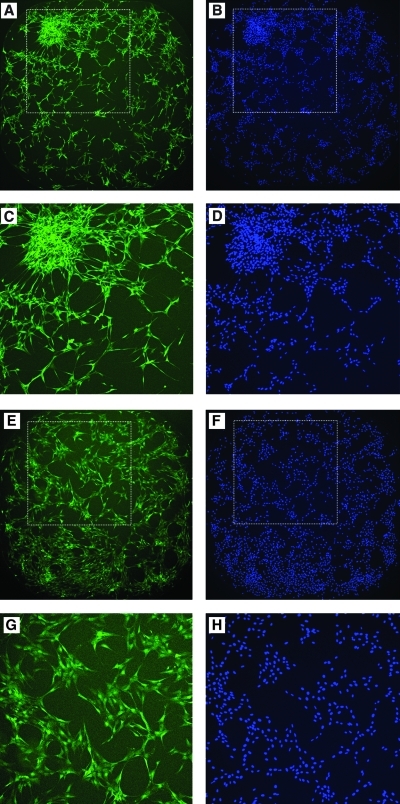
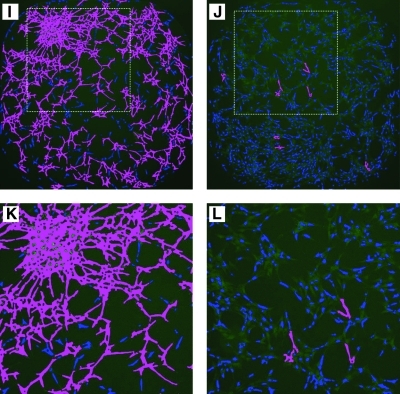
Oncogenic transformation often leads to the loss of cell-to-cell contact inhibition18,19 and results in cells piling up on top of each other forming transformed clusters as observed in Figure 1C and D. For the purposes of developing a simple and robust cell-based assay to screen for modulators of transformed phenotype, we opted to concentrate mainly on two striking features: (1) spacial distribution of nuclei as seen by Hoechst staining readout and (2) cellular population overlap/fusion and morphological changes as seen by GFP signal output. Combining both Hoechst staining and GFP signal output readouts, we could reproduce the reversal of KP-transformed phenotype depicted in Fig. 1A in 384-well microtiter plates imaged using the INCA2000 as follows: the imatinib (at a dose of 10 μM concentration in 1% DMSO [v/v])-treated KP cells exhibited flattened cell morphology with minimal overlap between cell populations as seen by GFP (Fig. 4E, G) and strikingly well scattered and individualized nuclei as seen by Hoechst staining (Fig. 4F, H); the residual phenotype post imatinib treatment is very similar to the untreated NIH-3T3 cells (Fig. 1B). By contrast, the untreated KP cells exhibited differential cluster sizes of overlapping cells as seen by GFP (Fig. 4A, C) and an uneven distribution and spacial scattering of the nuclei as seen by Hoechst staining (Fig. 4B, D).
Fig. 4.
Nuclei enrichment in KP cell clusters as a measure of reversal of transformed phenotype in KP assay. (A, B) Whole-well images of KP cells acquired by the INCA2000 of KP cells treated with 1% DMSO (v/v) in GFP (green) and Hoechst (blue) channels. (E, F) Whole-well images acquired by the INCA2000 of KP cells treated with 10 μM imatinib in 1% DMSO (v/v) in GFP (green) and Hoechst (blue) channels. (I, J) Images demonstrating segmentation analysis for GFP (green) channel done using Developer Toolbox 1.7 software depicted as an overlay (magenta) for both 1% DMSO (v/v) and 10 μM imatinib treated, respectively. (C, D, G, H, K, L) Magnified images of selected regions in the well (white dashed box). Color images available online at www.liebertonline.com/adt.
We developed a custom analysis method to quantify the differences between untreated and imatinib-treated KP cells. For this purpose, we investigated the use of several image analysis algorithms supplied as standalone methods within Developer Toolbox 1.7 software. We opted to use an analysis method based solely on the distribution of nuclei in clusters (untreated KP cells) versus parental phenotype (imatinib-treated KP cells) to quantify the reversal of transformed phenotype. The main difference between the nuclei distribution of the untreated KP cells (Fig. 4B, D) compared with the imatinib-treated cells (Fig. 4F, H) was the absence of overlapping nuclei postimatinib treatment. Indeed, important characteristics of the KP-transformed phenotype are in its features such as the piling up of cell clusters and the morphology of the cytoplasm, both of which could not be assessed using nuclei segmentation alone. The analysis method makes use of algorithms relying on both object-based segmentation of cell clusters in the green channel and a separate segmentation of the nuclei in the blue channel. In this combined segmentation method, clusters were identified using an exclusion parameter based on the cluster area (Fig. 4I–L), where the new method allows the extraction of several parameters such as the number of clusters, the size of the cluster area, and the number of nuclei present in the clusters. This method can easily be used in an automated and unsupervised fashion to analyze thousands of images from large screens. We introduce a new factor for reporting reversal of transformed phenotype as the NEF. NEF is defined as the resulting percentage nuclei enriched in the clusters multiplied by an arbitrary value of 100. Having established and optimized both image acquisition and data analysis, we performed an analysis of an ensemble of 65 wells containing either the compound carrier DMSO (1% (v/v)) as the high control or 10 μM imatinib in 1% DMSO (v/v) as the low control for our assay. The average NEF values for high control wells were 8,054 ± 28 (standard error) and a coefficient of variation of 3%, and for low control wells were 525 ± 37 and a coefficient of variation of 57%. A Z′ value of 0.79 was obtained with an S/N of 15 as a measure of good assay performance and S/N separation. The other added value of our high-content assay strategy allows for quantification of remaining nuclei per well postfixation as a measure of cytotoxicity; in the case of imatinib-treated KP cells, an average of 1,957 ± 29 nuclei was counted versus an average of 1,766 ± 26 in the DMSO control wells, suggesting that imatinib is not cytotoxic to the KP cells since our threshold for cytotoxic compounds is the standard 80% loss of nuclei count.20
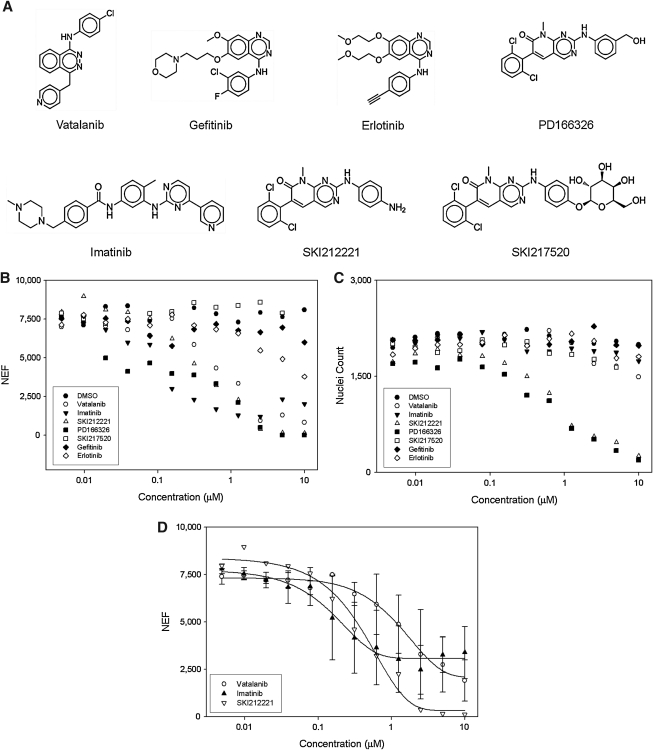
We next investigated the ability of our assay to identify reversers of the KP-transformed phenotype by performing dose–response studies against a panel of seven compounds: imatinib (a known PDGFRα inhibitor),14 vatalanib (a well-characterized PDGFRα inhibitor),15 PD166326 (a pyrido[2,3-d]pyrimidin-7-one compound and found to inhibit PDGFRα activity with an IC50 of 45 nM),16 SKI212221 and SKI217520 (two recently synthesized pyrido[2,3-d]pyrimidin-7-one derivatives and found to inhibit PDGFRα activity with IC50 values of 30 and 15 nM, respectively),16 and, finally, gefitinib and erlotinib (both phenylquinazolin-4-amine–based compounds and well-characterized inhibitors of EGFR).21 The chemical structures of each compound are summarized in Figure 5A. The obtained dose–response studies results are shown in Figure 5B. As expected vatalanib and imatinib reversed the KP-transformed phenotype with IC50 values of 1.50 ± 0.28 and 0.16 ± 0.03 μM, respectively (Fig. 5D). Two of the pyrido-pyrimidinones appear to reverse the phenotype with only SKI212221 exhibiting an IC50 value of 0.43 ± 0.06 μM (Fig. 5D). The apparent lack of potency of SKI217520 in the KP cells was probably due to its hydrophilic nature preventing cellular uptake.16 The EGFR inhibitors were predicted to have no effect on reversing the KP-transformed phenotype and found to have none (Fig. 5B). Assessment of compound cytotoxicity revealed that PD166326 and SKI212221 were found to be toxic to the KP cells at concentrations >1 μM (Fig. 5C).
Fig. 5.
Performance and concentration responses of a small panel of known compounds in the optimized KP assay. (A) Chemical structures of control kinase inhibitors. (B) NEF estimated at each concentration for each inhibitor of kinase panel. (C) Nuclei count assessment at each concentration of each inhibitor of kinase panel. (D) Dose–response curves for three PDGFR inhibitors. IC50 values (based on one data set) were calculated for: vatalanib 1.5 ± 0.28 μM, imatinib 0.16 ± 0.03 μM, and SKI212221 0.43 ± 0.06 μM. NEF, nuclear enrichment factor.
Screening the Focused Library for Reversers of the KP-Transformed Phenotype
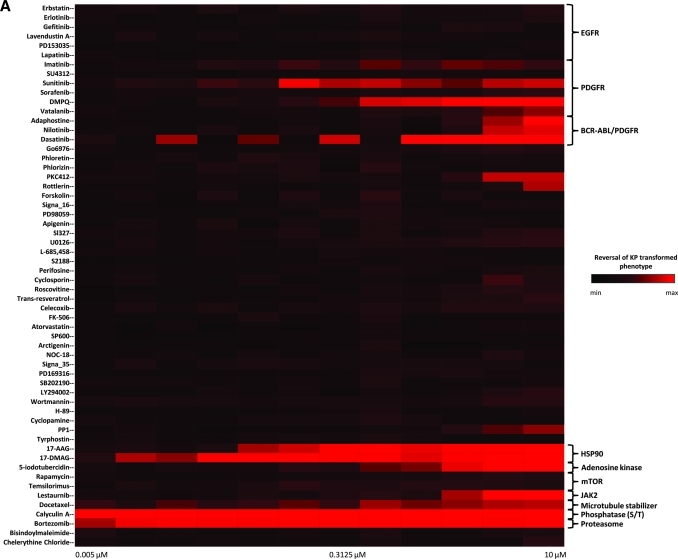
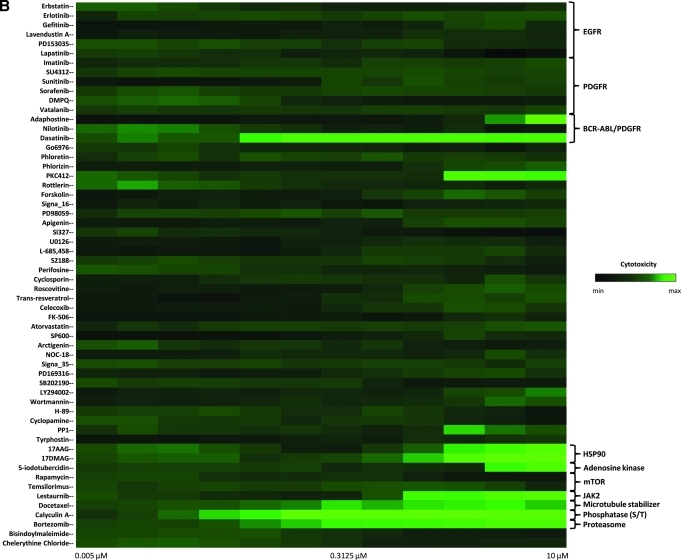
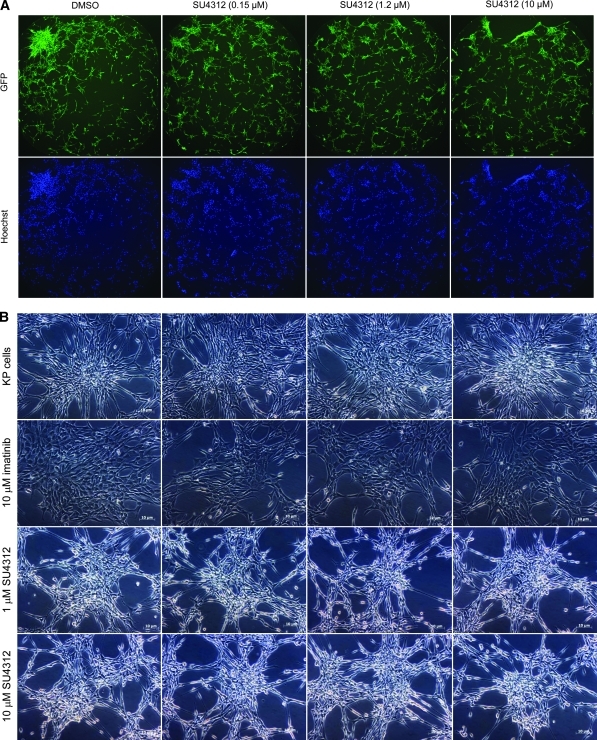
We performed a pilot screen as dose–response studies against a focused library of 58 compounds (Supplementary Table S1). All the dose–response screening studies are summarized as a heat map in Figure 6A. Among the nine PDGFR inhibitors in the library (imatinib, sunitinib, sorafenib, DMPQ, vatalanib, adaphostin, nilotinib, dasatinib, and SU4312), the assay identified eight compounds as reversers of the KP-transformed phenotype. Of interest, the 2 PDGFRα inhibitors (imatinib and vatalanib) expected to be identified were identified. The PDGFRα inhibitors DMPQ, sunitinib, and nilotinib showed complete reversal of the KP-transformed phenotype with IC50 values of 0.93 ± 0.11, 0.07 ± 0.57, and 2.50 ± 0.02 μM, respectively. The ninth PDGFR inhibitor, SU4312, was not picked up by our screen and its inactivity is probably due to the fact that it is a PDGFRβ antagonist22 with little or no inhibitory activity against PDGFRα (Fig. 7A). This was indeed found to be the case, as a secondary assay using brightfield microscopy was performed using SU4312 at either 1 or 10 μM concentration and confirmed it to be inactive at reversing the KP-transformed phenotype, whereas imatinib was active within the same experiment (Fig. 7B). Sensitivity and selectivity of the assay was further demonstrated by the finding that all the 6 EGFR inhibitors (erbastatin, erlotinib, gefitinib, lavendustin A, PD153035, and lapatinib) and the vascular endothelial growth factor inhibitor tyrphostin were not identified as reversers of the transformed phenotype (Fig. 6A). Assessment of compound cytotoxicity revealed that the PDGFRα inhibitor dasatinib, though found to reverse the KP-transformed phenotype, was cytotoxic to the KP cells at a concentration as low as 40 nM (Fig. 6B).
Fig. 6.
Dose–response studies result of the focused library for potential reverters of the transformed KP phenotype in the optimized assay. (A) The results are presented as a heat map readout showing the performance on the 58 compounds in the focused library that either reverse the KP-transformed phenotype (red) or have no effect on the KP clusters (black). (B) Resulting nuclear count assessment for the 58 compounds where cytotoxic compounds is depicted in increased green color. Color images available online at www.liebertonline.com/adt.
Fig. 7.
Concentration–response studies for the PDGFRβ inhibitor SU4312 as a reverser of the KP-transformed phenotype. (A) Representative images of KP cells treated with SU4312 at 0.15, 1.2, and 10 μM or 1% DMSO (v/v) acquired by the INCA2000 in GFP (green) and Hoechst (blue) channels, and showing no reversal of the KP phenotype. (B) Brightfield images of KP cells treated with 10 μM imatinib, 1 or 10 SU4312. Images clearly show the lack of activity of SU4312 in reversing the KP-transformed phenotype in contrast to the imatinib treatment. Scale bar: 10 μm. Color images available online at www.liebertonline.com/adt.
Besides the PDGFR inhibitors, we identified eight other compounds that partially reverse the KP-transformed phenotype and exhibit some cytotoxic effects on the KP cells (Fig. 6B). These compounds target downstream effectors of the PDGFR signaling pathway, such as MEK1/2, Erk, PI3K, PKC, and mTOR; this is an expected result in light of the signaling pathways activated by PDGFRα and clearly highlights the sensitivity of our assay in identifying compounds that only partially interfere with cell transformation with a potential for synergetic screens. Among these partial reverser compounds were PKC412, wortmannin, adaphostin, trans-resveratrol, 5-iodotubercidin, lestaurtinib, docetaxel, and protein phosphate 1 (PP1). Interestingly, among the 13 identified reversers of the KP phenotype that are cytotoxic to the KP cells were the HSP90 inhibitors 17-AAG and 17-DMAG23 and the proteasome inhibitor bortezomib.24
Discussion
HCS is emerging as an integrated process in HTS to efficiently integrate morphological and phenotypic cellular response into the drug discovery and development process.25–32 Aligning a high-content assay with the correct imaging platform is critical for its development and execution in HTS. The choice of an imaging platform would impact the overall quality of image acquisition for the recognition and quantification of objects by the analysis protocols. Thus, imaging platform validation, if at all feasible and available, would help ease the decision making process by means of comparative studies on different platforms. This certainly was the case for our cell-based assay development process where three imaging platforms the INCA1000, the INCA2000, and the INCA3000, all three available in our laboratory, were evaluated for the purpose of imaging clusters in 384-well microtiter plates. We show that imaging of multiple fields of view or tiles with the INCA1000 (4 fields/tiles) and INCA3000 (9 fields/tiles) were not of sufficient content to capture the entire cluster population distributions as observed by whole-well imaging on the INCA2000 (Fig. 2A).
One can argue that tile stitching can easily address the issues of multiple tiles per well resulting from the INCA1000 and the INCA3000 platforms, a process that has been successfully reported for a high-content wound healing siRNA screen involving twelve 96-well microtiter plates where the wound was imaged as 12 distinct tiles using an Applied Precision CellWorx (Cellomics) microscope equipped with a 10 × magnifying objective and later stitched as one to observe the entire wound.33 Tile stitching, though feasible in the case of this low-throughput wound healing assay, is hardly achievable in practice and not easily amenable to deal with stitching data from large screening campaigns (up to 400,000 compounds) where as many as 3,711,744 tiles from the INCA3000 would require stitching into 412,416 images for further analysis. Further, the obtained tiles typically cover predetermined areas of the well and the area covered by the different fields may not overlap with the region of interest, and in the case of our assay, stitching separate tiles may not accurately reflect the subtle changes in the KP-transformed phenotype due mainly to our discovery of random and differential growth of the clusters in the wells (Fig. 2B).
Our high-content assay using whole-well imaging as a platform provides an attractive alternative to multi-tile imaging and requiring stitching for applications involving cluster forming cells. The most significant advantages over (1) the multi-tile imaging and stitching33 and (2) the manual identification of spheroids and subsequent imaging by phase-contrast10 are the sensing time of 4 s per well, the “on the fly” 2D deconvolution option, and the rapid image analysis using this custom analysis method to quantify reversal as NEF. In addition, our assay recapitulated the observations made from brightfield imaging (Fig. 1A) for both vatalanib and imatinib on the reduction of cluster formation through inhibiting the PDGFRα kinase function (Fig. 5B), exhibited robust and acceptable screening parameters with a calculated Z′ value of 0.79 and an S/N ratio of 15. Due to the nature of the oncogenic transformation of the NIH-3T3 cells by expression of KP, in this case a fusion protein between PDGFRα and KDR proteins, our assay lived up to its expectations by identifying all those compounds affecting the PDGFRα signaling function, with the exception of SU4312, which was not identified as a reverse of the KP-transformed phenotype and it is believed to be due to its selectivity as a PDGFRβ antagonist.22 Secondary studies confirmed that SU4312 was inactive toward reversing the KP-transformed phenotype (Fig. 7B), further emphasizing the sensitivity and selectivity of our optimized assay together with its ability to identify the EGFR inhibitors as inactives. The high-content nature of the assay provide a measure of reversal of the transformed phenotype together with assessment of compound cytotoxicity against the KP cells and seem to overcome the limitations of the low-content viability assays.8–10 Hence, we could identify noncytotoxic compounds reversing the KP-transformed phenotype such as imatinib, sunitinib, sorafenib, DMPQ, vatalanib, and nilotinib that would have been overlooked in a low-content cytotoxicity screen.
In conclusion, we have shown the utility of whole-well imaging and its usefulness in developing and executing cell-based assays; to our knowledge, this is the first example of such a cell-based assay allowing for screening of modulators or reversers of oncogenic-induced transformed phenotype against chemical libraries, and with a broader applicability to RNAi screening and systems biology to study signaling pathways and connectivity nodes in the oncogenically addicted and rewired cells.
Supplementary Material
Abbreviations
- CCD
charged-coupled device
- DMSO
dimethyl sulfoxide
- GFP
green fluorescent protein
- HCS
high-content screening
- INCA1000
IN Cell Analyzer 1000
- INCA2000
IN Cell Analyzer 2000
- INCA3000
IN Cell Analyzer 3000
- NA
numerical aperture
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- S/N
signal-to-noise ratio
- 3D
three-dimensional
- 2D
two-dimensional
Acknowledgments
The authors wish to thank the members of the HTS Core Facility for their help during the course of this study, Dr. Guillaume Normand for all his contributions in the early phase of this project, and Mr. Tony J. Riley (Medical Graphics, MSKCC) for his help with Figure 2 in this article. The HTS Core Facility is partially supported by Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of the Memorial Sloan-Kettering Cancer Center, the William Randolph Hearst Fund in Experimental Therapeutics, the Lillian S. Wells Foundation, and by an NIH/NCI Cancer Center Support Grant 5 P30 CA008748-44. This study was supported by the Brain Tumor Center of the Memorial Sloan-Kettering Cancer Center.
Disclosure Statement
No competing financial interests exist.
References
- 1.Hanahan D. Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Dunham W. Report sees 7.6 million global 2007 cancer deaths. Reuters 2007. www.reuters.com/article/healthNews/idUSN1633064920071217. [Sep 01;2010 ]. www.reuters.com/article/healthNews/idUSN1633064920071217
- 3.Jemal A. Siegel R. Ward E, et al. Cancer statistics. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein IB. Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 5.Meyer N. Penn LZ. Reflecting on 25 years with MYC. Nat Rev. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 6.Greulich H. Chen TH. Feng W, et al. Oncogenic transformation by inhibitor-sensitive and–resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson FM. Ogasawara MA. Ye Z, et al. Imaging and analysis of 3D tumor spheroids enriched for a cancer stem cell phenotype. J Biomol Screen. 2010;15:820–829. doi: 10.1177/1087057110376541. [DOI] [PubMed] [Google Scholar]
- 8.Friedrich J. Eder W. Castaneda J, et al. A reliable tool to determine cell viability in complex 3-d culture: the acid phosphatase assay. J Biomol Screen. 2007;12:925–937. doi: 10.1177/1087057107306839. [DOI] [PubMed] [Google Scholar]
- 9.Nirmalanandhan VS. Duren A. Hendricks P, et al. Activity of anticancer agents in a three-dimensional cell culture model. Assay Drug Dev Technol. 2010;8:581–590. doi: 10.1089/adt.2010.0276. [DOI] [PubMed] [Google Scholar]
- 10.Friedrich J. Seidel C. Ebner R. Kunz-Schughart LA. Spheroid-based drug screen: considerations and practical approach. Nat Protoc. 2009;4:309–324. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- 11.Lee GY. Kenny PA. Lee EH. Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada KM. Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Ozawa T. Brennan CW. Wang L, et al. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24:2205–2218. doi: 10.1101/gad.1972310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deininger MW. Druker BJ. Specific targeted therapy of chronic myelogenous leukemia with imatinib. Pharmacol Rev. 2003;55:401–423. doi: 10.1124/pr.55.3.4. [DOI] [PubMed] [Google Scholar]
- 15.Jain RK. di Tomaso E. Duda DG, et al. Angiogenesis in brain tumours. Nat Rev Neurosci. 2007;8:610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 16.Antczak C. Veach DR. Ramirez CN, et al. Structure-activity relationships of 6-(2,6-dichlorophenyl)-8-methyl-2-(phenylamino)pyrido[2,3-d]pyrimidin-7-ones: toward selective Abl inhibitors: toward selective Abl inhibitors. Bioorg Med Chem Lett. 2009;19:6872–6876. doi: 10.1016/j.bmcl.2009.10.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang JH. Chung TDY. Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 18.Perucho M. Goldfarb M. Shimizu K, et al. Human-tumor-derived cell lines contain common and different transforming genes. Cell. 1981;27:467–476. doi: 10.1016/0092-8674(81)90388-3. [DOI] [PubMed] [Google Scholar]
- 19.Du Z. Podsypanina K. Huang S, et al. Introduction of oncogenes into mammary glands in vivo with an avian retroviral vector initiates and promotes carcinogenesis in mouse models. Proc Natl Acad Sci USA. 2006;14:17396–17401. doi: 10.1073/pnas.0608607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shum D. Smith JL. Hirsch AJ, et al. High content assay to identify inhibitors of dengue virus infection. Assay Drug Dev Technol. 2010;8:553–570. doi: 10.1089/adt.2010.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim ST. Lee J. Kim JH, et al. Comparison of gefitinib versus erlotinib in patients with non small cell lung cancer who failed previous chemotherapy. Cancer. 2010;116:3025–3033. doi: 10.1002/cncr.25130. [DOI] [PubMed] [Google Scholar]
- 22.Zaman GJ. Vink PM. van den Doelen AA, et al. Tyrosine kinase activity of purified recombinant cytoplasmic domain of platelet-derived growth factor beta-receptor (beta-PDGFR) and discovery of a novel inhibitor of receptor tyrosine kinases. Biochem Pharmacol. 1999;57:57–64. doi: 10.1016/s0006-2952(98)00271-8. [DOI] [PubMed] [Google Scholar]
- 23.Matei D. Satpathy M. Cao L, et al. The platelet-derived growth factor receptor alpha is destabilized by geldanamycins in cancer cells. J Biol Chem. 2007;282:445–453. doi: 10.1074/jbc.M607012200. [DOI] [PubMed] [Google Scholar]
- 24.Almond JB. Cohen GM. The proteasome: a novel target for cancer chemotherapy. Leukemia. 2002;16:433–443. doi: 10.1038/sj.leu.2402417. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman AF. Garippa RJ. A pharmaceutical company user's perspective on the potential of high content screening in drug discovery. Methods Mol Biol. 2007;356:19–31. doi: 10.1385/1-59745-217-3:19. [DOI] [PubMed] [Google Scholar]
- 26.Ivnitski-Steele I. Ramirez CN. Djaballah H. Human embryonic stem cells in drug discovery: are we there yet? Intern Drug Discov. 2010;5:24–28. [Google Scholar]
- 27.Desbordes SC. Placantonakis DG. Ciro A, et al. High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell. 2008;2:602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng L. Dai P. Ciro A, et al. Identification of novel antipoxviral agents: mitoxantrone inhibits vaccinia virus replication by blocking virion assembly. J Virol. 2007;81:13392–13402. doi: 10.1128/JVI.00770-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antczak C. Takagi T. Ramirez CN. Radu C. Djaballah H. Live-cell imaging of caspase activation for high-content screening. J Biomol Screen. 2009;14:956–969. doi: 10.1177/1087057109343207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haney SA. LaPan P. Pan J. Zhang J. High-content screening moves to the front of the line. Drug Discov Today. 2006;11:889–894. doi: 10.1016/j.drudis.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 31.Korn K. Krausz E. Cell-based high-content screening of small-molecule libraries. Curr Opin Chem Biol. 2007;11:503–510. doi: 10.1016/j.cbpa.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez CN. Antczak C. Djaballah H. Cell viability assessment: toward content-rich platforms. Exp Opin Drug Discov. 2010;5:223–233. doi: 10.1517/17460441003596685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson KJ. Selfors LM. Bui J, et al. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10:1027–1038. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.