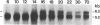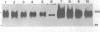Abstract
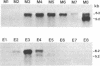
Genomic methylation patterns are established during maturation of primordial germ cells and during gametogenesis. While methylation is linked to DNA replication in somatic cells, active de novo methylation and demethylation occur in post-replicative spermatocytes during meiotic prophase (1). We have examined differentiating male germ cells for alternative forms of DNA (cytosine-5)-methyltransferase (DNA MTase) and have found a 6.2 kb DNA MTase mRNA that is present in appreciable quantities only in testis; in post-replicative pachytene spermatocytes it is the predominant form of DNA MTase mRNA. The 5.2 kb DNA MTase mRNA, characteristic of all somatic cells, was detected in isolated type A and B spermatogonia and haploid round spermatids. Immunobolt analysis detected a protein in spermatogenic cells with a relative mass of 180,000-200,000, which is close to the known size of the somatic form of mammalian DNA MTase. The demonstration of the differential developmental expression of DNA MTase in male germ cells argues for a role for testicular DNA methylation events, not only during replication in premeiotic cells, but also during meiotic prophase and postmeiotic development.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alcivar A. A., Hake L. E., Millette C. F., Trasler J. M., Hecht N. B. Mitochondrial gene expression in male germ cells of the mouse. Dev Biol. 1989 Oct;135(2):263–271. doi: 10.1016/0012-1606(89)90178-4. [DOI] [PubMed] [Google Scholar]
- Ariel M., McCarrey J., Cedar H. Methylation patterns of testis-specific genes. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2317–2321. doi: 10.1073/pnas.88.6.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellvé A. R., Cavicchia J. C., Millette C. F., O'Brien D. A., Bhatnagar Y. M., Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977 Jul;74(1):68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellvé A. R., Millette C. F., Bhatnagar Y. M., O'Brien D. A. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem. 1977 Jul;25(7):480–494. doi: 10.1177/25.7.893996. [DOI] [PubMed] [Google Scholar]
- Bestor T. H., Ingram V. M. Growth-dependent expression of multiple species of DNA methyltransferase in murine erythroleukemia cells. Proc Natl Acad Sci U S A. 1985 May;82(9):2674–2678. doi: 10.1073/pnas.82.9.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T. H., Ingram V. M. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T., Laudano A., Mattaliano R., Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988 Oct 20;203(4):971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Groudine M., Conkin K. F. Chromatin structure and de novo methylation of sperm DNA: implications for activation of the paternal genome. Science. 1985 May 31;228(4703):1061–1068. doi: 10.1126/science.2986289. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982 Feb 18;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- Hake L. E., Alcivar A. A., Hecht N. B. Changes in mRNA length accompany translational regulation of the somatic and testis-specific cytochrome c genes during spermatogenesis in the mouse. Development. 1990 Sep;110(1):249–257. doi: 10.1242/dev.110.1.249. [DOI] [PubMed] [Google Scholar]
- Husmann M., Görgen I., Weisgerber C., Bitter-Suermann D. Up-regulation of embryonic NCAM in an EC cell line by retinoic acid. Dev Biol. 1989 Nov;136(1):194–200. doi: 10.1016/0012-1606(89)90141-3. [DOI] [PubMed] [Google Scholar]
- Kleene K. C., Distel R. J., Hecht N. B. Translational regulation and deadenylation of a protamine mRNA during spermiogenesis in the mouse. Dev Biol. 1984 Sep;105(1):71–79. doi: 10.1016/0012-1606(84)90262-8. [DOI] [PubMed] [Google Scholar]
- Kleene K. C. Poly(A) shortening accompanies the activation of translation of five mRNAs during spermiogenesis in the mouse. Development. 1989 Jun;106(2):367–373. doi: 10.1242/dev.106.2.367. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Meistrich M. L. Separation of spermatogenic cells and nuclei from rodent testes. Methods Cell Biol. 1977;15:15–54. doi: 10.1016/s0091-679x(08)60207-1. [DOI] [PubMed] [Google Scholar]
- Monk M., Boubelik M., Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987 Mar;99(3):371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- NEBEL B. R., AMAROSE A. P., HACKET E. M. Calendar of gametogenic development in the prepuberal male mouse. Science. 1961 Sep 22;134(3482):832–833. doi: 10.1126/science.134.3482.832. [DOI] [PubMed] [Google Scholar]
- OAKBERG E. F. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am J Anat. 1956 Nov;99(3):391–413. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- OAKBERG E. F. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am J Anat. 1956 Nov;99(3):507–516. doi: 10.1002/aja.1000990307. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Drahovsky D. DNA methyltransferase polypeptides in mouse and human cells. Biochim Biophys Acta. 1986 Dec 18;868(4):238–242. doi: 10.1016/0167-4781(86)90059-x. [DOI] [PubMed] [Google Scholar]
- Rahe B., Erickson R. P., Quinto M. Methylation of unique sequence DNA during spermatogenesis in mice. Nucleic Acids Res. 1983 Nov 25;11(22):7947–7959. doi: 10.1093/nar/11.22.7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romrell L. J., Bellvé A. R., Fawcett D. W. Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev Biol. 1976 Mar;49(1):119–131. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- Sanford J. P., Clark H. J., Chapman V. M., Rossant J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev. 1987 Dec;1(10):1039–1046. doi: 10.1101/gad.1.10.1039. [DOI] [PubMed] [Google Scholar]
- Shaper N. L., Wright W. W., Shaper J. H. Murine beta 1,4-galactosyltransferase: both the amounts and structure of the mRNA are regulated during spermatogenesis. Proc Natl Acad Sci U S A. 1990 Jan;87(2):791–795. doi: 10.1073/pnas.87.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Sam J., Robinson M. O., Bellvé A. R., Simon M. I., Riggs A. D. Measurement by quantitative PCR of changes in HPRT, PGK-1, PGK-2, APRT, MTase, and Zfy gene transcripts during mouse spermatogenesis. Nucleic Acids Res. 1990 Mar 11;18(5):1255–1259. doi: 10.1093/nar/18.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R., Gruenbaum Y., Pollack Y., Razin A., Cedar H. Clonal inheritance of the pattern of DNA methylation in mouse cells. Proc Natl Acad Sci U S A. 1982 Jan;79(1):61–65. doi: 10.1073/pnas.79.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trasler J. M., Hake L. E., Johnson P. A., Alcivar A. A., Millette C. F., Hecht N. B. DNA methylation and demethylation events during meiotic prophase in the mouse testis. Mol Cell Biol. 1990 Apr;10(4):1828–1834. doi: 10.1128/mcb.10.4.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]