Abstract
RNA interference (RNAi) offers a powerful tool to specifically direct the degradation of complementary RNAs, and thus has great therapeutic potential for targeting diseases. Despite the reported preferences of RNAi, there is still a need for new techniques that will allow for a detailed mechanistic characterization of RNA-induced silencing complex (RISC) assembly and activity to further improve the biocompatibility of modified siRNAs. In contrast to previous reports, we investigated the effects of 2′-O-methyl (2′OMe) modifications introduced at specific positions within the siRNA at the early and late stages of RISC assembly, as well as their influence on target recognition and cleavage directly in living cells. We found that six to 10 2′OMe nucleotides on the 3′-end inhibit passenger-strand release as well as target-RNA cleavage without changing the affinity, strand asymmetry, or target recognition. 2′OMe modifications introduced at the 5′-end reduced activated RISC stability, whereas incorporations at the cleavage site showed only minor effects on passenger-strand release when present on the passenger strand. Our new fluorescence cross-correlation spectroscopy assays resolve different steps and stages of RISC assembly and target recognition with heretofore unresolved detail in living cells, which is needed to develop therapeutic siRNAs with optimized in vivo properties.
Introduction
RNA interference (RNAi) is an evolutionary conserved gene-silencing mechanism that uses short, double-stranded RNAs (dsRNAs) to identify complementary target RNAs for sequence-specific degradation (1). The RNAi mechanism requires a ribonucleoprotein complex known as the RNA-induced silencing complex (RISC), which is composed of an Argonaute (Ago) protein associated with a short, single-stranded RNA of 19–30 nucleotides (nt). These short guide RNAs originate from endogenous or exogenous dsRNA substrates of various secondary structures (2–5). To function as guiding cofactors, the short dsRNAs are bound by the RISC loading complex (RLC), which consists of Dicer, TRBP, and a member of the Argonaute protein family (4,6,7). The selection of the latter guide RNA is governed by the thermodynamic stability of the duplex termini (8). For target recognition, RISC is guided by single-stranded RNA on the basis of sequence complementarity. In the current two-state model, not all positions of the guide RNA participate equally in target recognition (9,10). Therefore, nucleotides 2–8 of the guide strand (termed the seed sequence) mediate the primary interaction with the target RNA. Once the seed sequence primes to the target, the duplex formation propagates toward the 3′-end, leading to a conformational change in Argonaute (11–13). Depending on the extent of complementarity in conjunction with the associated Argonaute family member, RISC can trigger gene expression by endonucleolytic cleavage, translational repression, deadenylation, decapping, and/or translocation into P-bodies (reviewed in Chekulaeva and Filipowicz (14)).
One class of small, silencing RNAs is composed of short interfering RNAs (siRNAs), which consist of a dsRNA stem of ∼19 nt containing a 5′-phosphate and 2 nt overhangs at both 3′-ends (3,5). The identification of siRNAs resulted in the development of chemically synthesized siRNAs to specifically trigger the degradation of targeted mRNA transcripts in mammalian cells (15). Recent advances in chemistry and oligonucleotide synthesis lead to the development of therapeutic siRNAs with improved pharmacokinetic and pharmacodynamic properties (16–19). Therefore, various modifications, such as the phosphorothioate (PTO) backbone modification and 2′-O-methyl (2′-OMe) and 2′-fluoro (2′F) sugar modification, have been comprehensively evaluated for their use in siRNA-based medical therapies (18–24). The PTO modification is compatible with siRNA function, leads to an increased half-life in serum, and facilitates cellular uptake; however, it also lowers thermal duplex stability, can display cytotoxic effects, and promotes nonspecific protein binding. This is illustrated by the strong affinity to polyanion-binding proteins, which results in accumulation in the nucleus and P-bodies and can potentially cause unwanted side effects, thereby limiting its use in siRNA-mediated silencing (20–23,25).
The incorporation of the 2′F modification on pyrimidines also stabilizes the siRNA duplex against nucleases without affecting the silencing activity, but also results in a strong localization in the nucleus (22–24).
The nontoxic 2′-OMe RNA modification is a naturally occurring RNA alteration that is found in mammalian tRNAs and rRNAs and does not alter siRNA subcellular localization (23). Furthermore, the substitution of a single 2′-OMe modification at position 2 of the guide strand reduces silencing of most off-target transcripts with complementarity to the seed region without affecting the silencing efficacy of the target mRNA (26). Increased 2′-OMe modification rates stabilize the duplex against nucleases, but can reduce or completely abolish silencing activity of siRNAs (22,23,27). 2′-OMe modifications on nucleotides 18–21 resulted in increased silencing rates after 48 h (23). Although the 2′-OMe modification can result in inactive duplexes or silencing of inhibited duplexes, it provides a natural and powerful tool that can be used to specifically increase the lifetime, specificity, and potency of siRNAs needed for RNAi-based pharmaceuticals (20,22,27). So far, investigators have studied the effects of 2′-OMe modification on siRNA-mediated silencing by classical silencing readouts, either by targeting an endogenous gene analyzed by Western blot or quantitative reverse transcription-polymerase chain reaction, or by standard reporter assays such as mRFP/EGFP expression or the dual luciferase assay (20–24,27). Still, little is known about the mechanism of silencing inhibition by 2′-OMe nucleotides, which can affect factors such as the affinity to RISC, strand separation, cleavage activity, and the stability of the incorporated guide strand. In addition, the ability to directly analyze the RNAi mechanism in living cells in real time at the early phase of RISC activation (1–6 h) (28) could result in a more detailed characterization of modified siRNAs, which is needed to further improve RNAi-based pharmaceuticals.
We recently established fluorescence cross-correlation spectroscopy (FCCS) assays that allowed us to investigate the origination of nuclear RISC in a living cell in real time with single-molecule sensitivity (28). The FCCS method has a high sensitivity and makes it possible to directly access strand separation, guide-strand incorporation (including the duration of incorporation), and RISC-target-RNA interaction independently of reporter systems, translational regulation, and knock-down of an mRNA or protein, which require longer incubation times and cannot locate the individual step of silencing inhibition.
Here, we report on the positional effects of 2′-OMe modifications in siRNAs on the 5′-end, 3′-end, and at the cleavage site of either the guide or passenger strand or in combination. By applying FCCS, we were able to perform this characterization in real time and in different cellular compartments. We show for the first time, to our knowledge, that stretches of modifications longer than 4 nt inhibit strand separation and increase the lifetime of the RISC-target interaction by inhibiting the cleavage activity of Ago2, and that modification of the 5′-end results in the destabilization of the Ago2–guide-strand interaction. Internal 2′-OMe modifications display only marginally inhibitory effects on strand separation and cleavage activity.
Materials and Methods
Cell culture
ER293 cells stably transfected with the pERV3 vector (Stratagene, Santa Clara, CA) were cultured at 37°C in DMEM (high glucose; Sigma. St. Louis, MO) with 10% fetal calf serum (FCS; PAA Laboratories GmbH, Pasching, Austria), 2 mM glutamine (Gibco, Carlsbad, Ca), and 0.3 mg/ml G418 (50 mg/ml; Gibco).
The cell line 10G, which stably expresses EGFP-Ago2 (introduced by Ohrt et al. (28)), was cultured at 37°C in Dulbecco's modified Eagle's medium (high glucose; Sigma) with 10% FCS, 2 mM glutamine, 0.3 mg/ml G418, and 0.4 mg/ml Hygromycin B. All cells were regularly passaged at subconfluency and plated at a density of 1–5 × 104 cells/ml.
Microinjection
For microinjection, 5–10 × 104 10G cells were transferred onto MatTek chambers coated with Fibronectin (25 μg/ml in phosphate-buffered saline including CaCl2 and MgCl2; Roche, Basel, Switzerland) 24 h before microinjection. The micropipette (Femtotip 2; Eppendorf, Hamburg, Germany) was loaded with 1.5–4 μM of labeled siRNAs in 110 mM K-gluconate, 18 mM NaCl, 10 mM HEPES pH 7.4, and 0.6 mM MgSO4. The micromanipulator consisted of a FemtoJet and InjectMan NI2 mounted directly on a microscope. The working pressure for injection was 20–40 hPa for 0.1 s and a holding pressure of 15 hPa.
FCCS setup
FCCS and laser scanning microscopy were carried out on a commercial system consisting of an LSM510 and a ConfoCor3 (Zeiss, Oberkochen, Germany). The 488 nm line of an Ar-Ion laser and the 633 nm line of a HeNe laser were attenuated by an acousto-optical tunable filter to 3.5 kW/cm2 and 1.05 kW/cm2, and directed via a 488/633 dichroic mirror onto the back aperture of a Zeiss C-Apochromat 40×, N.A. = 1.2, water immersion objective. Fluorescence emission light was collected by the same objective and split into two spectral channels by a second dichroic (LP635). To remove any residual laser light, a 505–610 nm bandpass or 655 nm longpass emission filter, respectively, was employed. The fluorescence was recorded by avalanche photodiode detectors (APDs) in each channel. For EGFP-Ago2 autocorrelation measurements, a mirror was substituted for the second dichroic and a 505 nm longpass emission filter was used in a one-channel setup. Out-of-plane fluorescence was reduced with a 70 μm pinhole. The fluorescence signals were software-correlated and evaluated with MATLAB (The MathWorks, Natick, MA) via a weighted Marquardt nonlinear least-square fitting routine.
Laser scanning microscopy was performed with the APDs of the ConfoCor3 on the same setup. Cell measurements were performed in air-buffer (150 mM NaCl, 20 mM HEPES pH 7.4, 15 mM glucose, 150 μg/ml bovine serum albumin, 20 mM Trehalose, 5.4 mM KCl, 0.85 mM MgSO4, 0.6 mM CaCl2) at room temperature.
Results
Establishment of delivery and FCCS compatibility conditions of modified siRNAs
We investigated 2′-OMe, PTO, and 2′F RNA modifications (the most commonly used and characterized RNA modifications) for their compatibility with our recently published intracellular FCCS assay (28). Differently modified Cy5-labeled siRNAs were microinjected into ER293 cells, and their subcellular localization and diffusion was studied in living cells. As described previously (23), the PTO and 2′F modifications alter the subcellular localization of siRNAs from a predominantly cytoplasmic localization to a preferentially nuclear localization, as illustrated by a strong and punctuate accumulation of these siRNAs in the nucleus (see Fig. S1 A, lower panel in the Supporting Material). In previous experiments, we showed that unmodified siRNAs were excluded from the nucleus in an Exportin-5 (Exp5)-dependent process (29). To rule out the possibility that the predominantly nuclear localization was not due to reduced binding affinities of the modified duplexes to Exp5, we developed a two-color fluorescence RNA electrophoretic mobility shift assay (REMSA). Here, the affinity of modified siRNA duplexes (Cy5-labeled; red) to Exp5 in the presence of increasing amounts of unmodified competitor siRNA (Alexa488-labeled; green) is determined. The advantage of the two-color REMSA is that the amount of shifted competitor siRNA intensity can be measured and quantified simultaneously with the fluorescence intensity of the modified duplexes. We found that modified and unmodified siRNA duplexes decreased in shifted fluorescence intensity in almost identical competitor-dependent manner (Fig. S2). This result indicates that the alteration of the subcellular localization in the case of 2′F- and PTO-modified siRNAs was not caused by changed affinities to Exp5, and most probably resulted from the previously reported affinity of PTO and 2′F modifications to polyanion-binding proteins concentrated in this organelle (25).
Intracellular FCCS analysis of 3′–4-nt-long PTO- and 2′F-modified siRNAs displayed strong bleaching rates of up to 70% for the Cy5-labeled siRNAs, especially in the nucleus, which made it impossible to obtain quantitative results (Fig. S1 B, lower panel). This strong photobleaching may arise from the increased dwell times of the siRNAs in the excitation volume caused by the aforementioned increased affinity of PTO and 2′F modifications to polyanion-binding proteins, which greatly reduces their mobility (25). In contrast, the 2′-OMe-modified siRNAs conserved the preferentially cytoplasmic localization independently of the position and quantity of the 2′-OMe modifications (Fig. S1 A, upper panel and Fig. S3) with moderate bleaching rates of 10–20% after 4 min (Fig. S1 B, upper panel).
Therefore, the 2′-OMe modification is suitable for a detailed FCCS analysis of modified siRNAs in living cells to investigate position- and quantity-dependent effects on siRNA affinity to RISC, strand separation, asymmetry, guide-strand incorporation, the stability of the guide RNA-RISC interaction, target RNA recognition, and cleavage.
FCCS assays for investigating the RNAi pathway in living cells
To study the incorporation of the guide strand, asymmetry, and the stability of the RISC-guide RNA complex, we microinjected Cy5-labeled siRNAs into an EGFP-Ago2 stable expressing cell line with endogenous expression levels (28) and measured the cross-correlation amplitudes after different incubation times. The incorporation of the labeled strand resulted in increased cross-correlation amplitudes (Fig. 1 A, red curve), whereas the incorporation of the nonlabeled strand led to low cross-correlation amplitudes (Fig. 1 A, black curve).
Figure 1.
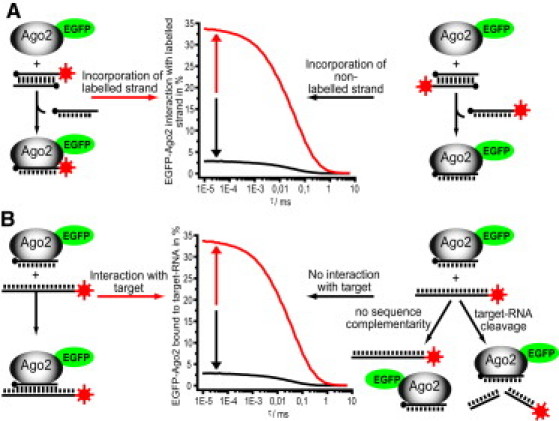
Illustration of the FCCS assays used in this study. (A) Incorporation assay. An intracellular FCCS assay was used to study the incorporation of the guide strand. The incorporation of the labeled guide strand results in increased cross-correlation amplitudes (red curve), whereas the incorporation of the nonlabeled passenger strand results in low cross-correlation amplitudes (black curve). (B) Target interaction assay. An intracellular FCCS assay was used to study the interaction of EGFP-RISC with its fluorescently labeled target RNA. A stable interaction can be observed for miRNAs that do not display RNA-cleavage activity, resulting in increased cross-correlation values (red curve), whereas no stable interaction can be observed for RISC complexes loaded with a guide RNA showing no sequence similarities or a guide strand leading to RISC-mediated target RNA cleavage (black curve).
To investigate the RISC-target interaction, the EGFP-Ago2 expressing cell line was transfected with nonlabeled siRNAs and microinjected with a 50-nt-long, Cy5-labeled target RNA 24 h after transfection (Fig. 1 B). The stable interaction of RISC with the target RNA resulted in increased cross-correlation amplitudes (Fig. 1 B, red curve), whereas RISC-mediated cleavage or no interaction led to low cross-correlation amplitudes (Fig. 1 B, black curve) (28).
Increased 2′-OMe modifications on the 3′-end inhibit strand separation and target RNA cleavage
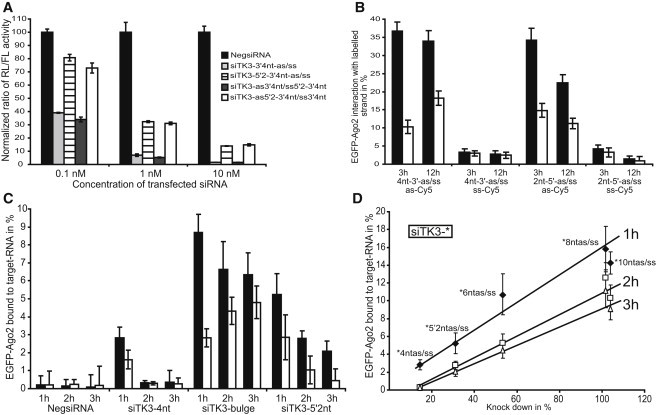
To study the incorporation and strand separation of 2′-OMe-modified siRNAs intracellularly, we used siTK3 siRNA, which targets the mRNA of Renilla luciferase encoded on the plasmid pRL-TK but does not show any sequence homology with endogenous mRNA transcripts. We calculated the 5′-end hybridization energy of siTK3 as described previously (8) to define the guide and passenger strand of the siTK3 duplex (Fig. 2 A). We used siTK3 siRNA because it displays a strictly asymmetric incorporation of the guide strand (23) and high levels of Renilla luciferase silencing (Fig. 2 A).
Figure 2.

Effects of 3′-2′-OMe modifications on silencing, guide-strand incorporation, strand separation, and asymmetry. (A) Illustration of the siTK3 siRNA with 2′OMe nt positions of the various modified strands. ER293 cells were transfected with the indicated amounts of differently modified siTK3 together with the fixed concentration of the pGL2-Control and pRL-TK reporter plasmids. After 48 h, the ratios of target to control luciferase concentration were normalized to the NegsiRNA control (black bar). Unmodified siTK3, lined bar; siTK3 3′4nt, light gray; siTK3 3′6nt, gray; siTK3 3′8nt, dark gray; siTK3 3′10nt, open bar. The plotted data were averaged from six independent experiments ± SD. (B) ER293 cells were transfected with the indicated amounts of siTK3 modified on the 3′-end with either 4 or 8 2′-OMe nt as illustrated in the graph. After 48 h, the ratios of target to control luciferase concentration were normalized to the NegsiRNA control. The plotted data were averaged from six independent experiments ± SD. (C and D) Cross-correlation amplitudes in the cytoplasm (solid bars) and nucleus (open bars) after 3 and 12 h of incubation for the indicated siRNAs. Mean values ± SE.
To investigate the positional effects of 2′-OMe modifications within small RNAs, we generated siRNAs with constantly increasing numbers of 2′-OMe modifications on the 3′-end (Fig. 2 A). The introduction of 4 2′-OMe-modified nucleotides at the 3′-end resulted in increased silencing activity compared with unmodified siRNAs as detected in a dual-luciferase silencing readout. On the other side, the introduction of 6 2′-OMe nt resulted in strongly reduced silencing activity and was completely abolished with 8 and 10 2′-OMe nt on the 3′-end (Fig. 2 A). To investigate whether the inhibition was caused by modification of the guide or passenger strand, we measured the silencing activity of only partially modified siRNA duplexes. We detected identical silencing rates for the siRNA duplex modified on the passenger strand with 8 2′-OMe nt compared with the control, whereas the modification on the guide strand resulted in an almost complete loss of silencing activity (Fig. 2 B). In agreement with the results for the siTK3 duplex, a second siRNA modified on the 3′-end with 8 2′-OMe nt targeting Renilla luciferase completely lost its silencing activity (data not shown).
To further characterize the mechanism of inhibition, we tested the interaction of the differently modified siRNAs with Ago2 by FCCS in living cells. To that end, we microinjected guide-strand-labeled siTK3 duplexes into EGFP-Ago2 cells and analyzed the amount of loaded Ago2 by FCCS after 3 and 12 h (Fig. 2 C). We specifically chose these two time points because the 3 h value represents the early (still rising RISC-loading level) and 12 h represents the late RISC-activation phase (already declining RISC-loading level) (28). Therefore, they are ideally suited to investigate strand separation and RISC stability, as both time points display almost identical loading levels in the cytoplasm.
Under our experimental conditions with guide-strand-labeled siTK3 duplexes, we detected no differences in the guide-strand affinity to Ago2 between a silencing active 4 nt modified duplex and the silencing impaired duplexes containing 6–10 2′OMe nt. All labeled guide strands were incorporated equally well with cross-correlation amplitudes of ∼30–35% (Fig. 2 C). This indicates that the silencing inhibition is not a result of reduced affinities to the RLC resulting from the 2′-OMe modification. Therefore, the 2′-OMe modification on the 3′-end does not influence the affinity to the RLC/RISC.
To study the effect of 2′-OMe modifications on strand separation and asymmetry, we microinjected the aforementioned modified siTK3 duplexes labeled on the passenger strand and compared the resulting cross-correlation amplitudes with the guide-strand-labeled duplexes (Fig. 2 C). The silencing active duplex containing 4 2′-OMe modifications on the 3′-end displayed almost no interaction of RISC with the labeled passenger strand. With increasing amounts of introduced 2′-OMe nt, the interaction of the labeled passenger strand with RISC increased in the cytoplasm as well as in the nucleus. For the completely inhibited duplexes containing 8 and 10 2′-OMe nt, we obtained almost identical incorporation levels in the cytoplasm after 3 h compared with guide-strand-labeled duplexes, whereas after 12 h we detected a clear reduction of RISC-interacting passenger strands (Fig. 2 C). Because we detected silencing inhibition by 2′-OMe modifications only on the guide strand (Fig. 2 B), we microinjected siRNAs that were modified with 8 nt 2′-OMe modifications on the guide or passenger strand only, and investigated passenger-strand release after 3 and 12 h (Fig. 2 D). In agreement with the silencing experiments, we found that strand separation was mainly inhibited when the guide strand of the duplex was modified, whereas the modified passenger strand showed only marginal effects (Fig. 2 D). These results indicate that 2′-OMe-modified duplexes do not interfere with the asymmetry or the binding affinity to RISC, but clearly inhibit the strand separation during the activation of RISC. A selection of the measured cross-correlation curves is shown in Fig. S4 A.
We note that the level of nuclear-loaded RISC constantly increased with larger amounts of remaining passenger strand in RISC after 3 h. A similar effect was previously observed for nuclear RISC targeting 7SK snoRNA inhibited in target-mediated cleavage by secondary structures in the target RNA or bulges between the target guide strand at the cleavage site (28). In addition, it has been shown that hAgo2 containing RISC cleaves the passenger strand during activation to facilitate strand separation (30). Therefore, we tested the hAgo2-mediated cleavage activity of RISC loaded with a guide strand modified with either 4 or 8 2′-OMe nt in vitro (Fig. 3 A). Consistent with the dual luciferase silencing assays, only siTK3-4nt was able to specifically cleave the siTK3 target RNA, whereas siTK3-8nt displayed no detectable cleavage activity. Therefore, the 8 2′OMe nt on the 3′-end clearly inhibited Ago2-mediated cleavage, thereby interfering with passenger-strand cleavage and release, as indicated by the increased levels of passenger strand compared with siTK3-4nt after microinjection (Fig. 2 C). Of interest, the level of passenger-strand-labeled duplex bound to RISC strongly decreased after 12 h compared with 3 h. This indicates that in addition to the cleavage-mediated passenger-strand release, a bypass mechanism exists that also results in strand separation and passenger-strand removal (Fig. 2 C). The possibility that the duplex was released during the incorporation period can be excluded because we did not obtain reduced interaction levels of the guide-labeled duplex. This bypass mechanism is much slower and time-consuming than the cleavage-mediated mechanism.
Figure 3.
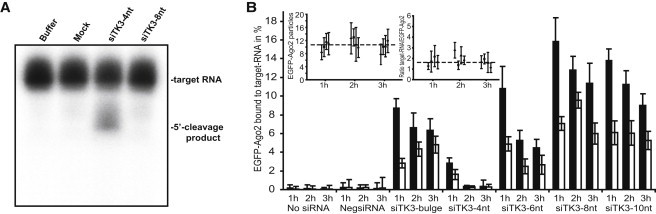
Effects of 3′-2′-OMe modifications on target interaction and cleavage. (A) The cleavage activity of siTK3-4nt or -8nt was analyzed in vitro. Only siTK3-4nt was able to cleave the target RNA; no cleavage product can be observed in buffer only, mock treated cells, or siTK3-8nt. (B) EGFP-Ago2 serves as a fluorescent label for RISC that can interact with the Cy5-labeled target RNA. The interaction with the target RNA is determined by FCCS in living cells. Cross-correlation measurements were performed 1, 2, or 3 h after microinjection. Data are represented as mean ± SE; solid and open bars indicate measurements obtained in the cytoplasm and nucleus, respectively. The number of particles of EGFP-Ago2 and the ratio of target RNA/EGFP-Ago2 for the experiments with the siRNAs modified with 4, 6, 8, and 10 2′-OMe nt are shown.
In a previous study (28) we showed that it is possible to investigate RISC-target interactions via FCCS in living cells by microinjecting a 50-nt-long, single-stranded, and fluorescently labeled RNA containing the target sequence for siTK3, which is stabilized by 7 2′-OMe nt on both ends. FCCS measurements in nontransfected cells and cells transfected with a control siRNA sharing no sequence complementarity with the target RNA (NegsiRNA) displayed no interaction between EGFP-Ago2/RISC and the target RNA after 1–3 h (Fig. 3 B). This demonstrates the absence of miRNA-binding sites within the target RNA and the specificity of the RISC-target RNA interaction assay, as the presence of siRNAs leading to an miRNA-like interaction results in increased levels of target RNA bound to RISC (Fig. 3 B). The fraction of bound target RNA by RISC was strongly decreased in the presence of a perfect matching guide strand (siTK3-4nt) due to the cleavage of the target RNA followed by the release of the cleavage products from RISC (Fig. 3 B). The levels of RISC bound to the target RNA increased with the amount of incorporated 2′-OMe modifications and correlated with the aforementioned silencing activities of the siRNAs containing 6–10 nt at the 3′-end (Fig. 3 B). A selection of the measured cross-correlation curves is shown in Fig. S4 B. To rule out any concentration-mediated effects or changed EGFP-Ago2 expression levels, the average number of particles obtained from the autocorrelation curves is shown in the inset of Fig. 3 B, which clearly demonstrates homogeneous levels for both species over the time of the experiment. Of interest, RISC loaded with the modified guide RNAs containing 8–10 2′-OMe nt displayed even higher interaction levels compared with RISC loaded with a bulge-forming guide strand. These higher amplitudes may result from a stabilization effect of the 2′-OMe modification on the RISC-target interaction. This can be caused by the increased affinity due to the complete hybridization of the guide-target RNA duplex or by the protection of the guide strand against nucleases.
2′-OMe modifications on the 5′-end lead to destabilization of the RISC-guide RNA interaction
After characterizing the effect of 2′-OMe modifications on the 3′-end, we wanted to test the influence of two 2′-OMe modified nucleotides on the 5′-end of siRNAs. In the silencing readouts, a strong decrease of silencing activity was detected when the 2′-OMe modifications were present on the 5′-end (Fig. 4 A). To identify whether the inhibition results from the guide or the passenger strand only, we also analyzed duplexes with either the guide or the passenger strand. The duplex with the 5′-end guide-strand modification showed the same silencing inhibition as the dual-modified siRNA, whereas the modification on the passenger strand had no impact on the silencing activity (Fig. 4 A). We then analyzed the affinity of the 5′-end modified siRNAs to RISC by microinjecting the guide-strand-labeled duplex. We detected the same amount of guide-strand-loaded RISC compared with the unmodified duplex after 3 h (Fig. 4 B). From this result we can conclude that the affinity to the RLC/RISC is not affected. However, after 12 h we detected a strong decrease in guide-strand-loaded RISC by approximately one-third (34% down to 22%), whereas the levels of unmodified duplexes remained stable (Fig. 4 B). This result shows that the 5′-2′OMe modification destabilizes the interaction of RISC with the modified guide strand and not with the duplex, as we could not detect increased levels of passenger-strand–RISC interaction. This destabilization may cause the relatively strong silencing inhibition measured in the silencing assay (Fig. 4 A). The asymmetry of the 5′-end 2′OMe-modified siRNAs was not affected, as the passenger-strand-labeled duplex displayed values comparable to those obtained in the unmodified control duplex (Fig. 4 B).
Figure 4.
Effects of 5′-2′-OMe modifications on silencing, guide-strand incorporation, strand separation, asymmetry, and RISC-target interaction. (A) ER293 cells were transfected with the indicated amounts of differently modified siTK3 together with the fixed concentration of the pGL2-Control and pRL-TK reporter plasmids. After 48 h, the ratios of target to control luciferase concentration were normalized to the NegsiRNA control (indicated in black). siTK3-3′4nt-as/ss, light gray; siTK3-5′2-3′4nt-as/ss, lined bar; siTK3-as3′4nt/ss5′2-3′4nt, dark gray; siTK3-as5′2-3′4nt/ss3′4nt, open bar. The plotted data were averaged from six independent experiments ± SD. (B) Cross-correlation amplitudes in the cytoplasm (solid bars) and nucleus (open bars) after 3 and 12 h of incubation for the indicated siRNAs (mean ± SE). (C) Target interaction levels with EGFP-RISC determined by FCCS in living cells. Cross-correlation measurements were performed 1, 2, or 3 h after microinjection of labeled target RNA. Data are represented as the mean ± SE; solid and open bars indicate measurements obtained in the cytoplasm and nucleus, respectively. (D) The levels of target RNA bound to EGFP-Ago2 are plotted against the silencing values obtained for the siRNAs at 1 nM concentration after 48 h: 1 h, black diamonds; 2 h, open squares; 3 h, open triangles.
We microinjected the fluorescently labeled target RNA in 5′-2′-OMe-transfected 10G cells to elucidate the impact of the 5′-2′-OMe modification on target interaction. We clearly detected increased interaction levels of RISC with its target RNA compared with the negative control (NegsiRNA) or a cleavage active siRNA (siTK3-4nt; Fig. 4 C). Although the levels are marginally lower compared with the siTK3-bulge positive control, this could result from the destabilized interaction of the guide strand with RISC. Nevertheless, the 2′-OMe modification on the 5′-end has an impact on the stability and cleavage activity of RISC, but does not interfere with the loading machinery and asymmetry of the duplex.
So far, we were able to detect increased interaction levels of silencing inhibited RISC loaded with differently modified guide RNAs on the 3′- or 5′-end with its target RNA. We plotted the fraction of target RNA bound RISC versus the silencing activity for 1, 2, and 3 h to see whether a correlation exists between silencing activity and target interaction (Fig. 4 D). Of interest, we detected a linear relationship between the silencing activity and target RNA bound by RISC at different time points. By applying this assay, were able to calculate the cleavage activities of modified siRNAs by the level of interaction with their target RNA. However, the reduction of the slope may be due to degradation of the target RNA with increasing incubation times or translocation into P-bodies.
2′-OMe modifications on positions 9–11 have only minor effects on silencing activity and strand separation
It has been shown in vitro that 2′-OMe modifications can inhibit passenger-strand cleavage and strand separation at position 9 of the passenger strand (31). We therefore sought to determine how 2′-OMe modifications modulate RISC activity at the cleavage site in living cells. To that end, we modified the guide and passenger strand at positions 9–11 starting from each 5′-end with 2′-OMe (siTK3-Int). In the dual luciferase silencing assay, we could only detect minor changes in silencing activity (<8%) for the double and guide-modified duplexes (Fig. S5). Within the accuracy of our FCCS assay, the internal modification had no effect on affinity to RISC and asymmetry (Fig. 5 A). However, we could detect increased interaction levels of labeled passenger strand (ss-Cy5) with the dual-modified (3nt-Int-as/ss) and passenger-strand-modified (3nt-Int-ss) duplexes compared with the siTK3-3′4nt duplex (4nt-3′-as/ss; Fig. 5 A).
Figure 5.
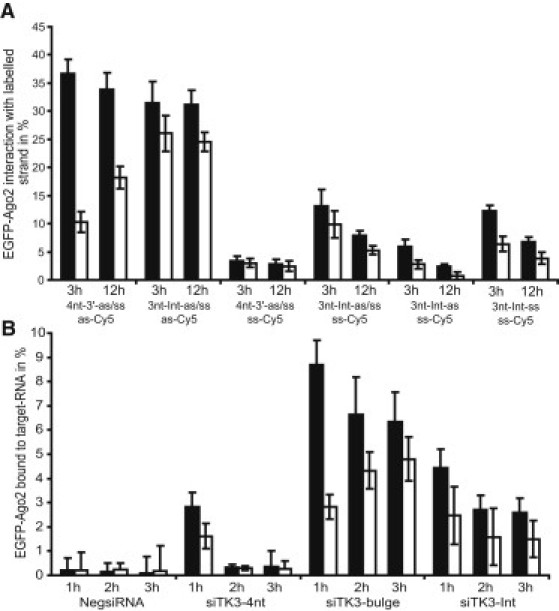
Effect of internal 2′-OMe modifications on guide-strand incorporation, strand separation, asymmetry, and RISC-target interaction. (A) Cross-correlation amplitudes in the cytoplasm (solid bars) and nucleus (open bars) after 3 and 12 h of incubation for the indicated siRNAs (mean ± SE). (B) Target interaction levels with EGFP-RISC determined by FCCS in living cells. Cross-correlation measurements were performed 1, 2, or 3 h after microinjection. Data are represented as the mean ± SE; solid and open bars indicate measurements obtained in the cytoplasm and nucleus, respectively.
In contrast, the guide-strand-modified duplex (3nt-Int-as) displayed no detectable effect on strand separation, and showed almost identical interaction levels compared with the passenger-strand-labeled siTK3-3′4nt duplex (4nt-3′-as/ss).
The levels of siTK3-Int guide-strand-loaded RISC bound to the target RNA were slightly increased compared with the siTK3-3′4nt displaying cleavage activity, and lower in relation to the bulge-forming guide strand (Fig. 5 B).
Consistent with the moderate effects measured in the silencing assays, the intracellular FCCS analysis detected only small inhibitory changes in strand separation and target interaction. Nevertheless, it also demonstrated the high sensitivity of our FCCS assays in living cells, as even small changes in silencing activity of <8% were clearly visible and allowed for the clear detection of individual inhibited steps.
Discussion
Up to now, the initial steps of RISC formation (i.e., those occurring in the first 1–6 h after siRNA delivery (28)) have been mainly inaccessible with conventional assays. To better understand the inhibitory effect of 2′OMe or other introduced modifications on siRNA-mediated silencing, we need to develop new intracellular assays that can analyze RISC formation and target interactions in their initial and late phase. By using a combination of an EGFP-Ago2 reporter cell line, specific delivery of fluorescent siRNAs via microinjection, and FCCS, we were able to analyze differently 2′OMe-modified siRNA duplexes with respect to RISC affinity, strand asymmetry and separation, activated RISC stability, target interaction, and RISC-mediated target RNA cleavage in living cells at the early and late phases of RISC activation.
In contrast to previous reports, we were interested in the individual mechanistic effects of 2′OMe modifications at different positions within siRNAs on RISC assembly and activity intracellularly. We were able to show that 2′OMe at different positions within the siRNA leads to several ways of RISC activity inhibition. We specifically decided to investigate the 2′OMe modification, as PTO and 2′F modifications were incompatible with our intracellular FCCS assay. This was due to intense photobleaching caused by the strong nonspecific affinity for cellular proteins and structures. This is consistent with the changed subcellular localization of PTO and 2′F-modified duplexes reported previously (23). Therefore, such modified duplexes or single-stranded PTO or 2′F modified RNAs cannot be investigated by standard FCS/FCCS intracellularly. Furthermore, 2′OMe nt are very interesting siRNA modifications because they can be used to specifically increase the lifetime, specificity, and potency of siRNAs, as well as inhibit RNA-mediated gene silencing.
In standard dual luciferase assays, we investigated the effect of increasing amounts of 2′OMe modifications on the 3′-end and found that 4 2′OMe nt led to increased silencing efficiency after 48 h, whereas 6 2′OMe strongly inhibited and 8–10 2′OMe insertions completely abolished gene silencing. This is consistent with previous reports demonstrating the inhibition of fully or highly 2′OMe-modified siRNAs (22–24,27,32). For the first time (to our knowledge), we were able to unravel the mechanism of 2′OMe-mediated gene silencing inhibition on the 3′-end. By applying FCCS in living cells, we showed that neither the affinity nor the asymmetry of guide-strand incorporation was affected. By contrast, we demonstrated that removal of the passenger strand, a step that is strictly required for RISC activation and maturation, was strongly inhibited. Furthermore, the inhibitory effect on passenger-strand release occurred only when 2′OMe nt were present on the guide strand. Although we found that the cleavage activity of RISC, which is needed for efficient passenger strand removal after 3 h, was mainly affected, we also detected passenger-strand release of up to 50% of bound siRNA after 12 h of siRNA supply. This indicates that a less-efficient bypass mechanism exists without passenger-strand cleavage, resulting in guide-strand-loaded RISC (activated RISC). This demonstrates the advantage of the FCCS assay, which allows RISC assembly to be measured at early and late stages after siRNA supply. In addition, we showed that RISCs loaded with cleavage-impaired 2′OMe guide strands were still capable of recognizing and binding their target RNA. Surprisingly, they associated to higher extent with the target RNA compared with a bulge-forming guide strand, which should also inhibit target RNA cleavage. We can only speculate as to why they associate with higher amplitudes with the target RNA, but most probably this reflects a stabilization effect of the 2′OMe modification on the RISC target interaction or increased affinity of the RISC-target interaction caused either by the increased affinity due to the complete hybridization of the guide-target RNA duplex or by protection of the guide strand against nucleases.
Although Chiu and Rana (22) previously showed that 2′OMe inhibits target-RNA cleavage in vitro, they could not exclude the possibility that passenger-strand release was also affected or sufficient.
Our results showing silencing inhibition with 2′OMe-modified nt from 6–10 from the 3′end are in agreement with the results of Wang et al. (13), who showed that hAgo2-mediated cleavage occurs only with guide/target RNA duplexes larger than 16 nt. Therefore, the 3′-RNA region from 15–21 of the guide strand seems to be important for the transition into or stabilization of the active confirmation, which can be inhibited by 2′OMe-modified nt.
The insertion of 2 2′OMe nt on the 5′-end inhibited gene silencing activity when it was present on the guide strand. We showed that 3 h after siRNA was supplied, in similarity to the early stages of RISC assembly, neither the affinity nor the asymmetry was affected. In contrast to the 3′-end modification, the 5′-end 2′OMe nt did not interfere with strand separation. Of interest, after 12 h we detected a strong decrease in loaded RISC levels, indicating that the 2 2′OMe nt on the 5′-end somehow destabilized the RISC-guide strand complex. We also showed that this 5′-end modification led to increased RISC-target RNA levels, thereby reducing the level of target RNA cleavage efficiency. We propose that 2′OMe on the 5′-end leads to reduced hAgo2-guide strand stability in similarity to the destabilization with a guide strand lacking the 5′-phosphate (33). This would also explain why passenger-strand removal is unaffected, whereas target RNA cleavage seems to be less efficient. Nevertheless, the effect of 2′OMe nucleotides on the 5′-end may be sequence-dependent, as other groups also found no inhibitory function of 1–2 2′OMe-modified nucleotides on the 5′-end (20,26).
To our surprise, we could only detect marginal effects with inserted 2′OMe nt at the cleavage site at positions 9–11. In our standard dual luciferase assay, we detected a reduction in silencing activity by <8% for the double- and guide-modified duplexes. Nevertheless, our FCCS assay was able to detect minor reductions in strand separation. This is in agreement with a previous study by Leuschner et al. (31), who showed passenger-strand cleavage inhibition with 2′OMe on position 9 of the passenger strand. The minor inhibition detected may reflect the fact that we performed our measurements in living cells with optimal siRNA processing conditions and in the presence of an RISC loading complex, whereas Leuschner et al. used purified minimal RISC, which represents the single hAgo2 protein alone. Nevertheless, we were able to detect slightly higher levels of RISC-target RNA interaction, indicating a slightly inhibited target RNA cleavage process.
This study shows that our intracellular FCCS assays are very well suited for investigating the individual steps of RISC assembly with very high specificity and sensitivity in real time. We were able to show a direct correlation between cleavage activity and target interaction, which can also be used to determine the cleavage properties of guide RNAs, thereby allowing for multiparameter measurements. Our results also demonstrate that various positions of 2′OMe nt affect RISC assembly and activity in different ways. Therefore, 2′OMe-modified 3′-end siRNAs with up to 8–10 nt can be used to mimic miRNA-like interactions of RISC with its target RNA, increasing the stability of the RISC-target-RNA complex. These results can lead to the development of a new type of therapeutic small RNAs because they allow one to apply miRNA-like duplexes with increased affinities to their target RNA.
Acknowledgments
We thank J. Martinez for providing the hAgo2 plasmid, C. Gwizdek and C. Dargemont for providing the Exp5-6xHis and RanQ69L-6xHis plasmids, Cenix BioSciences GmbH for discussions, Z. Petrasek and E. Petrov for advice and discussions, and W. Querbes and S. Mende for critically reading the manuscript. Author contributions: T.O. and P.S. designed the research; T.O., W.S., J.M., K.C., and M.L. performed the research; T.O., W.S., M.L., and P.S. analyzed the data; and T.O., W.S., and P.S. wrote the article.
This work was supported by the Deutsche Forschungsgemeinschaft (SPP 1128) and the Max-Planck Society.
Supporting Material
References
- 1.Fire A., Xu S.Q., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein E., Caudy A.A., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ketting R.F., Fischer S.E.J., Plasterk R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe T., Totoki Y., Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 6.Gregory R.I., Chendrimada T.P., Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Hutvágner G., McLachlan J., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 8.Schwarz D.S., Hutvágner G., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 9.Ameres S.L., Martinez J., Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 10.Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Juranek S., Patel D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan Y.R., Pei Y., Patel D.J. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y.L., Juranek S., Patel D.J. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chekulaeva M., Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Elbashir S.M., Harborth J., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 16.de Fougerolles A., Manoharan M., Vornlocher H.P. RNA interference in vivo: toward synthetic small inhibitory RNA-based therapeutics. Methods Enzymol. 2005;392:278–296. doi: 10.1016/S0076-6879(04)92016-2. [DOI] [PubMed] [Google Scholar]
- 17.de Fougerolles A., Vornlocher H.P., Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Layzer J.M., McCaffrey A.P., Sullenger B.A. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soutschek J., Akinc A., Vornlocher H.P. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 20.Amarzguioui M., Holen T., Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braasch D.A., Jensen S., Corey D.R. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 22.Chiu Y.L., Rana T.M. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohrt T., Schwille P. siRNA modifications and sub-cellular localization: a question of intracellular transport? Curr. Pharm. Des. 2008;14:3674–3685. doi: 10.2174/138161208786898798. [DOI] [PubMed] [Google Scholar]
- 24.Prakash T.P., Allerson C.R., Bhat B. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 25.Krieg A.M., Stein C.A. Phosphorothioate oligodeoxynucleotides: antisense or anti-protein? Antisense Res. Dev. 1995;5:241. doi: 10.1089/ard.1995.5.241. [DOI] [PubMed] [Google Scholar]
- 26.Jackson A.L., Burchard J., Linsley P.S. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czauderna F., Fechtner M., Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohrt T., Mütze J., Schwille P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36:6439–6449. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohrt T., Merkle D., Schwille P. In situ fluorescence analysis demonstrates active siRNA exclusion from the nucleus by Exportin 5. Nucleic Acids Res. 2006;34:1369–1380. doi: 10.1093/nar/gkl001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matranga C., Tomari Y., Zamore P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 31.Leuschner P.J., Ameres S.L., Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraynack B.A., Baker B.F. Small interfering RNAs containing full 2′-O-methylribonucleotide-modified sense strands display Argonaute2/eIF2C2-dependent activity. RNA. 2006;12:163–176. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivas F.V., Tolia N.H., Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


