Abstract
Pancreatic cancer studies have shown that inhibition of glycogen synthase kinase-3β (GSK-3β) leads to decreased cancer cell proliferation and survival by abrogating nuclear factor κB (NFκB) activity. In this investigation, various citrus compounds, including flavonoids, phenolic acids, and limonoids, were individually investigated for their inhibitory effects on GSK-3β by using a luminescence assay. Of the 22 citrus compounds tested, the flavonoids luteolin, apigenin, and quercetin had the highest inhibitory effects on GSK-3β, with 50% inhibitory values of 1.5, 1.9, and 2.0 μM, respectively. Molecular dockings were then performed to determine the potential interactions of each citrus flavonoid with GSK-3β. Luteolin, apigenin, and quercetin were predicted to fit within the binding pocket of GSK-3β with low interaction energies (−76.4, −76.1, and −84.6 kcal·mol−1, respectively) and low complex energies (−718.1, −688.1, and −719.7 kcal·mol−1, respectively). Our results indicate that several citrus flavonoids inhibit GSK-3β activity and suggest that these have potential to suppress the growth of pancreatic tumors.
Key Words: Glycogen synthase kinase-3β, GSK-3β, citrus compounds, flavonoids, luminescence assay, molecular docking
Introduction
Pancreatic cancer is the fourth most common cause of cancer-related death among both men and women in the United States, with a 5-year survival rate of only 4%.1 An estimated 43,140 new patients will be diagnosed with pancreatic cancer in 2010, with about half of these likely to die within 6 months of diagnosis.2 Because of this poor prognosis, there is a great need to expand our knowledge on the mechanisms causing pancreatic cancer and to develop new preventive and treatment strategies.
One potential therapeutic target is glycogen synthase kinase 3 (GSK-3), a serine/threonine kinase that has become of interest because of its implications in many diseases, including diabetes,3–5 Alzheimer's disease,6–8 and cancer.9–13 This kinase has 2 homologous mammalian isoforms, GSK-3α and GSK-3β, that are closely related (85% identical) but not functionally identical.14 This is especially apparent in studies linking GSK-3β, but not GSK-3α, to pancreatic cancer.15–17 It is now known that GSK-3β is overexpressed in the nucleus of pancreatic cancer cells, where it stimulates nuclear factor κB (NFκB) activity and, as a consequence, activates the inflammatory response cascade.11 Although the mechanism by which GSK-3β regulates NFκB is unknown, several studies have shown that inhibition of GSK-3β activity decreases NFκB activity and leads to decreased cancer cell proliferation and survival.11,12
Citrus fruits are of interest in cancer research because of the substantial amounts of bioactive compounds that they contain and the health benefits that these compounds confer. The bioactive compounds found in citrus fruits are fiber, folate, potassium, ascorbic acid (vitamin C), and phytochemicals (monoterpenes, limonoids, flavonoids, carotenoids, and hydroxycinnamic acid).18 In vitro studies conducted with flavonoids and limonoids have shown that they inhibit the proliferation of human pancreatic cancer cells.19,20 Epidemiologic studies have reported an inverse association between the consumption of citrus fruits and the risk for pancreatic cancer.21–31 Further investigation of the effects of citrus compounds on this type of cancer is certainly warranted. The objective of our present study was to identify specific citrus compounds that inhibit GSK-3β activity. Inhibitor data collected by using biochemical luminescence assays and computational molecular dockings provide direct evidence that several flavonoids in citrus fruit inhibit GSK-3β activity and predict binding modes for these compounds.
Materials and Methods
Reagents
Human recombinant GSK-3β and phosphoglycogen synthase peptide-2 substrate were purchased from Millipore (Billerica, Massachusetts, USA). Kinase-Glo Luminescent Kinase Assay™ was provided by Promega (Madison, Wisconsin, USA). Citrus compounds purchased from Sigma-Aldrich (St. Louis, Missouri, USA) included luteolin (>98%), apigenin (>95%), quercetin (>98%), kaempferol (>97%), rutin hydrate (>94%), naringenin (>95%), neohesperidin (>90%), flavone (97%), naringin (>90%), hesperidin (>80), caffeic acid (>98%), chlorogenic acid (>95%), and L-ascorbic acid (>99%). Hesperetin (>95%) was purchased from SAFC (Wicklow, Ireland) and limonin (>90%), from MP BioMedicals (Solon, Ohio, USA). Nobiletin (94.9%), tangeretin (96.4%), narirutin (93.9%), nomilin (87.7%), eriocitrin (97.4%), obacunone (85.8%), and azadirachtin (90.7%) were purchased from Chromadex (Irvine, California, USA). UltraPure water was purchased from Cayman Chemical (Ann Arbor, Michigan, USA). Adenosine triphosphate (ATP) and all other reagents were purchased from Sigma-Aldrich or Fisher Scientific (Pittsburgh, Pennsylvania, USA). Assay buffer contained 50 mM 4-(2-hydroxyethyl) piperazine-1-ethanesulfonic acid (HEPES) (pH, 7.5), 15 mM magnesium acetate, 1 mM EDTA, and 1 mM EGTA. Enzyme buffer contained 50 mM Tris/HCl (pH, 7.5), 150 mM NaCl, 0.l mM EGTA, 0.03% Brij-35, 270 mM sucrose, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM benzamidine, and 0.1% 2-mercaptoethanol.
GSK-3β biochemical assay
GSK-3β activity was determined by using the Kinase-Glo Luminescent Kinase Assay, as optimized by Baki et al.32 In a typical assay, the test inhibitor was dissolved in dimethylsulfoxide at a 10 mM concentration and then diluted to the desired concentrations (0.1, 1, 10, 25, 50, 100, 200, and 300 μM) using the assay buffer. The test inhibitor was then mixed in a black 96-well plate with 10 μL (20 ng) of GSK-3β and 20 μL of assay buffer containing 25 μM substrate and 1 μM ATP. The mixture was incubated at 30°C for 30 minutes, and the reaction was stopped by adding 40 μL of Kinase-Glo reagent. After an additional 10 minutes of incubation at 30°C, the luminescence was recorded by using the luminescence option on the Synergy 2 multimode microplate reader (BioTek Instruments, Inc., Winooski, Vermont, USA). All plates carried negative controls (100% inhibition), which were achieved by adding 5 μM of the known GSK-3β inhibitor SB 415286 (Tocris Bioscience, Ellisville, Michigan, USA), and positive controls (0% inhibition), which contained no test inhibitors and only assay buffer. The percentage inhibition was calculated for each test inhibitor as follows:
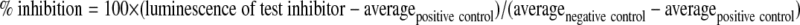 |
Each citrus compound was assayed in at least 2 independent trials with 3 replicates of each concentration per trial. Their inhibitory activities were expressed as the concentration of that particular citrus compound inhibiting GSK-3β activity by 20% and 50% (IC20 and IC50).
Molecular modeling
Preparation of GSK-3β structure
Of the 35 GSK-3β crystal structures available in the Protein Data Bank (http://www.rcsb.org), only 1 structure (PDB 1I09)33 had no ligand bound in the active site; our initial docking experiments were initiated with this structure. Another structure (PDB 1H8F)34 had the smallest ligand (HEPES) in the active site and was used to fill in 2 large internal gaps in the ligand-free structure. In the 1I09 file, these missing loop regions include residues 120–126 and 286–300 and the missing terminal regions include 24 residues at the N-terminus and 36 residues at C-terminus (Fig. 1). The complete 420–amino acid GSK-3β sequence was obtained from the Protein Knowledgebase (http://www.uniprot.org) and used to build 10 different models of the combined GSK-3β structure in the Molecular Operating Environment program35 using its HOMOLOGY function. The model with the best residue packing score calculated by this program's residue packing quality function was selected and hydrogen atoms were added to the structure before it was energy-minimized using the CHARMM22 forcefield36 and docked with inhibitors.
FIG. 1.
Sequence alignments. Amino acid sequences of the glycogen synthase kinase-3β (GSK-3β) structures reported in PDB 1H8F34 and 1I0933 and the complete 420 amino acid coding sequence of GSK-3β are aligned. Both 1H8F and 1I09 are missing residues from their N- and C-termini; 1I09 is missing 2 loop regions between residues 120-126 and 286-300.
Preparation of structures of citrus compounds and synthetic inhibitor SB 415286
The chemical structures of most citrus compounds were downloaded from the Kyoto Encyclopedia of Genes and Genomes database (http://www.genome.jp/kegg/). Some of the rare citrus compounds and the GSK-3β inhibitor SB 41528637 were built in the Molecular Operating Environment program using the MOLECULE BUILDER function and then energy-minimized using the MMFF94 forcefield.38
Docking experiment
Ligands were docked within the active site of the energy-minimized GSK-3β protein model by using the DOCK function within the Molecular Operating Environment program, which docks each ligand by initially placing it in the identified binding cavity and allowing it to vary through Monte Carlo simulations that remove biases due to manual placement. For each ligand, 50 possible conformations were generated while maintaining rigid protein side chains. The interaction energies between the ligand conformations and the protein were then calculated by using the potential energy function in the Molecular Operating Environment program and the 5 with the lowest energies were selected as the optimal conformations for each ligand and subjected to further energy minimization using the MMFF94 forcefield, while allowing full protein side chain relaxation. The interaction energies between the minimized protein and the ligands were calculated as the difference between the total potential energy of the minimized complex and the sum of the individual protein and ligand components of the complex. The potential energy function contains the sum of the ligand/protein internal energy, van der Waals and electrostatic energy, and terms the conformation with the lowest sum.39,40 The conformation with the lowest calculated interaction energy was selected as the most possible binding conformation.
Statistical analysis
The sigmoidal dose–response analysis of the IC20 and IC50 values of each compound on GSK-3β activity was performed by nonlinear regression (curve fit) using GraphPad Prism software.41 Pearson correlation analyses were performed by using SAS 9.2 statistical software.42
Results and Discussion
Citrus compounds inhibit GSK-3β activity
Among the citrus compounds tested (flavonoids, limonoids, phenolic acids, and ascorbic acid), the flavonoids (Fig. 2A presents the basic chemical structure of flavonoids) had the overall highest inhibitory activity on GSK-3β (Tables 1 and 2). In particular, the flavonoids luteolin (IC50, 1.5 μM), apigenin (IC50, 1.9 μM), quercetin (IC50, 2.0 μM), kaempferol (IC50, 3.5 μM), rutin (IC50, 10.3 μM), hesperetin (IC50, 26.9 μM), and naringenin (IC50, 45.7 μM) all inhibited GSK-3β activity by at least 50% at concentrations of 50 μM or lower. On the basis of the structures of the flavonoids (Fig. 2B and C) tested, it is apparent that side group substitutions affect their capability to inhibit GSK-3β activity and that flavonoids with larger side groups have lower inhibitory activity. For instance, hesperidin, narirutin, eriocitrin, naringin, and neohesperidin, all of which have sugar substitutions, have low inhibitory activities ranging from 3% to 34% at concentrations of 100 or 300 μM. Other classes of citrus compounds, such as the limonoids (nomilin, obacunone, limonin, and azadirachtin), phenolic acids (caffeic acid and chlorogenic acid), and ascorbic acid, also have less of an effect on GSK-3β activity with inhibition ranging from 1% to 40% at concentrations of 100, 200, or 300 μM (data not shown).
FIG. 2.
Flavonoids and known glycogen synthase kinase-3β (GSK-3β) inhibitor structures. (A) Basic flavonoid structure. The A-ring is commonly hydroxylated at positions 5 and 7, and the B-ring is hydroxylated at positions 4′, 3′4′ or 3′4′5′. (B) Structures of flavonoids presented in Table 1. (C) Some of the flavonoid structures and the 2 known inhibitor structures presented in Table 2.
Table 1.
Physicochemical Properties, Glycogen Synthase Kinase-3β Inhibition, and Predicted Interaction and Complex Energies of Citrus Flavonoids
| Compound | Molecular Mass (mg/mmol) | Free Hydroxyl Groups | IC20 Value (μM)a | IC50 Value (μM)a | Interaction Energy (kcal·mol−1)b | Complex Energy (kcal·mol−1)b |
|---|---|---|---|---|---|---|
| Flavonoids | ||||||
| Luteolin | 286.24 | 4 | 0.48±0.03 | 1.51±0.05 | −76.4 | −718.1 |
| Apigenin | 270.24 | 3 | 0.48±0.03 | 1.91±0.06 | −76.1 | −688.1 |
| Quercetin | 302.24 | 5 | 0.68±0.00 | 2.04±0.00 | −84.6 | −719.7 |
| Kaempferol | 286.24 | 4 | 0.83±0.03 | 3.47±0.11 | −67.1 | −626.7 |
| Hesperetin | 302.28 | 3 | 5.89±0.38 | 26.92±0.88 | −56.1 | −710.2 |
| Naringenin | 272.26 | 3 | 10.97±0.36 | 45.71±0.00 | −60.7 | −688.1 |
| Flavone | 222.24 | 0 | 44.68±1.45 | >100 | −24.1 | −675.0 |
| Methoxyflavones | ||||||
| Nobiletin | 402.39 | 0 | 12.30±0.00 | 52.48±0.00 | −51.8 | −586.8 |
| Tangeretin | 372.37 | 0 | >100 (13.2%, 100 μM) | >100 | −48.6 | −592.1 |
Data are expressed as the mean±standard deviation. The lower the IC value, the higher the potency.
The interaction energies and complex energies were calculated by using the potential energy function in the Molecular Operating Environment program. The lower the energy, the stronger the binding.
IC20 and IC50, concentration inhibiting glycogen synthase kinase-3β activity by 20% and 50%, respectively.
Table 2.
Physicochemical Properties, Glycogen Synthase Kinase-3β Inhibition and Predicted Interaction and Complex Energies of Citrus Flavonoids and Synthetic Inhibitors SB 415286 and I-5
| Compound | Molecular Mass (mg/mmol) | Free Hydroxyl Groups | IC20 Value (μM)a | IC50 Value (μM)a | Interaction Energy (kcal·mol−1)b | Complex Energy (kcal·mol−1)b |
|---|---|---|---|---|---|---|
| Glycosylated flavonoids | ||||||
| Rutin | 610.52 | 10 | 2.58±0.34 | 10.28±1.33 | −98.0 | −576.8 |
| Hesperidin | 610.57 | 8 | 133.79±15.22 | >300 | −136.2 | −560.5 |
| Narirutin | 580.53 | 8 | >100 (11.3%, 100 μM) | >100 | −142.9 | −556.5 |
| Eriocitrin | 596.53 | 9 | >100 (10.3%, 100 μM) | >100 | −88.9 | −606.0 |
| Naringin | 580.53 | 8 | >300 (15.0%, 300 μM) | >300 | −134.6 | −566.5 |
| Neohesperidin | 610.56 | 8 | >100 (2.8%, 100 μM) | >100 | −122.4 | −529.8 |
| Synthetic inhibitors | ||||||
| SB 415286 | 359.73 | 1 | 0.02±0.00 | 0.07±0.00 | −94.0 | −641.4 |
| I-5 | 377.18 | 1 | N/A | 0.160c | −99.0 | −682.5 |
Data are expressed as the mean±standard deviation. The lower the IC value, the higher the potency.
The interaction energies and complex energies were calculated by using the potential energy function in the Molecular Operating Environment program. The lower the energy, the stronger the binding.
Value reported in literature.43
IC20 and IC50, concentration inhibiting glycogen synthase kinase-3β activity by 20% and 50%, respectively.
Molecular interactions of GSK-3β and citrus compounds
The citrus flavonoids were selected for further molecular docking analysis because of their high inhibitory activity on GSK-3β. Because no crystal structure of GSK-3β bound with SB 415286 or a flavonoid exists, the first step in our docking process was to identify the potential binding site for citrus flavonoids. For this, the crystal structure of GSK-3β bound to another anilinomalemide with a size similar to that of SB 415286, 2-chloro-5-{[4-(3-chloro-phenyl)-2,5-dioxo-2,5-dihydro-1H-pyrrol-3-yl]-amino}benzoic acid (I-5) (PDB 1Q4L),43 was used to locate the probable binding site for flavonoids within the active site of the GSK-3β structure. Figure 3 shows the orientation of I-5 bound in the active site of the GSK-3β crystal structure, located between the N-terminal β-strand domain and the C-terminal α-helical domain and bordered by a glycine-rich loop and a hinge region. For each citrus flavonoid, the conformation closest to the binding position of I-5 and with the least predicted interaction energy was selected as the most possible binding conformation.
FIG. 3.
Predicted structure of glycogen synthase kinase-3β (GSK-3β) complexed with I-5. The predicted protein backbone of GSK-3β is shown in ribbon format, with α-helices shown in red, β-sheets in yellow, and loops in blue. The active site is located between the β-rich N-terminus and the α-rich C-terminus, and the predicted binding mode of I-5 is shown in space-filling format.
Analysis of the predicted binding site for flavonoids (Fig. 4A) showed a planar cleft with a narrow opening of about 11 Å in length and about 4.5 Å in width. The top and bottom surfaces of the cleft were formed by hydrophobic residues, and the edges were formed by polar residues from the hinge and the glycine-rich loop of the GSK-3β structure. The edge directly opposite to the cavity opening had fewer polar residues. This architecture appears to make the site uniquely suitable for planar aromatic ligands with polar substituted groups. All the flavonoids were predicted to bind in almost the same orientation with the B-ring hydroxyls stabilized by hydrogen bonding with Arg141 and Tyr134 in the hinge; A-ring hydroxyls stabilized by hydrogen bonding with Asn64, Gly65, Lys85, and Asp200 residues in the glycine-rich loop; and the carbonyl oxygen on the C-ring facing toward the back of the cavity.
FIG. 4.
Predicted docking mode for luteolin in the binding cavity of glycogen synthase kinase-3β (GSK-3β). (A) Surface representation of the binding cavity of GSK-3β is shown with the predicted mode for luteolin binding and the interacting amino acid residues. Binding cavity residues are shown with acidic residues in red, basic residues in dark blue, hydrophobic residues in green, and hydrophilic residues in light blue. (B) Two-dimensional representation of the luteolin interacting residues following the method of Clark and Labute.54 Binding cavity residues that are shown with green circles indicate residues with no polar or charged side chains. Residues with light mauve circles indicate polar side chains that can be acidic, indicated by a red ring, or basic, indicated by a blue ring. The arrows indicate hydrogen bonds to side chain residues in green and backbone residues in blue.
Tables 1 and 2 compare the IC20, IC50, interaction energy (IE), and complex energy (CE) values of the flavonoids tested and the 2 known GSK-3β inhibitors, as well as their physicochemical properties. Luteolin, apigenin, quercetin, kaempferol, and flavone all have an unsaturated C-ring (Fig. 2B) that allows them to remain planar in the binding cavity of the enzyme, thereby avoiding unfavorable steric interactions and allowing them to minimize their CEs (−626.7 to −719.7 kcal·mol−1). The fact that, among these 5 flavonoids, flavone has no hydroxyl residues capable of hydrogen bonding with residues in the active site causes its IE to be quite high (−24.1 kcal·mol−1) compared with the others (IE, −67.1 to −84.6 kcal·mol−1). Further analysis showed that there is a negative correlation (r=−0.83) between the number of hydroxyl substituted groups on the flavonoids (includes all 15 flavonoids tested) and the predicted IE values of the flavonoids. The lower IE values calculated for the various hydroxylated flavonoids indicate that their interactions are comparatively more favorable and likely to more effectively block GSK-3β activity, as observed in our biochemical assays.
Several other studies have reported the importance of the presence of hydroxyl groups on the flavonoid structure for some of its biochemical activities. Particularly relevant to our discussion of cancer prevention, a study conducted by Ueda et al.44 determined that flavonoids with hydroxyl groups on the A-5, A-7, and B-4′ positions, such as luteolin, apigenin, quercetin, and kaempferol, had the highest inhibitory effect on tumor necrosis factor α production in vitro. Several studies have also demonstrated that the total number and location of hydroxyl groups on flavonoids greatly influence their impact on several mechanisms of antioxidant activity.45–49
Both hesperetin and naringenin contain hydroxyl groups on their A- and B-rings that form hydrogen bonds with residues in the active site and have lower inhibitory capacities (IC50, 26.9 μM for hesperetin and 45.7 μM for naringenin) and slightly higher IE values (IE, −56.1 kcal·mol−1 for hesperetin and −60.7 kcal·mol−1 for naringenin) than luteolin (IC50, 1.5 μM and IE, −76.4 kcal·mol−1). Comparison of their structures (Fig. 2B) shows that the C-ring of luteolin is unsaturated, while the C-rings in both hesperetin and naringenin are saturated, causing bulging in the middle of these molecules and unfavorable steric interactions in the narrow binding site. A study conducted by Chen et al.50 assaying for proteasome inhibition also found that flavonoids having saturated C-rings are much less potent than flavonoids having unsaturated C-rings. In particular, unsaturated luteolin and apigenin were approximately 11- and 21-fold more effective at inhibiting 20S and 26S proteasome activities than their saturated counterparts eriodictyol and naringenin, respectively.
Both nobiletin and tangeretin contain multiple methoxy side groups (Fig. 2B) that decrease their ability to form hydrogen bonds in the active site, as well as decrease their inhibitory capacities. In addition to the lower number of potential hydrogen bonds, the large methyl groups on these molecules lead to steric interactions that increase their IEs and CEs. Analyses of their docking conformations did not explain why the inhibitory activity of nobiletin (IC50, 52.5 μM) with 6 methoxy groups was so much higher than that of tangeretin (IC50>100 μM) with 5 methoxy groups. The IC50 values of flavonoids that were found to inhibit GSK-3β activity, including luteolin, apigenin, quercetin, kaempferol, rutin, hesperetin, naringenin, and nobiletin correlated with their IE values (r=0.71).
In comparing the nonglycosylated citrus flavonoids (Table 1) with the glycosylated citrus flavonoids (Table 2), it was determined that conjugation of bulky sugar groups to the flavonoid core structure drastically decreased the inhibitory capacity of the tested flavonoids. Docking results indicate that the complex energies calculated for GSK-3β bound to these glycosylated flavonoids were generally higher than for flavonoid aglycones, causing the interactions to be less stable and have shorter lifetimes. These results are similar to those of a study conducted by Lin et al.51 that found, through molecular modeling and docking, that flavonoids conjugated with sugars had weaker interactions with xanthine oxidase, an enzyme that causes gout and is responsible for oxidative damage to tissues. In both our work and that of Lin et al.,51 glycosylation probably decreased the hydrophobicity of the flavonoids, thereby reducing hydrophobic interactions between these ligands and their target proteins. Rutin is the exception to this conclusion in that it has a much greater inhibitory capacity (IC50, 10.3 μM) compared with the other glycosylated flavonoids. Examination of the rutin structure (Fig.2C) indicates that its sugar group is conjugated to the C-ring of the flavonoid skeleton rather than to the A-ring, as in all other tested glycosylated flavonoids. This orientation is much more favorable for binding to the GSK-3β active site because it causes the sugar group to orient to the middle of the opening in the binding site (Fig. 5) rather than very close to the edge of the opening, as is the case for narirutin (also shown in Fig. 5) and other glycosylated flavonoids (not shown).
FIG. 5.
Predicted docking modes for rutin and narirutin in the binding cavity of glycogen synthase kinase-3β (GSK-3β). Surface representation of the binding cavity of GSK-3β is shown with the predicted modes for rutin in blue and for narirutin in orange. The binding cavity residues are shown with acidic residues in red, basic residues in dark blue, hydrophobic residues in green, and hydrophilic residues in light blue.
To the best of our knowledge, this is the first report of the inhibition of GSK-3β activity by citrus bioactive compounds evaluated biochemically and computationally. In 2009, Bustanji et al.52 studied the effects of curcumin, another polyphenol, on GSK-3β activity. They used a different software program, FRED software, and a different GSK-3β crystal structure, 1Q5K.53 However, both studies found that either ligand, curcumin52 or flavonoids (present study), helped stabilize themselves within the GSK-3β active site by hydrogen bonding with the amino acid residues Lys85 and Arg141. The researchers of the curcumin study also validated their docking results by in vitro studies, which showed curcumin potently and more effectively inhibited GSK-3β (IC50, 66.3 nM) than the GSK-3β known inhibitor TDZD-8 (IC50, 1.5 μM). Additional in vivo analyses by these researchers showed that curcumin significantly increased liver glycogen reserves in fasting Balb/c mice in a dose-dependent manner, possibly as the result of GSK-3β inhibition. These results, along with our findings, provide critical evidence documenting the need for further investigation into the mechanisms of inhibition of GSK-3β and the downstream effects this may cause.
A limitation of our study is that the findings are not of physiologic relevance at this time. However, in our laboratory we are studying the effects of citrus compounds in pancreatic cancer cells to determine whether inhibition of GSK-3β activity is indeed part of their mechanism of action. Future studies will consider bioavailability and metabolism of these flavonoids.
In conclusion, our study demonstrated that a variety of citrus flavonoids can inhibit GSK-3β activity directly by binding in the active site of the enzyme. Flavonoids with hydroxyl side groups that are available for hydrogen bonding with the amino acid residues in the enzyme were the most favorable. Flavonoids with large side groups (i.e., methoxy groups or sugar conjugations) were much more unfavorable because of the drastic alterations the enzyme had to make in order to accommodate them into its binding site.
Acknowledgments
The work resulting in this publication was supported by a U.S. Department of Agriculture National Needs Predoctoral Fellowship, a Division of Nutritional Sciences Margin of Excellence Student Research Award, and a National Institutes of Health grant covering the modeling programs (1RO1 GM079530).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Jemal A. Siegel R. Xu J. Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.National Cancer Institute. What you need to know about cancer of the pancreas. http://www.cancer.gov/cancertopics/wyntk/pancreas. [Jun 10;2010 ]. http://www.cancer.gov/cancertopics/wyntk/pancreas
- 3.Eldar-Finkelman H. Glycogen synthase kinase 3: an emerging therapeutic target. Trends Mol Med. 2002;8:126–132. doi: 10.1016/s1471-4914(01)02266-3. [DOI] [PubMed] [Google Scholar]
- 4.Eldar-Finkelman H. Schreyer S. Shinohara M. LeBoeuf R. Krebs E. Increased glycogen synthase kinase-3 activity in diabetes- and obesity-prone C57BL/6J mice. Diabetes. 1999;48:1662–1666. doi: 10.2337/diabetes.48.8.1662. [DOI] [PubMed] [Google Scholar]
- 5.Wagman A. Johnson K. Bussiere D. Discovery and development of GSK3 inhibitors for the treatment of type 2 diabetes. Curr Pharm Des. 2004;10:1105–1137. doi: 10.2174/1381612043452668. [DOI] [PubMed] [Google Scholar]
- 6.Hanger D. Hughes K. Woodgett J. Brion J. Anderton B. Glycogen synthase kinase-3 induces Alzheimer's disease-like phoshorylation of tau: generation of paired helical filament epitopes and neuronal localisation of the kinase. Neurosci Lett. 1992;147:58–62. doi: 10.1016/0304-3940(92)90774-2. [DOI] [PubMed] [Google Scholar]
- 7.Hernández F. Avila J. The role of glycogen synthase kinase 3 in the early stages of Alzheimers' disease. FEBS Lett. 2008;582:3848–3854. doi: 10.1016/j.febslet.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 8.Martin L. Magnaudeix A. Esclaire F. Yardin C. Terro F. Inhibition of glycogen synthase kinase-3β downregulates total tau proteins in cultured neurons and its reversal by the blockade of protein phosphatase-2A. Brain Res. 2009;1252:66–75. doi: 10.1016/j.brainres.2008.11.057. [DOI] [PubMed] [Google Scholar]
- 9.Erdal E. Ozturk N. Cagatay T. Eksioglu-Demiralp E. Ozturk M. Lithium-mediated downregulation of PKB/Akt and cyclin E with growth inhibition in hepatocellular carcinoma cells. Int J Cancer. 2005;115:903–910. doi: 10.1002/ijc.20972. [DOI] [PubMed] [Google Scholar]
- 10.Mazor M. Kawano Y. Zhu H. Waxman J. Kypta R. Inhibition of glycogen synthase kinase-3 represses androgen receptor activity and prostate cancer cell growth. Oncogene. 2004;23:7882–7892. doi: 10.1038/sj.onc.1208068. [DOI] [PubMed] [Google Scholar]
- 11.Ougolkov A. Fernandez-Zapico M. Bilim V, et al. Aberrant nuclear accumulation of glycogen synthase kinase-3β in human pancreatic cancer: association with kinase activity and tumor dedifferentiation. Clin Cancer Res. 2006;12:5074–5081. doi: 10.1158/1078-0432.CCR-06-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ougolkov A. Fernandez-Zapico M. Savoy D. Urrutia R. Billadeau D. Glycogen synthase kinase-3β participates in nuclear factor κB-mediated gene transcription and cell survival in pancreatic cancer cells. Cancer Res. 2005;65:2076–2081. doi: 10.1158/0008-5472.CAN-04-3642. [DOI] [PubMed] [Google Scholar]
- 13.Shakoori A. Ougolkov A. Yu Z, et al. Deregulated GSK3β activity in colorectal cancer: its association with tumor cell survival and proliferation. Biochem Biophys Res Commun. 2005;334:1365–1373. doi: 10.1016/j.bbrc.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 14.Doble B. Woodgett J. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beg A. Sha W. Bronson R. Ghosh S. Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 16.Hoeflich K. Luo J. Ruble E, et al. Requirement for glycogen synthase kinase-3β in cell survival and NF-κB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 17.Li Q. Van Antwerp D. Mercurio F. Lee K. Verma I. Severe liver degeneration in mice lacking the IκB kinase 2 gene. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- 18.Economos C. Clay W. Nutritional and health benefits of citrus fruits. Food Nutr Agric. 1999;24:11–18. [Google Scholar]
- 19.Patil J. Jayaprakasha G. Murthy K. Chetti M. Patil B. Characterization of Citrus aurantifolia bioactive compounds and their inhibition of pancreatic cancer cells through apoptosis. Microchem J. 2010;94:108–117. [Google Scholar]
- 20.Patil J. Murthy K. Jayaprakasha G. Chetti M. Patil B. Bioactive compounds from Mexican lime (Citrus aurantifolia) juice induce apoptosis in human pancreatic cells. J Agric Food Chem. 2009;57:10933–10942. doi: 10.1021/jf901718u. [DOI] [PubMed] [Google Scholar]
- 21.Bueno de Mesquita H. Maisonneuve P. Runia S. Moerman C. Intake of foods and nutrients and cancer of the exocrine pancreas: a population-based case-control study in the Netherlands. Int J Cancer. 1991;48:540–549. doi: 10.1002/ijc.2910480411. [DOI] [PubMed] [Google Scholar]
- 22.Chan J. Wang F. Holly F. Vegetable and fruit intake and pancreatic cancer in a population-based case-control study in the San Francisco Bay area. Cancer Epidemiol Biomarkers Prev. 2005;14:2093–2097. doi: 10.1158/1055-9965.EPI-05-0226. [DOI] [PubMed] [Google Scholar]
- 23.Coughlin S. Calle E. Patel A. Thun M. Predictors of pancreatic cancer mortality among a large cohort of United States adults. Cancer Causes Control. 2000;11:915923. doi: 10.1023/a:1026580131793. [DOI] [PubMed] [Google Scholar]
- 24.Ji B. Chow W. Gridley G, et al. Dietary factors and the risk of pancreatic cancer: a case-control study in Shanghai, China. Cancer Epidemiol Biomarkers Prev. 1995;4:885–893. [PubMed] [Google Scholar]
- 25.Larsson S. Håkansson N. Näslund I. Bergkvist L. Wolk A. Fruit and vegetable consumption in relation to pancreatic cancer risk: a prospective study. Cancer Epidemiol Biomarkers Prev. 2006;15:301–305. doi: 10.1158/1055-9965.EPI-05-0696. [DOI] [PubMed] [Google Scholar]
- 26.Lin Y. Kikuchi S. Tamakoshi A, et al. Dietary habits and pancreatic cancer risk in a cohort of middle-aged and elderly Japanese. Nutr Cancer. 2006;56:40–49. doi: 10.1207/s15327914nc5601_6. [DOI] [PubMed] [Google Scholar]
- 27.Nöthlings U. Murphy S. Wilkens L, et al. Dietary glycemic load, added sugars, and carbohydrates as risk factors for pancreatic cancer: the multiethnic cohort study. Am J Clin Nutr. 2007;86:1495–1501. doi: 10.1093/ajcn/86.5.1495. [DOI] [PubMed] [Google Scholar]
- 28.Olsen G. Mandel J. Gibson R. Wattenberg L. Schuman L. Nutrients and pancreatic cancer: a population-based case-control study. Cancer Causes Control. 1991;2:291–297. doi: 10.1007/BF00051668. [DOI] [PubMed] [Google Scholar]
- 29.Polesel J. Talamini R. Negri E, et al. Dietary habits and risk of pancreatic cancer: an Italian case-control study. Cancer Causes Control. 2010;21:493–500. doi: 10.1007/s10552-009-9480-2. [DOI] [PubMed] [Google Scholar]
- 30.Silverman D. Swanson C. Gridley G, et al. Dietary and nutritional factors and pancreatic cancer: a case-control study based on direct interviews. J Natl Cancer Inst. 1998;90:1710–1719. doi: 10.1093/jnci/90.22.1710. [DOI] [PubMed] [Google Scholar]
- 31.Stolzenberg-Solomon R. Pietinen P. Taylor P. Virtamo J. Albanes D. Prospective study of diet and pancreatic cancer in male smokers. Am J Epidemiol. 2002;155:783–792. doi: 10.1093/aje/155.9.783. [DOI] [PubMed] [Google Scholar]
- 32.Baki A. Bielik A. Molnár L. Szendrei G. Keserü G. A high throughput luminescent assay for glycogen synthase kinase-3β inhibitors. Assay Drug Dev Technol. 2007;5:75–83. doi: 10.1089/adt.2006.029. [DOI] [PubMed] [Google Scholar]
- 33.Haar E. Coll J. Austen D, et al. Structure of GSK3β reveals a primed phosphorylation mechanism. Nat Struct Biol. 2001;8:593–596. doi: 10.1038/89624. [DOI] [PubMed] [Google Scholar]
- 34.Dajani R. Fraser E. Roe S, et al. Crystal structure of glycogen synthase kinase 3β: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105:721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 35.Molecular Operating Environment Program, Version 2009.10. Montreal, Quebec, Canada: Chemical Computing Group Inc.; 2009. [Google Scholar]
- 36.MacKerell A. Bashford D. Bellott M, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 37.Coghlan M. Culbert A. Cross D, et al. Selective small molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem Biol. 2000;7:793–803. doi: 10.1016/s1074-5521(00)00025-9. [DOI] [PubMed] [Google Scholar]
- 38.Halgren T. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17:490–519. [Google Scholar]
- 39.Baudry J. Li W. Pan L. Berenbaum M. Schuler M. Molecular docking of substrates and inhibitors in the catalytic site of CYP6B1, an insect cytochrome P450 monooxygenase. Protein Engineering. 2003;16:577–587. doi: 10.1093/protein/gzg075. [DOI] [PubMed] [Google Scholar]
- 40.Wang W. Rupasinghe S. Schuler M. Gonzalez de Mejia E. Identification and characterization of topoisomerase II inhibitory peptides from soy protein hydrolysates. J Agric Food Chem. 2008;56:6267–6277. doi: 10.1021/jf8005195. [DOI] [PubMed] [Google Scholar]
- 41.GraphPad Prism Software, Version 4.0. La Jolla, California: GraphPad Software Inc., USA; 2003. [Google Scholar]
- 42.Statistical Analysis SAS/STAT Software, Version 9.2. Cary, North Carolina, USA: SAS Institute Inc.; 2009. [Google Scholar]
- 43.Bertrand J. Thieffine S. Vulpetti A, et al. Structural characterization of the GSK-3β active site using selective and non-selective ATP-mimetic inhibitors. J Mol Biol. 2003;333:393–407. doi: 10.1016/j.jmb.2003.08.031. [DOI] [PubMed] [Google Scholar]
- 44.Ueda H. Yamazaki C. Yamazaki M. A hydroxyl group of flavonoids affects oral anti-inflammatory activity and inhibition of systemic tumor necrosis factor-α production. Biosci Biotechnol Biochem. 2004;68:119–125. doi: 10.1271/bbb.68.119. [DOI] [PubMed] [Google Scholar]
- 45.Burda S. Oleszek W. Antioxidant and antiradical activities of flavonoids. J Agric Food Chem. 2001;49:2774–2779. doi: 10.1021/jf001413m. [DOI] [PubMed] [Google Scholar]
- 46.Cao G. Sofic E. Prior R. Antioxidant and prooxidant behavior of flavonoids: structure-activity relationships. Free Radic Biol Med. 1997;22:749–760. doi: 10.1016/s0891-5849(96)00351-6. [DOI] [PubMed] [Google Scholar]
- 47.Haenen G. Paquay J. Korthouwer R. Bast A. Peroxynitrite scavenging by flavonoids. Biochem Biophys Res Commun. 1997;236:591–593. doi: 10.1006/bbrc.1997.7016. [DOI] [PubMed] [Google Scholar]
- 48.Pannala A. Chan T. O'Brien P. Rice-Evans C. Flavonoid B-ring chemistry and antioxidant activity: fast reaction kinetics. Biochem Biophys Res Commun. 2001;282:1161–1168. doi: 10.1006/bbrc.2001.4705. [DOI] [PubMed] [Google Scholar]
- 49.Yokozawa T. Chen C. Dong E, et al. Study on the inhibitory effect of tannins and flavonoids against the 1,1-diphenyl-2-picrylhydrazyl radical. Biochem Pharmacol. 1998;56:213–222. doi: 10.1016/s0006-2952(98)00128-2. [DOI] [PubMed] [Google Scholar]
- 50.Chen D. Chen M. Cui Q. Yang H. Dou Q. Structure-proteasome-inhibitory activity relationships of dietary flavonoids in human cancer cells. Front Biosci. 2007;12:1935–1945. doi: 10.2741/2199. [DOI] [PubMed] [Google Scholar]
- 51.Lin C. Chen C. Chen C. Liang Y. Lin J. Molecular modeling of flavonoids that inhibits xanthine oxidase. Biochem Biophys Res Commun. 2002;294:167–172. doi: 10.1016/S0006-291X(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 52.Bustanji Y. Taha M. Almasri I, et al. Inhibition of glycogen synthase kinase by curcumin: investigation by simulated molecular docking and subsequent in vitro/in vivo evaluation. J Enzyme Inhib Med Chem. 2009;24:771–778. doi: 10.1080/14756360802364377. [DOI] [PubMed] [Google Scholar]
- 53.Bhat R. Xue Y. Berg S, et al. Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-014418. J Biol Chem. 2003;278:45937–45945. doi: 10.1074/jbc.M306268200. [DOI] [PubMed] [Google Scholar]
- 54.Clark A. Labute P. 2D depiction of protein-ligand complexes. J Chem Inf Model. 2007;47:1933–1944. doi: 10.1021/ci7001473. [DOI] [PubMed] [Google Scholar]







