The nitrate transporter NRT1.9 is expressed in root companion cells and has an important role in the transport of nitrate in plant phloem tissue. Mutants of NRT1.9 show reduced downward nitrate transport in phloem and increased root-to-shoot nitrate transport in xylem. This suggests that phloem transport serves as fine-tuning for nitrate redistribution.
Abstract
This study of the Arabidopsis thaliana nitrate transporter NRT1.9 reveals an important function for a NRT1 family member in phloem nitrate transport. Functional analysis in Xenopus laevis oocytes showed that NRT1.9 is a low-affinity nitrate transporter. Green fluorescent protein and β-glucuronidase reporter analyses indicated that NRT1.9 is a plasma membrane transporter expressed in the companion cells of root phloem. In nrt1.9 mutants, nitrate content in root phloem exudates was decreased, and downward nitrate transport was reduced, suggesting that NRT1.9 may facilitate loading of nitrate into the root phloem and enhance downward nitrate transport in roots. Under high nitrate conditions, the nrt1.9 mutant showed enhanced root-to-shoot nitrate transport and plant growth. We conclude that phloem nitrate transport is facilitated by expression of NRT1.9 in root companion cells. In addition, enhanced root-to-shoot xylem transport of nitrate in nrt1.9 mutants points to a negative correlation between xylem and phloem nitrate transport.
INTRODUCTION
Nitrate is one of the major nitrogen sources for the synthesis of amino acids and nucleic acids in higher plants. Nitrate assimilation starts with the reduction of nitrate to nitrite by nitrate reductase (NR) in the cytoplasm and the subsequent reduction of nitrite to ammonium by nitrite reductase in chloroplasts (or plastids in the roots). The resulting ammonium is subsequently incorporated into carbon skeletons via the Gln synthetase–Glu synthase cycle. Nitrate is mainly taken up from the soil by the plant root, but its assimilation can take place in both the root and shoot. In addition to being assimilated in the cytoplasm, nitrate can be stored in vacuoles, and it has therefore been proposed that nitrate may be an important osmotic solute (Smirnoff and Stewart, 1985; Song et al., 2006). Besides serving as a nutrient source and osmoticum, nitrate also functions as a signal molecule regulating nitrogen and carbon metabolism and coordinating whole-plant development (Forde, 2002; Sakakibara et al., 2006; Vidal and Gutiérrez, 2008). Since nitrate is a key component in the regulation of plant development and growth, the regulation of nitrate distribution, for example, root-to-shoot nitrate transport, is important for plants to modulate their growth in response to various environmental conditions.
The amount of nitrate transported from the root to the shoot depends on the plant species, temperature, external nitrate concentration, and light intensity (Smirnoff and Stewart, 1985). Some plants (e.g., species of Xanthium, Gossypium, and Cucumis) appear to lack root NR activity, and all the nitrate absorbed is transported to the shoot (Pate, 1980, 1983). Other species, such as barley (Hordeum vulgare), soybean (Glycine max), and Arabidopsis thaliana (Aslam and Huffaker, 1982; Crafts-Brandner and Harper, 1982; Cheng et al., 1991), reduce nitrate in both root and shoot. In terms of environmental factors, it has been shown that a greater portion of absorbed nitrate is assimilated in the root at low temperature, low external nitrate concentration, or under low light intensity (Beevers and Hageman, 1980; Deane-Drummond et al., 1980; Clarkson and Deane-Drummond, 1983; Pate, 1983). In the leaf, the reductants and ATP required for nitrate assimilation come from photosynthetic electron transport (Abrol et al., 1983; Wallsgrove et al., 1983); whereas, in the root, they come from mitochondrial respiration, the pentose phosphate pathway, and malate oxidation (Lee, 1980). Since nitrate assimilation in the root requires carbohydrate and malate to be transported from the shoot to the root, it has been proposed that, if the amount of light is sufficient, leaf assimilation has a lower energy cost (Smirnoff and Stewart, 1985). However, when the light intensity is low, no advantage will be gained in assimilating nitrate in the leaf because nitrate and carbon dioxide compete for the reductants and ATP (Canvin and Atkins, 1974). Thus, nitrate transfer from root to shoot is not only regulated by intrinsic factors that differ between species but also by environmental conditions. However, little is known about how root-to-shoot nitrate transport is regulated at the molecular level.
CHL1 (NRT1.1), NRT1.2, NRT2.1, and NRT2.2, members of the NRT1 and NRT2 families of nitrate transporters, are known to be involved in nitrate uptake from the soil solution into root cells (Tsay et al., 1993; Wang et al., 1998; Huang et al., 1999; Liu et al., 1999; Cerezo et al., 2001; Filleur et al., 2001; Little et al., 2005; Orsel et al., 2006; Li et al., 2007). In Arabidopsis, there are 53 NRT1 transporters. One of these, NRT1.5, which can mediate nitrate efflux and is expressed in root pericycle cells close to the xylem, is responsible for loading nitrate into the xylem for root-to-shoot nitrate transport (Lin et al., 2008). NRT1.8, expressed in root xylem parenchyma cells, is responsible for nitrate retrieval from the xylem sap (Li et al., 2010). In contrast with amino acids, which can be circulated between the roots and shoots through the xylem and phloem, it is generally believed that nitrate can be translocated in the xylem, but not the phloem (Daniel-Vedele and Chaillou, 2005). However, a recent study of another NRT1 transporter, NRT1.7, showed that nitrate can be remobilized from older leaves to younger leaves and that source-to-sink remobilization of nitrate is mediated by the phloem (Fan et al., 2009). In this study, we report that mutation of the Arabidopsis nitrate transporter NRT1.9 reduced nitrate content in root phloem and increased shoot nitrate content. The phenotypes of nrt1.9 mutants suggest that NRT1.9-mediated phloem loading of nitrate in roots influences nitrate distribution between the root and shoot.
RESULTS
Sequence Analysis of NRT1:9
NRT1:9 (At1g18880) is a member of the Arabidopsis NRT1 (PTR) transporter family (Tsay et al., 2007). According to phylogenetic analysis of the Arabidopsis NRT1 family, NRT1.9 is classified, together with NRT1.6 and NRT1.7, in subgroup IV, while CHL1, NRT1.2, NRT1.3, and NRT1.4 are in subgroup I, and NRT1.5 is in subgroup II (Tsay et al., 2007). The predicted protein sequence of NRT1:9 shows 45, 42, 35, 34, 33, 31, and 28% identity with that of NRT1.7, NRT1.6, CHL1, NRT1.3, NRT1.4, NRT1.5, and NRT1.2, respectively. Similar to other reported NRT1 transporters, NRT1.9 also contains 12 putative transmembrane domains and a long hydrophilic loop between transmembrane domains 6 and 7 (see Supplemental Figure 1 online).
NRT1.9 Is a Low-Affinity Nitrate Transporter
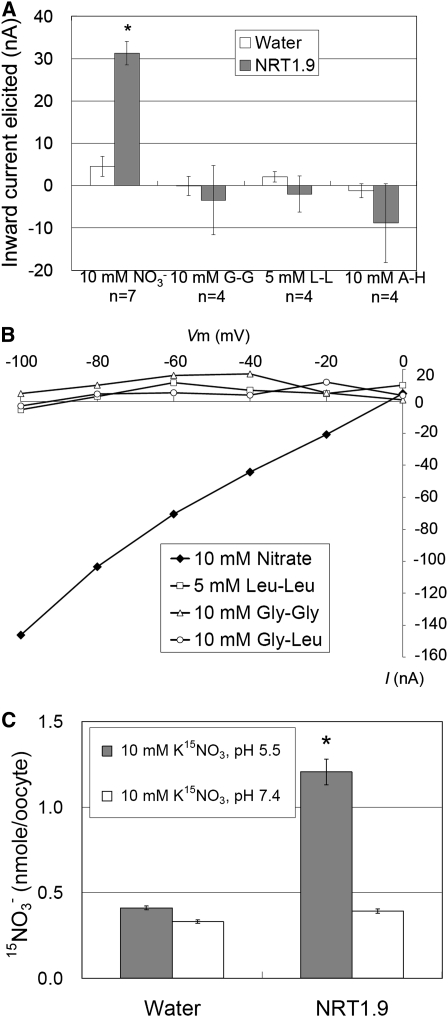
Some members of the NRT1 family are nitrate transporters, examples being CHL1 (Tsay et al., 1993) in Arabidopsis and Bn NRT1.2 (Zhou et al., 1998) in Brassica napus, while others, such as At PTR1 (Dietrich et al., 2004) and At PTR2 (Rentsch et al., 1995), are di/tripeptide transporters. To elucidate the function of NRT1.9, it was heterologously expressed in Xenopus laevis oocytes and the substrate specificity analyzed by two-electrode whole-cell clamping. As shown in Figure 1A, compared with water-injected oocytes, at pH 5.5, a larger current was elicited by 10 mM HNO3 in NRT1.9-injected oocytes, but no current was elicited by 5 mM Leu-Leu, 10 mM Gly-Gly, or 10 mM Ala-His. At all membrane potentials tested (from −100 to 0 mV), nitrate, but not any of the dipeptides tested, could elicit an inward current in NRT1.9-injected oocytes (Figure 1B). These data indicate that NRT1.9 is a nitrate transporter, but not a dipeptide transporter. Nitrate uptake activity of NRT1.9 was further confirmed by enhanced 15NO3− uptake activity of NRT1.9-injected oocytes. As expected for a proton-coupled nitrate transporter with a proton/nitrate ratio larger than one, an inward current was elicited by the negatively charged nitrate in NRT1.9-injected oocytes (Figures 1A and 1B), and the 15NO3− transport activity of NRT1.9 was pH dependent, being higher at pH 5.5 and lower at pH 7.4 (Figure 1C).
Figure 1.
NRT1.9 Is a Nitrate Transporter.
(A) Substrate specificity of NRT1.9. The elicited currents were recorded at pH 5.5 with two-electrode whole-cell clamping as described in Methods. The currents shown are the differences between those in the presence and absence of the indicated substrate (nitrate or dipeptides). The values are the mean ± se for the indicated number of oocytes recorded from three different frogs. An asterisk indicates a significant difference (P < 0.05) compared with the water-injected control. G-G, Gly-Gly; L-L, Leu-Leu; A-H, Ala-His.
(B) Current-to-voltage curves for NRT1.9. Oocytes were voltage clamped at −60 mV and stepped into a test voltage between 0 and −100 mV for 300 ms, in −20-mV increments. The currents (I) shown here are the difference between the currents flowing at +300 ms in the presence and absence of the indicated substrates at pH 5.5. The curves presented here were recorded from a single oocyte. Similar results were obtained from four other oocytes from three different frogs.
(C) pH dependence of NRT1.9 uptake activity. The injected oocytes were incubated with 10 mM K15NO3 buffer at pH 5.5 or 7.4 for 2 h, and their 15N content was determined as described in Methods. The values are the mean ± se (n = 8 for all four experiments). An asterisk indicates a significant difference (P < 0.05) compared with the pH 7.4 value. Similar results were obtained using another two batches of oocytes.
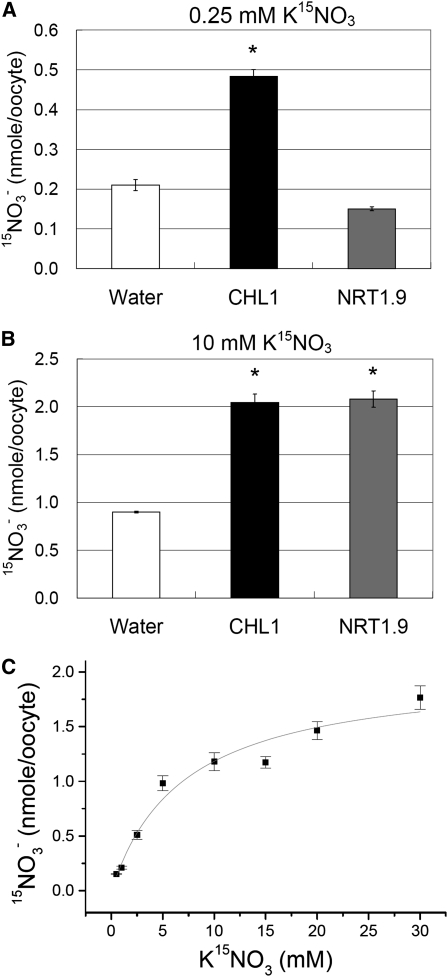
Different from CHL1, which shows dual-affinity nitrate transport activity with high-affinity nitrate transport activity when assayed with 250 μM 15NO3− (Figure 2A) and low-affinity transport with 10 mM 15NO3− (Figure 2B), NRT1.9 only displayed low-affinity nitrate transport activity (Figures 2A and 2B). These results indicate that, like most nitrate transporters in the NRT1 (PTR) family, such as NRT1.2 (Huang et al., 1999), NRT1.4 (Chiu et al., 2004), NRT1.5 (Lin et al., 2008), and NRT1.6 (Almagro et al., 2008), NRT1.9 shows nitrate transport activity in the low affinity range. To determine the affinity of NRT1.9 for nitrate, uptake activity of NRT1.9-injected oocytes at pH 5.5 was measured using different concentrations of 15N-labeled nitrate ranging from 0.5 to 30 mM, and the Km for nitrate, calculated by fitting to the Michaelis-Menten equation, was estimated as ~7 mM (Figure 2C). Taken together, these functional studies in Xenopus oocytes indicate that NRT1.9 is a low-affinity nitrate transporter with a Km of ~7 mM.
Figure 2.
NRT1.9 Is a Low-Affinity Nitrate Transporter with a Km of ~7 mM.
(A) and (B) Nitrate uptake activity of NRT1.9- or CHL1-injected oocytes at pH 5.5. High-affinity (A) or low-affinity (B) nitrate uptake activity was examined by incubating oocytes with 0.25 or 10 mM K15NO3 for 3 h, respectively, then measuring the 15N in the oocytes. The values are the mean ± se (n = 9, 9, or 10 for the water-, CHL1-, and NRT1.9-injected oocytes, respectively, in [A]; n = 8 for all oocytes in [B]). An asterisk indicates a significant difference (P < 0.05) compared with the water-injected control. Similar results were obtained using another two batches of oocytes.
(C) Uptake kinetics of NRT1.9. NRT1.9-injected oocytes were incubated with different concentrations of K15NO3 at pH 5.5 for 1.5 h, and then their 15N content was determined. The values are the mean ± se (n = 9 to 12 oocytes for each concentration). In this particular experiment, the Km, calculated by fitting to the Michaelis-Menten equation using a nonlinear least squares methods in the ORIGIN 5.0 program (Microcal Software; GE Healthcare), was 7 mM. Similar results were obtained using three different batches of oocytes.
NRT1.9 Is Mainly Expressed in the Root
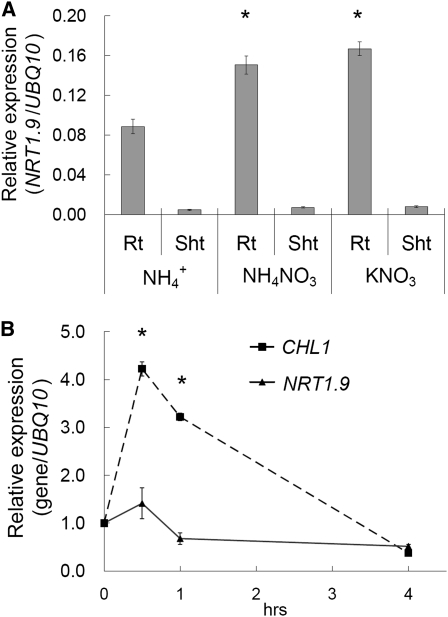
Quantitative RT-PCR analyses of plants grown vertically on plates or hydroponically showed that NRT1.9 was mainly expressed in the root (Figure 3A; see Supplemental Figure 2A online). The expression of NRT1.9 in 9-d-old plants grown in ammonium nitrate or potassium nitrate was higher than that in plants grown with ammonium as the sole nitrogen source (Figure 3A), indicating that NRT1.9 is upregulated by nitrate after long time exposure.
Figure 3.
NRT1.9 Is Dominantly Expressed in Roots.
(A) Quantitative PCR analysis of NRT1.9 expression in plants grown with three different nitrogen sources. Total RNA was extracted from the roots (Rt) and shoots (Sht) of 9-d-old wild-type (Col-0) plants grown on agar plates with 5 mM ammonium succinate (NH4+), 5 mM ammonium nitrate (NH4NO3), and 10 mM potassium nitrate (KNO3), respectively. NRT1.9 expression was analyzed with gene-specific primers and is shown as the relative expression normalized to UBQ10 level. The values are mean ± se. Three biological replicates were analyzed, and each replicate contained ~50 seedlings. The asterisk indicates a significant difference (P < 0.05) compared with the values of root grown in NH4+.
(B) Quantitative PCR analysis of short-term nitrate induction. Col-0 plants were grown on agar plates with 5 mM ammonium succinate for 9 d and then shifted to 5 mM ammonium nitrate for 0.5, 1, and 4 h. Total RNA was isolated from roots, and expression levels of CHL1 and NRT1.9 were analyzed. Relative expression was normalized to the level of UBQ10. The values are mean ± se for triplicate samples, with each replicate containing ~50 seedlings. The asterisk indicates a significant difference (P < 0.05) compared with the values at zero time point.
It is known that nitrate can rapidly induce the expression of several nitrate-related genes, including CHL1 (Tsay et al., 1993). This rapid transcriptional response has been referred to as the primary nitrate response (Redinbaugh and Campbell, 1991; Hu et al., 2009). Short-term nitrate induction analysis showed that, although CHL1 expression was indeed rapidly induced by nitrate within 30 min, NRT1.9 expression was unchanged by incubation with nitrate for at least 4 h (Figure 3B). In summary, NRT1.9 is preferentially expressed in root tissues, and its expression is not rapidly induced by nitrate but is increased by long-term nitrate exposure.
NRT1.9 Is Expressed in Companion Cells in Arabidopsis Roots
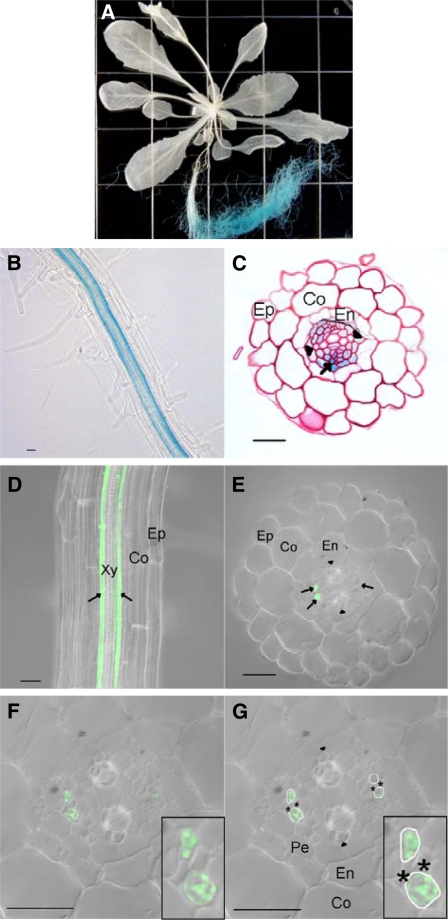
To elucidate the expression pattern in more detail, promoter-GUS (β-glucuronidase) reporter analysis was performed. The 1.6-kb upstream region of NRT1.9 was used to drive the expression of GUS, and the NRT1.9p:GUS construct was transformed into wild-type (Columbia-0 [Col-0]) Arabidopsis plants. Whole-mount staining confirmed that NRT1.9 is mainly expressed in roots (Figure 4A). If stained for an extended period (overnight), weak GUS staining could also be detected in the major veins of the leaves (see Supplemental Figure 2B online). Enlarged images of the root revealed two strands of GUS staining in the central stele (Figure 4B), while a cross section of the GUS-stained root showed that GUS was expressed in the phloem and in the phloem pole pericycle cells of the root vascular stele (Figure 4C). To confirm the tissue expression pattern, NRT1.9 protein localization in planta was determined by expression of a NRT1.9-GFP (green fluorescent protein) fusion driven by the NRT1.9 promoter. A 4-kb genomic fragment of NRT1.9, including the 1.6-kb promoter and coding region, was fused upstream of the coding region for GFP and introduced into nrt1.9-1 mutant. As shown in Figure 4D, consistent with the GUS pattern, a confocal section of root expressing NRT1.9-GFP showed that NRT1.9 was located in two rows of cells. In cross section, the GFP signal was detected specifically in companion cells (Figures 4E to 4G). Compared with the specific expression of NRT1.9-GFP in companion cells, GUS staining was more diffuse, with an additional signal in the phloem pole pericycle cells. Since diffusion is a common problem of GUS staining, it is likely that NRT1.9 is expressed in the companion cells of the root phloem.
Figure 4.
NRT1.9 Is Expressed in Companion Cells of Roots.
(A) to (C) Histochemical localization of GUS activity in transgenic plants. Transgenic plants expressing GUS fused with the first 74 amino acids of NRT1.9 and driven by NRT1.9 promoter were grown hydroponically with 1 mM NH4NO3 for 4 weeks (A) or vertically on plates with 5 mM NH4NO3 for 6 d ([B] and [C]). Images are the whole plant (A), the root (B), and a 3-μm-thick section of the root counterstained with periodic acid-Schiff reagent (C) of a transgenic seedling. GUS staining patterns were consistent in five independent transgenic lines. The squares in (A) are 2 cm × 2 cm.
(D) to (G) Confocal laser scanning microscope pictures of NRT1.9pro:NRT1.9-GFP transgenic plants. Images are the root (D) and 150-μm-section of the root ([E] to [G]) of a transgenic seedling grown vertically on plates with 5 mM NH4NO3 for 6 d. Arrowheads, the position of xylem axis; arrows, companion cells; asterisk, sieve element. Ep, epidermis; Co, cortex; En, endodermis; Pe, pericycle. GFP localization patterns were consistent in six independent transgenic lines. Insets in (E) and (F) show enlarged pictures of companion cells and sieve elements. Bars = 20 μm.
NRT1.9 Is Localized in the Plasma Membrane
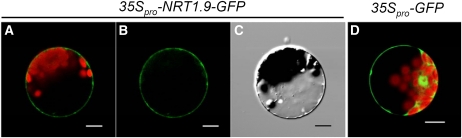
To determine the subcellular localization of NRT1.9, the GFP fusion protein driven by the cauliflower mosaic virus 35S promoter was transiently expressed in Arabidopsis mesophyll protoplasts, and the fluorescence signal was found to be confined to a ring external to the chloroplast signal (Figures 5A and 5B). Thus, NRT1.9 is a nitrate transporter localized in the plasma membrane.
Figure 5.
NRT1.9 Is Located in the Plasma Membrane.
(A) to (C) Subcelluar localization of NRT1.9-GFP fusion protein in Arabidopsis mesophyll protoplasts. NRT1.9-GFP was driven by the cauliflower mosaic virus 35S promoter and transiently expressed in Arabidopsis mesophyll protoplasts. The overlap image of the GFP (green) and chlorophyll (red) fluorescence (A), images of the GFP fluorescence (green) only (B), and the bright-field image (C) are presented.
(D) Photo of a control protoplast expressing GFP alone.
Bars = 10 μm.
Phloem Nitrate Content and Downward Nitrate Transport Is Reduced in nrt1.9 Mutants
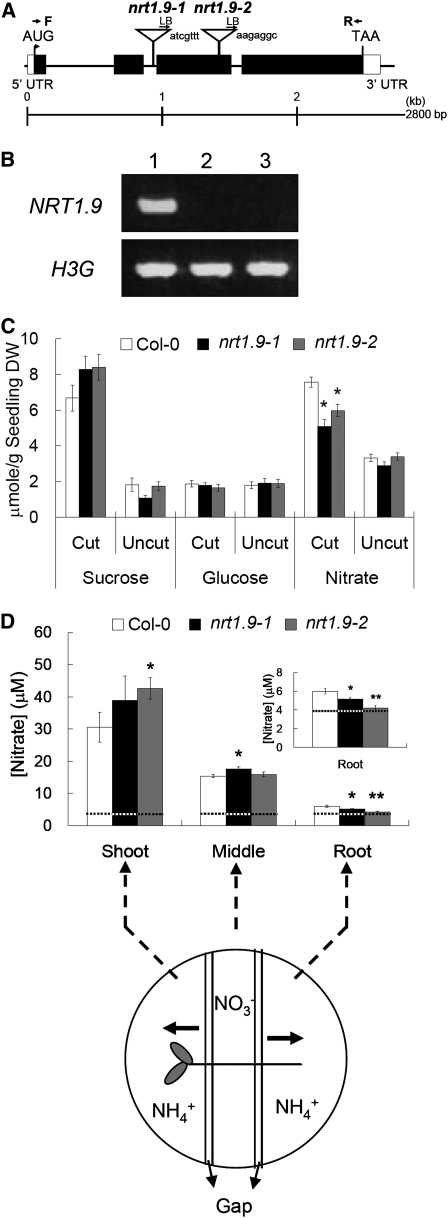
To determine the in vivo functions of NRT1.9, two independent T-DNA insertion lines, nrt1.9-1 and nrt1.9-2, generated by GABI (Genomanalyse im biologischen System Pflanze, http://www.gabi-kat.de/), were obtained. In the nrt1.9-1 and nrt1.9-2 mutants, the T-DNA was inserted in the second intron or the third exon of NRT1.9, respectively (Figure 6A). No RNA transcript was detected in either mutant by RT-PCR using gene-specific primers (Figure 6B), showing that both mutants are null mutants. Since NRT1.9 is a nitrate transporter and is expressed in the companion cells of root phloem, we speculated that NRT1.9 mediates nitrate loading into root phloem. To test this hypothesis, the nitrate content of Arabidopsis root phloem exudates was measured in the wild type and nrt1.9 mutants. The roots of 11-d-old seedlings were cut in the middle and placed in the exudation buffer. Root phloem exudates were collected in humid CO2-saturated boxes for 6 h. As a control for nitrate efflux and apoplastic contamination of the previous growth medium, roots of uncut seedlings were also placed in the exudation buffer. Higher amounts of Suc and nitrate, but not Glc, were detected in phloem exudates of cut seedlings than the uncut seedlings, suggesting that contamination of damaged cells from cut seedlings is low and, like Suc, nitrate is also present in the phloem exudates (Figure 6C). Furthermore, compared with the wild type, similar Suc but lower nitrate content was found in the phloem exudates from nrt1.9 mutants, suggesting that NRT1.9 facilitates nitrate loading into root phloem (Figure 6C).
Figure 6.
NRT1.9 Mediates Nitrate Loading into Phloem.
(A) Schematic map of the T-DNA insertion sites in nrt1.9-1 and nrt1.9-2 mutants. Mutants nrt1.9-1 and nrt1.9-2 carried T-DNA insertions in the second intron and the third exon of NRT1.9 gene, respectively. Black boxes, coding regions; white boxes untranslated regions (UTRs); F, forward primer used for RT-PCR; R, reverse primer used for RT-PCR; LB, left border primer of T-DNA.
(B) NRT1.9 expression in mutant lines. Total RNA from 6-d-old seedlings grown in liquid medium with 5 mM NH4NO3 of the wild type (lane 1), nrt1.9-1 (lane 2), and nrt1.9-2 (lane 3) was used for RT-PCR analysis. H3G was used as an internal control. Both NRT1.9 and H3G were amplified for 35 cycles.
(C) Nitrate content of root phloem exudates in the wild type and nrt1.9 mutants. Root phloem exudates of 11-d-old seedlings were collected as described in Methods. Sugar content was measured by enzymatic method, and nitrate content relative to dry weight (DW) was analyzed by HPLC. The values are mean ± se for 18 replicates collected from two independent experiments, and each replicate contained four seedlings. An asterisk indicates a significant difference (P < 0.05) compared with the values of the wild type. Cut: the seedlings were cut in the middle of roots; uncut: intact roots were used as negative controls.
(D) Downward nitrate transport in the wild type and nrt1.9 mutants. Top: nitrate concentration in the three parts of the seedlings was analyzed in the wild type and nrt1.9 mutants. Bottom: schematic drawing illustrates the experimental design. Eleven-day-old seedlings grown with ammonium succinate were treated with 30 mM ammonium nitrate at the middle part of root (Middle) for 24 h. The nitrate concentration in shoot (Shoot; root-to-shoot xylem transfer of nitrate) and untreated root (Root; downward phloem transfer of nitrate) was analyzed by HPLC. The values are mean ± se for the results obtained from 15 seedlings. An asterisk indicates a significant difference (*P < 0.05; **P < 0.001) compared with the values of the wild type. The inset is an enlarged view of the root. The dashed lines indicate the nitrate concentration before NH4NO3 treatment.
Since root phloem is responsible for downward transport, the influence of NRT1.9 on upward and downward nitrate transport was examined. NH4+-grown seedlings were placed on segmented NH4+ agar, and the middle part of the root was treated with 30 mM NH4NO3 for 24 h. The nitrate concentration in the shoot and the untreated bottom part of the root was measured to monitor root-to-shoot xylem transport of nitrate and downward phloem transport of nitrate, respectively. As shown in Figure 6D, nitrate concentration in the shoot was slightly higher in nrt1.9 mutants, whereas the nitrate concentration in untreated bottom root was lower in the mutants. This result suggests that phloem loading of nitrate mediated by NRT1.9 may facilitate the downward phloem transport of nitrate and affect upward xylem transport of nitrate.
Root-to-Shoot Nitrate Transport Is Increased in nrt1.9 Mutants
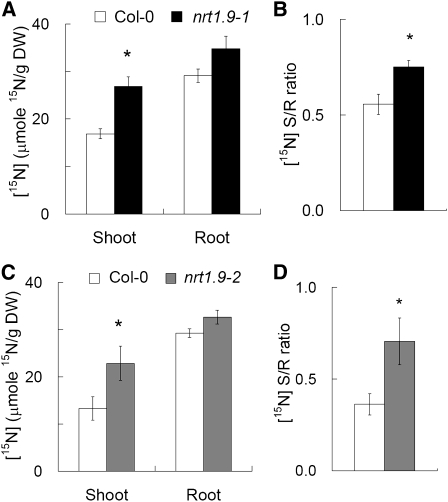
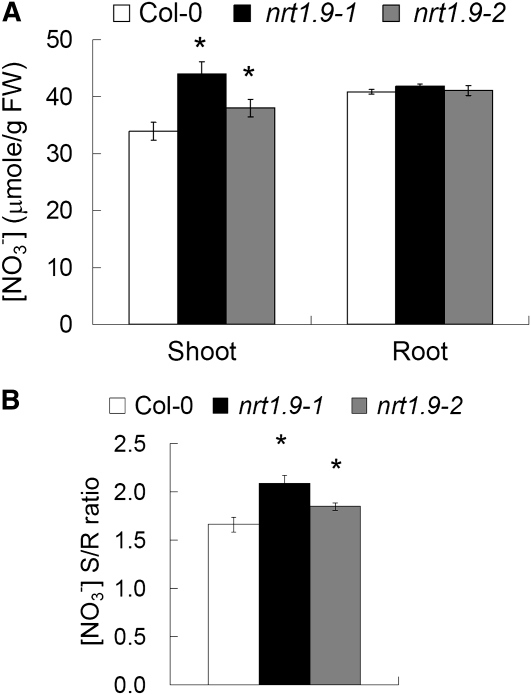
To confirm that the upward xylem transport of nitrate was increased in nrt1.9 mutants (Figure 6D), root-to-shoot nitrate translocation was analyzed by submerging wild-type (Col-0) and nrt1.9 roots in 10 mM NH415NO3 for 30 min. The plants used were first grown in liquid medium with 5 mM NH4NO3 for 10 d, at which stage no statistically significant difference in growth was detected between the wild type (Col-0) and the mutants. After 30 min of NH415NO3 labeling, the concentration of 15N in the shoot was higher in both nrt1.9 mutants than in the wild type (Col-0) (Figures 7A and 7C), resulting in a higher shoot-to-root ratio of 15N concentration in the mutants (Figures 7B and 7D). Consistent with this short-term 15N labeling result, the shoot nitrate content of 10-d-old plants grown in 10 mM NH4NO3 was higher in nrt1.9 mutants than in the wild type (Col-0), but the root nitrate contents were comparable (Figure 8A). The shoot-to-root nitrate content ratio in nrt1.9 mutants was also higher than that in wild type (Col-0) (Figure 8B), but under this condition, the total N content was not changed in nrt1.9 mutants (see Supplemental Figure 3 online). It has been shown that NRT1.5 is involved in root-to-shoot nitrate transport by loading nitrate into xylem (Lin et al., 2008). NRT1.5 expression is similar in the wild type and in nrt1.9-1 mutant (see Supplemental Figure 4 online), suggesting that enhanced root-to-shoot nitrate transport in nrt1.9 mutants is not due to enhanced expression of NRT1.5 in the mutants. Taken together, these data indicate that nonfunctional mutations of NRT1.9, which is expressed in companion cells of the root phloem, enhance root-to-shoot transport of nitrate.
Figure 7.
Root-to-Shoot Nitrate Transport Is Increased in nrt1.9 Mutants.
(A) and (C) 15N content in shoot and root of the wild type and nrt1.9 mutants.
(B) and (D) Shoot-to-root 15N ratio in the wild type and nrt1.9 mutants. The wild type and nrt1.9 mutants were grown in liquid medium with 5 mM NH4NO3 for 10 d and then labeled with 10 mM NH415NO3 for 30 min.
15N concentration and ratio between shoots and roots (S/R) were analyzed and calculated relative to dry weight (DW) as described in Methods. Values in (A) to (D) are mean ± se. An asterisk indicates a significant difference (P < 0.05) compared with the values of the wild type. Similar results were shown in three independent experiments. Each experiment was performed in four replicates, and each replicate contained 30 seedlings.
Figure 8.
Nitrate Content in Shoot Is Increased in nrt1.9 Mutants.
(A) Nitrate content relative to fresh weight (FW) of shoots and roots in the wild type and nrt1.9 mutants.
(B) The shoot-to-root ratio of the nitrate content. Wild-type plants and nrt1.9 mutants were grown in liquid medium with 10 mM NH4NO3 for 10 d, and the nitrate contents in both shoot and root were measured by HPLC.
Values in (A) and (B) are mean ± se for 4 replicates, and each replicate contained 30 seedlings. An asterisk indicates a significant difference (P < 0.05) compared with the values of the wild type. Similar results were shown in three independent experiments.
When Grown in High Nitrate, nrt1.9 Shows Increased Growth Compared with the Wild Type
Unexpectedly, under high nitrate conditions, nrt1.9 mutants showed enhanced growth when plants were grown at low density on plate to examine the growth differences in individual plants (Figure 9). The enhanced growth phenotype of the nrt1.9-1 mutant could be restored to the wild-type level by the NRT1.9pro:NRT1.9-GFP construct (see Supplemental Figure 5 online), further confirming that the growth phenotype is associated with NRT1.9. Furthermore, this result shows that the promoter used in this construct, and NRT1.9-GFP protein, are functional and can represent endogenous NRT1.9.
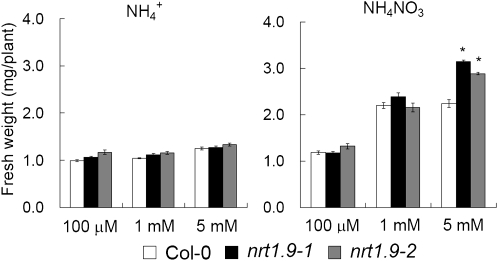
Figure 9.
nrt1.9 Mutants Show Enhanced Growth When Nitrate Concentration Is High in the Medium.
The fresh weight of 6-d-old wild-type and nrt1.9 mutant seedlings grown vertically on plates with different concentrations of NH4+ or NH4NO3 were measured. The values are mean ± se; n = 10. An asterisk indicates a significant difference (P < 0.01) compared with the values of the wild type. Similar results were obtained in three independent experiments.
When the medium contained ammonium as the sole nitrogen source or contained only a low concentration (100 μM and 1 mM) of NH4NO3, no growth differences were detected between the wild type (Col-0) and nrt1.9 mutants. However, when the medium contained higher concentrations of NH4NO3 (5 mM) or KNO3 (10 mM), the nrt1.9 mutants exhibited increased growth (Figure 9; see Supplemental Figure 6 online). Since no growth difference was seen between the wild type (Col-0) and the mutants when ammonium was the sole nitrogen source, the growth difference observed in NH4NO3 was due to the presence of nitrate. The fresh weight of nrt1.9 mutants grown on 5 mM NH4NO3 was 30 to 40% higher than that of the wild type (Col-0). In the wild type, the fresh weight increased when the medium nitrate concentration was increased from 100 μM to 1 mM, but no further increase was seen when the medium nitrate concentration was increased from 1 to 5 mM. By contrast, in the mutants, growth increased with increasing nitrate concentration up to at least 5 mM nitrate. Thus, the increased growth of nrt1.9 mutants was nitrate dependent and concentration dependent. These data suggest that NRT1.9-regulated long-distance nitrate transport has an influence on plant growth in Arabidopsis.
DISCUSSION
Arabidopsis NRT1.9 Mediates Nitrate Loading into Root Phloem
Functional analysis in Xenopus oocytes showed that, like most NRT1 transporters, NRT1.9 exhibits low-affinity nitrate transport activity with a Km of ~7 mM (Figures 1 and 2). Besides nitrate, some transporters in the NRT1 (PTR) family can also transport peptides (Yamashita et al., 1997; Chiang et al., 2004; Dietrich et al., 2004; Komarova et al., 2008), histidine (Yamashita et al., 1997; Zhou et al., 1998), dicarboxylate (Jeong et al., 2004), and auxin (Krouk et al., 2010). No dipeptide transport activity was detected in NRT1.9-injected oocytes (Figures 1A and 1B). Although the possibility that NRT1.9 might transport other substrates cannot be excluded, the nitrate transport activity observed in NRT1.9-injected oocytes and nitrate-associated phenotypes found in nrt1.9 mutants suggests that NRT1.9 mediates nitrate transport in Arabidopsis.
Protein localization analysis using a GFP fusion protein indicated that NRT1.9 is a plasma membrane transporter (Figure 5). Promoter reporter assays using GUS and GFP showed that NRT1.9 is expressed in the companion cells of the root phloem (Figure 4). This spatial expression pattern, together with the nitrate transport activity of NRT1.9, suggests that it is responsible for loading nitrate into root phloem. This hypothesis is further supported by the fact that nitrate content in root phloem exudates and downward nitrate translocation were reduced in nrt1.9 mutants (Figures 6C and 6D). Although, in general, nitrate has been thought to be solely transported by xylem, several studies have shown that nitrate at the concentrations of 0.59 to 8.1 mM is found in phloem sap of rice (Oryza sativa), wheat (Triticum aestivum), Ricinus communis, and broccoli (Brassica oleracea; Hayashi and Chino, 1985, 1986; Allen and Smith, 1986; Shelp, 1987). Our previous study of another NRT1 homolog, NRT1.7, showed that nitrate transport in phloem is important for nitrate remobilization from older to younger leaves (Fan et al., 2009). In this study, our data provide additional evidence that phloem nitrate transport can also occur in roots. In several cases, when both phloem and xylem nitrate contents were measured, the phloem nitrate concentration was about one-tenth to one-fourth of the xylem level (Allen and Smith, 1986; Shelp, 1987). Taken together, these findings suggest that xylem provides the major route for long-distance nitrate transport, while phloem transport serves as fine-tuning for local nitrate redistribution.
NRT1.9-facilitated nitrate loading into companion cells and downward nitrate transport through phloem. It is still unclear, however, why a mechanism that essentially sends nitrate back to where it came from would be needed. Since nitrate is not evenly distributed in the natural soil, in some cases, the lower parts of the root may not be exposed to nitrate at all. We can postulate that in such cases, downward nitrate transport through phloem might be required to allocate nitrate harvested from more established roots to the younger roots.
Phloem flow is driven by differences in hydrostatic pressure between source organs and sink organs (Patrick et al., 2001; Lalonde et al., 2003; Knoblauch and Peters, 2010). In shoots, Suc, potassium, and amino-nitrogen compounds are the major osmotica driving phloem flow (Allen and Smith, 1986; Shelp, 1987; Patrick et al., 2001; Lalonde et al., 2003). Our study showed that in root phloem exudates, nitrate content is compatible with sucrose content, suggesting that compositions of phloem exudates collected from different tissues could be different and that nitrate, working in coordination with Suc, K+, and amino-nitrogen compounds, may contribute as an osmoticum in root phloem.
NRT1.9-Facilitated Nitrate Loading in Root Phloem Exerts Influence on Nitrate Translocation and Distribution
In nrt1.9 mutants, more nitrate is transported and accumulated in shoot (Figures 7 and 8), suggesting that nitrate loaded by NRT1.9 into root phloem may affect root-to-shoot nitrate partition in plants. It is possible that the enhanced root-to-shoot nitrate transport in nrt1.9 mutants is caused indirectly by the increased nitrate uptake (see Supplemental Figure 7 online). Alternatively, enhanced root-to-shoot nitrate transport in nrt1.9 mutants might be affected more directly by phloem loading. NRT1.9 is responsible for loading nitrate into root phloem and apoplastic nitrate source for NRT1.9-mediated loading may come from efflux of vascular parenchyma cells or leakage of xylem stream. Similar to NRT1.8 expressed in xylem parenchyma cells (Li et al., 2010), NRT1.9 expressed in root companion cells may also participate in retrieving nitrate from xylem sap.
Xylem-to-phloem transfer has been documented for K+, Na+, Mg2+, Cl−SO42−PO43−glutathione, and amino acids (Biddulph, 1956; Da Silva and Shelp, 1990; Jeschke and Pate, 1991; Schneider et al., 1994). This pathway is important for delivering nutrients to sink organs, such as developing leaves and fruits, and recycling Na+ from shoots to roots under salt stress (Atkins et al., 1980; Dickson et al., 1985; Da Silva and Shelp, 1990; Schupp et al., 1992; Schneider et al., 1994; Berthomieu et al., 2003; Tanaka et al., 2008). Despite the importance of this transport pathway, only a few transporters, such as NIP6;1 and AAP2, have been shown to mediate xylem-to-phloem transfer (Tanaka et al., 2008; Zhang et al., 2010). In nrt1.9 mutants, reduced nitrate content in root phloem exudates and enhanced root-to-shoot nitrate transport support the possible role of NRT1.9 in xylem-to-phloem transfer of nitrate, suggesting that xylem-to-phloem transfer may also occur in the root, and in addition to organic amino acids, inorganic nitrogen nitrate may also be transferred from xylem to phloem.
NRT1.9 Affects Plant Growth under High Nitrate Conditions
As shown in Figure 9 and Supplemental Figure 6 online, nrt1.9 mutants showed enhanced growth compared with the wild type (Col-0) in the presence of the higher concentrations of nitrate (5 and 10 mM) but not at lower concentrations of nitrate (100 μM and 1 mM) or in the absence of nitrate (ammonium only). Consistent with the growth differences, increased nitrate content in the nrt1.9 mutant seedlings was only observed in the presence of higher concentration of nitrate (see Supplemental Figure 8 online). These results indicate that, probably due to its high Km value, instead of playing a housekeeping function, the influence of NRT1.9 on nitrate distribution is more dominant at higher concentrations of nitrate. Under high nitrate conditions, NRT1.9 might facilitate the flow of excess nitrate from xylem into phloem to regulate the nitrate partition between roots and shoots.
Phenotypes of nrt1.9 mutants suggest that NRT1.9 has a negative impact on root-to-shoot nitrate transport, shoot nitrate content, and plant growth. From the evolutionary point of view, this raises an interesting question about what kind of advantage plants would gain from this negative effect of NRT1.9 on plant growth. It is possible that under certain conditions, high nitrate content in the shoot might have no beneficial effect on plant growth, thus suggesting NRT1.9 may serve as a safety valve to ensure proper distribution of nitrate between root and shoot.
Nitrate Transport in Root Phloem Plays a Role in Nitrate Partition
The phloem delivers different signals or nutrients in response to intrinsic cues and fast-changing environmental conditions, such as florigen FLOWER LOCUS T, miRNA399, and plant hormones (Vlot et al., 2008; Robert and Friml, 2009; Liu et al., 2010) With respect to nutrients, similar to NRT1.9, mutations of phloem-localized transporters for Suc, sodium, sulfate, borate, and amino acids show altered substrate distribution and affect plant development under certain conditions (Truernit and Sauer, 1995; Gottwald et al., 2000; Berthomieu et al., 2003; Yoshimoto et al., 2003; Srivastava et al., 2008; Tanaka et al., 2008; Zhang et al., 2010). Although mass nitrate transport is mediated by xylem, the mutant phenotypes of phloem-localized nitrate transporter NRT1.7 in our previous study and NRT1.9 in this study show that phloem nitrate transport is also important for modulating nitrate distribution in the shoot and root. Therefore, coordination between xylem and phloem transport of nitrate may ensure proper distribution of nitrate at different developmental stages to counteract fluctuating nitrogen status in the soil.
METHODS
Functional Analysis of NRT1.9 in Xenopus laevis Oocytes
The 1.8-kb NRT1.9 cDNA was cloned into the oocyte expression vector pGEMHE (Liman et al., 1992) and linearized with NheI. Capped mRNA was transcribed in vitro using mMESSAGE mMACHINE kits (Amion). Oocytes were isolated and injected with 50 to 100 ng of NRT1.9 cRNA in 50 nL of water, as described previously (Tsay et al., 1993), except that the Barth solution was replaced with ND96 solution (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, and 5 mM HEPES, pH 7.4) (Bröer et al., 2000). The oocytes were then incubated for 2 d in ND96 solution containing 10 mg/L of gentamycin before further experiments. Current measurements were recorded as described previously (Huang et al., 1999). For the analysis of uptake properties by 15N measurements, oocytes were incubated for the indicated time in a solution containing 230 mM mannitol, 0.3 mM CaCl2, 10 mM MES-Tris, pH 5.5 or 7.4, and the required concentration of K15NO3 and then were rinsed five times with ND96 buffer and dried at 80°C for 24 h. The retained 15N was then measured on a continuous-flow isotope ratio mass spectrometer coupled to a carbon nitrogen elemental analyzer (ANCA-GSL MS; PDZ Europa).
RT-PCR and Quantitative PCR
Total RNA was extracted from roots, shoots, or whole seedlings from the wild type (Col-0) and mutants using TRIzol reagent (Gibco BRL). The first-strand cDNAs were synthesized using oligo(dT) primers and ImProm-II reverse transcriptase (Promega). Specific primers for NRT1.9 and H3G were used to amplify the transcripts; these were NRT1.9 (forward, 5′-GGGTCTAGAATGGAGGTTGAGAAGACA-3′; reverse, 5′-CCCGGATCCCTGACACCTTATCAAACT-3′) and H3G (forward, 5′-AACCACTGGAGGAGTCAA GA-3′; reverse, 5′-AGAGGAGAACGTGCTTAATTG-3′). Quantitative PCR was performed using Power CYBR Green Master Mix (Applied Biosystems). The initial denaturing step at 94°C for 10 min was followed by 40 cycles of 94°C for 15 s and 60°C for 1 min. After the PCR cycles, the melting temperature of the PCR product was measured. The expression level of UBQ10 was used for normalization by 7500 software v2.0 (Applied Biosystems). The primers used for quantitative PCR were CHL1 (forward, 5′-GATGAGCACGGGTCTATTGTTGA-3′; reverse, 5′-CACGAGAACCGAGCTGAAGAA-3′), NRT1.9 (forward, 5′-CAATCCGATAGACGCCTTGG-3′; reverse, 5′-CAACCACATACCGGACATCG-3′), NRT1.5 (forward, 5′-TGGAGCGTTTCTCAGCGATT-3′; reverse, 5′-TCCATCATGGA ATGTGAACCAC-3′), and UBQ10 (forward, 5′-AGAAGTTCAATGTTTCGTTTCAT GTAA-3′; reverse, 5′-GAACGGAAACATAGTAGAACACTTAT TCA-3′).
Subcellular Localization by Arabidopsis thaliana Protoplast Transformation
NRT1.9 cDNA was amplified by PCR using the forward primer 5′-GGGTCTAGAATGGAGGTTGAGAAGACA-3′ with an XbaI site and the reverse primer 5′-CCCGGATCCCTGACACCTTATCAAACT-3′ with a BamHI site. The amplified fragment was then subcloned in frame in front of the GFP coding region in the vector 326-GFP (Rus et al., 2001), resulting in the NRT1.9-GFP construct under the control of the 35S promoter. The NRT1.9-GFP fusion construct or the vector 326-GFP was then transiently expressed in Arabidopsis protoplasts. Arabidopsis protoplast transformation was performed following the protocol described by Sheen (2001). Arabidopsis protoplasts were isolated from leaf tissues of 3- to 4-week-old plants grown on soil. GFP-fused plasmids were purified on Qiagen columns according to the manufacturer’s procedure and transformed into protoplasts by the polyethylene glycol–mediated method. After incubation in W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 5.7, and 5 mM Glc) under light for 9 to 12 h, fluorescent cells were imaged by confocal microscopy (Zeiss LSM510) with excitation at 488 nm and emission at 505 to 530 nm for GFP or at 650 nm for chloroplast autofluorescence.
Plant Transformation and Promoter-GUS Analysis
A 2.3-kb genomic fragment containing the promoter, first exon, first intron, and partial second exon of NRT1.9 was isolated by PCR using Pyrobest DNA polymerase (TaKaRa) and the forward primer 5′-GGGAAGCTTAGAAAGGCGGGAATTGAGA-3′ with a HindIII site and the reverse primer 5′-CCCGGATCCGCTTGTGCCACC GTAGAT-3′ with a BamHI site. After the sequence was confirmed, the fragment was cloned in front of, and in frame with, the GUS gene in the binary vector pBI101. The binary vector was introduced into wild-type (Col-0) plants using the Agrobacterium tumefaciens (strain GV3101)–mediated floral dip method (Clough and Bent, 1998). Putative transformants were selected on half-strength Murashige and Skoog plates containing 50 mg/L of kanamycin. For GUS staining, 6-d-old transgenic seedlings grown on plates with 5 mM NH4NO3 were used, vacuum-infiltrated in prefix solution (0.5% formaldehyde, 0.05% Triton X-100, and 50 mM sodium phosphate, pH 7.0), and incubated in prefix solution at room temperature for 1.5 h. After three rinses with 50 mM sodium phosphate, pH 7.0, the seedlings were incubated for 30 min at 37°C in X-Gluc staining solution (50 mM sodium phosphate, pH 7.0, 0.05% Triton X-100, 1 mM potassium ferrocyanide, 1 mM potassium ferricyanide, and 1 mM 5-bromo-4-chloro-3-indoyl- β-d-glucuronide). After three washes with 50 mM sodium phosphate, pH 7.0, the seedlings were fixed in 2% formaldehyde, 0.5% glutaraldehyde, and 100 mM sodium phosphate, pH 7.0, and GUS staining was visualized on an AxioImager-Z1 (Zeiss). For sections, the seedlings were furthered dehydrated in a graded series of ethanol and embedded in LR White medium-grade resin (London Resin Company). Then, 3-μm semifine sections were cut, mounted on glass slides, and counterstained with periodic acid-Schiff reagent (Sigma-Aldrich).
NRT1.9-GFP Localization in Transgenic Plants
The 4-kb genomic fragment of NRT1.9 including the 1.6 kb upstream of the translation start site and the coding region was amplified from wild-type Col-0 genomic DNA by PCR using the forward primer 5′-GGGAAGCTTAGAAAGGCGGGAATTGAGA-3′ with a HindIII site and the reverse primer 5′-CCCGGATCCCTGACACCTTATCAAACT-3′ with a BamHI site. The amplified fragment was cloned in frame with GFP in the vector 326-GFP (Lee et al., 2001) at the HindIII and BamHI sites. After the sequence was confirmed, the NRT1.9pro:NRT1.9-GFP fragment was subcloned into the pCambia binary vector at the HindIII and EcoRI sites. The resulting plasmid was transformed into the nrt1.9-1 mutant using the Agrobacterium-mediated floral dip method. Putative transformants were selected on half-strength Murashige and Skoog plates containing 25 mg/L hygromycin. For NRT1.9-GFP protein observation, 6-d-old transgenic seedlings grown on plates with 5 mM NH4NO3 were mounted with normal medium on slides and observed directly under a confocal microscope (Zeiss LSM510META) with excitation at 488 nm, and the emission signals for GFP was detected at 505 to 530 nm. For sections, the roots were embedded in 5% (w/v) agarose dissolved in 1× PBS and cut into 150-μm cross sections with a Vibratome Series 1000 (Technical Products International Inc.). The slices of agarose were mounted in normal medium on slides before observation by confocal microscope (Zeiss LSM510META).
Isolation of Two T-DNA–Tagged Mutants, nrt1.9-1 and nrt1.9-2
The nrt1.9-1 (284B09 for GABI) mutant was obtained from GABI (Genomanalyse im Biologischen System Pflanze) (Rosso et al., 2003), and the nrt1.9-2 (N378000 for NASC; 099B01 for GABI) mutant was generated by GABI and obtained from NASC (European Arabidopsis Stock Centre) (Scholl et al., 2000). The primers used for PCR screening were the T-DNA left border primer 5′-CCCATTTGGAC GTGAATGTAGACAC-3′ and the NRT1.9 reverse primer 5′-ACACTGACAAG GTTCTTTCAAATATCCAA-3′, which amplify a 2- and 1.6-kb fragment in the nrt1.9-1 or nrt1.9-2 mutant, respectively. Gene expression was confirmed by RT-PCR as described above.
Analysis of Root-to-Shoot Nitrate Transport Using 15NO3−
Wild-type (Col-0) and mutant seeds were surface sterilized with 70% ethanol for 2 min and with sterilization solution (0.5% SDS and 20% bleach) for 15 min. Approximately 100 seeds (30 seeds for each line) were sown on nylon netting supported by a raft in a culture vessel (Magenta) as described previously (Touraine and Glass, 1997) in medium containing 5 mM NH4NO3, 10 mM K2HPO4-KH2PO4, 2 mM MgSO4, 1 mM CaCl2, 0.1 mM FeSO4-EDTA, 50 μM H3BO3, 12 μM MnSO4·H2O, 1 μM ZnCl2, 1 μM CuSO4·5H2O, 0.2 μM NaMoO4·2H2O, 1 g/L MES, and 0.5% Suc, pH 6.0. The plants were grown under continuous illumination and rotated at 80 rpm at 24°C for 9 d; care was taken to make sure that shoots were not exposed to the liquid medium. After two changes of the same medium (overnight and 3 h on the next day), plants were labeled for 30 min in 10 mM NH415NO3 medium with a 99% atom excess of 15N, pH 6.0, then washed in 0.1 mM CaSO4 for 1 min. The roots and the shoots were separated and dried at 80°C for 3 d. 15N content was analyzed on a continuous-flow isotope ratio mass spectrometer coupled to a carbon nitrogen elemental analyzer (ANCA-GSL MS; PDZ Europa).
Nitrate Content Analysis by HPLC
Surface-sterilized seeds were grown for 9 d in culture boxes as described above, except that the concentration of NH4NO3 in the medium was 10 mM to ensure a sufficient nitrogen supply. The medium was replaced with fresh medium overnight, then the roots and shoots were collected separately and the nitrate extracted in boiling distilled water for 30 min. The nitrate content was then determined by HPLC (Thayer and Huffaker, 1980) using a PARTISIL 10 SAX (strong anion exchanger) column (Whatman), with 50 mM KH2PO4 buffer, pH 3.0, as the mobile phase.
Collection of Root Phloem Exudates
Sterilized seeds were grown vertically on 5 mM NH4NO3 with 0.8% Difco agar for 11 d. The seedlings were washed several times with large amount of 10 mM K2HPO4-KH2PO4 and cut at the middle of the root in the exudation buffer (10 mM K2HPO4-KH2PO4, 0.5 mM EDTA, and 100 mM sorbitol, pH 7.0). The cut seedlings were rinsed once in the fresh exudation buffer, and then the end of the cut roots of four seedlings were dipped into 200 μL of exudation buffer in a micrtube cap. During the collection of phloem exudates, the rest of the seedlings were placed on agarose medium with 20 mM NH4NO3 to enhance nitrate content in phloem. The root phloem exudates were collected in a humid and CO2-saturated box (Deeken et al., 2008) for 6 h. Suc was first catalyzed by invertase (Sigma-Aldrich) in 100 mM citric acid/K2HPO4, pH 4.5, at 55°C for 40 min. Then the resulting Glc was measured by Glc oxidase (Sigma-Aldrich) and peroxidase (Sigma-Aldrich) in 50 mM NaOAc, pH 5.9, at 37°C for 30 min. Detect the OD490 after stopping the reation with 2 n H2SO4. The nitrate content was measured by HPLC.
Measurement of Downward Transport of Nitrate
Sterilized seeds were grown vertically on 5 mM (NH4)2 succinate with 0.8% Difco agar for 10 d to ensure that there was little or no nitrate in plants. Shift the seedlings to the segmented agar with 5 mM (NH4)2 succinate and let them grow for 1 d. A sterilized and sharp knife was used to remove the agar to generate the gaps with width of 0.5 cm. Drop the concentrated NH4NO3 on the middle agar (1 cm in width) to final concentration of 30 mM, and move the plates in a humid and CO2-saturated box for 24 h. When collecting the tissues, cut the seedling into three parts and wash them by water. Tissue was boiled in 200 μL of water for 20 min to extract nitrate. The nitrate concentration of different parts of seedlings was measured by HPLC.
Phenotypic Analysis on Plates
To analyze the total fresh weight of seedlings, ~30 surfaced-sterilized seeds were sown on 15-cm plates containing the desired concentration of (NH4)2 succinate or NH4NO3 in 0.45% agarose. After stratification at 4°C in the dark for 3 d, the seedlings were grown under continuous illumination at 24°C for 6 d and the fresh weight per seedling measured.
Accession Numbers
The sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At1g18880 (NRT1.9), At12110 (CHL1, NRT1.1), At1g69850 (NRT1.2), At2g26690 (NRT1.4), At1g32450 (NRT1.5), At1g27080 (NRT1.6), At1g69870 (NRT1.7), At4g21680 (NRT1.8), At5g62680 (NRT1.10), At1g52190 (NRT1.11), At1g08090 (NRT2.1), U17987 (Bn NRT1.2), At1g08100 (NRT2.2), At1g80760 (NIP6;1), At1g22710 (SUC2); At4g10310 (HKT1), At1g22150 (Sultr1;3), At5g09220 (AAP2) At4g05320 (UBQ10), and At4g40040 (Histone 3G).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignment of NRT1 Homologs in Arabidopsis.
Supplemental Figure 2. RNA Expression Pattern of NRT1.9.
Supplemental Figure 3. Total N Content in nrt1.9 Mutants.
Supplemental Figure 4. RNA Expression Levels of NRT1.5.
Supplemental Figure 5. Complementation of Growth Phenotype of nrt1.9 by NRT1.9pro:NRT1.9-GFP.
Supplemental Figure 6. Fresh Weight of nrt1.9 Seedlings in Nitrate Medium.
Supplemental Figure 7. Net Nitrate Uptake in nrt1.9 Mutants.
Supplemental Figure 8. Nitrate Content in 6-d-Old Seedlings of nrt1.9 Mutants.
Supplemental Data Set 1. Text File of the Alignment Presented in Supplemental Figure 1.
Acknowledgments
We thank Sue-Pin Li from our confocal core facility for help with the GFP and GUS images, Shan-Hua Lin for help with the 15N analysis, Chi-Chou Chiu for help with the amino acid sequence alignment, and Tom Barkas for English editing. The 326-GFP vector was a gift from Inhwan Hwang (Pohang University of Science and Technology). This work was supported by grants from the National Science Council (NSC 97-2311-B-001-009-MY3) and the Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan.
References
- Abrol Y.P., Sawhney S.K., Naik M.S. (1983). Light and dark assimilation of nitrate in plants. Plant Cell Environ. 6: 595–599 [Google Scholar]
- Allen S., Smith J.A.C. (1986). Ammonium nutrition in Ricinus communis: Its effect on plant growth and the chemical composition of the whole plant, xylem and phloem saps. J. Exp. Bot. 37: 1599–1610 [Google Scholar]
- Almagro A., Lin S.H., Tsay Y.F. (2008). Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20: 3289–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam M., Huffaker R.C. (1982). In vivo nitrate reduction in roots and shoots of barley (Hordeum vulgare L.) seedlings in light and darkness. Plant Physiol. 70: 1009–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins C.A., Pate J.S., McNeil D.L. (1980). Phloem loading and metabolism of xylem-borne amino compounds in fruiting shoots of a legume. J. Exp. Bot. 31: 1509–1520 [Google Scholar]
- Beevers L., Hageman R.H. (1980). Nitrate and nitrite reduction. In The Biochemistry of Plants, Stumpf P.K., Conn E.E., eds (London: Academic Press; ), pp. 115–168 [Google Scholar]
- Berthomieu P., et al. (2003). Functional analysis of AtHKT1 in Arabidopsis shows that Na(+) recirculation by the phloem is crucial for salt tolerance. EMBO J. 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddulph S.F. (1956). Visual indications of S35 and P32 translocation in the phloem. Am. J. Bot. 43: 143–148 [Google Scholar]
- Bröer A., Wagner C., Lang F., Bröer S. (2000). Neutral amino acid transporter ASCT2 displays substrate-induced Na+ exchange and a substrate-gated anion conductance. Biochem. J. 346: 705–710 [PMC free article] [PubMed] [Google Scholar]
- Canvin D.T., Atkins C.A. (1974). Nitrate, nitrite and ammonia assimilation by leaves: Effect of light, carbon dioxide and oxygen. Planta 116: 207–224 [DOI] [PubMed] [Google Scholar]
- Cerezo M., Tillard P., Filleur S., Muños S., Daniel-Vedele F., Gojon A. (2001). Major alterations of the regulation of root NO(3)(-) uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol. 127: 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.-L., Acedo G.N., Dewdney J., Goodman H.M., Conkling M.A. (1991). Differential expression of the two Arabidopsis nitrate reductase genes. Plant Physiol. 96: 275–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C.S., Stacey G., Tsay Y.F. (2004). Mechanisms and functional properties of two peptide transporters, AtPTR2 and fPTR2. J. Biol. Chem. 279: 30150–30157 [DOI] [PubMed] [Google Scholar]
- Chiu C.C., Lin C.S., Hsia A.P., Su R.C., Lin H.L., Tsay Y.F. (2004). Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 45: 1139–1148 [DOI] [PubMed] [Google Scholar]
- Clarkson D.T., Deane-Drummond C.E. (1983). Thermal adaptation of nitrate transport. In Nitrogen as an Ecological Factor, Lee J.A., McNeill S., Rorison I.H., eds (Oxford: Blackwell; ), pp. 211–224 [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crafts-Brandner S.J., Harper J.E. (1982). Nitrate reduction by roots of soybean (Glycine max [L.] Merr.) seedlings. Plant Physiol. 69: 1298–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel-Vedele F., Chaillou S. (2005). Nitrogen. In Plant Nutritional Genomics, Broadley M.R., White P.J., eds (Oxford, UK: Wiley-Blackwell; ), pp. 1–25 [Google Scholar]
- Da Silva M.C., Shelp B.J. (1990). Xylem-to-phloem transfer of organic nitrogen in young soybean plants. Plant Physiol. 92: 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane-Drummond C., Clarkson D., Johnson C. (1980). The effect of differential root and shoot temperature on the nitrate reductase activity, assayed in vivo and in vitro in roots of Hordeum vulgare (barley). Planta 148: 455–461 [DOI] [PubMed] [Google Scholar]
- Deeken R., Ache P., Kajahn I., Klinkenberg J., Bringmann G., Hedrich R. (2008). Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J. 55: 746–759 [DOI] [PubMed] [Google Scholar]
- Dickson R.E., Vogelmann T.C., Larson P.R. (1985). Glutamine transfer from xylem to phloem and translocation to developing leaves of Populus deltoides. Plant Physiol. 77: 412–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich D., Hammes U., Thor K., Suter-Grotemeyer M., Flückiger R., Slusarenko A.J., Ward J.M., Rentsch D. (2004). AtPTR1, a plasma membrane peptide transporter expressed during seed germination and in vascular tissue of Arabidopsis. Plant J. 40: 488–499 [DOI] [PubMed] [Google Scholar]
- Fan S.-C., Lin C.-S., Hsu P.-K., Lin S.-H., Tsay Y.-F. (2009). The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21: 2750–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filleur S., Dorbe M.F., Cerezo M., Orsel M., Granier F., Gojon A., Daniel-Vedele F. (2001). An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 489: 220–224 [DOI] [PubMed] [Google Scholar]
- Forde B.G. (2002). Local and long-range signaling pathways regulating plant responses to nitrate. Annu. Rev. Plant Biol. 53: 203–224 [DOI] [PubMed] [Google Scholar]
- Gottwald J.R., Krysan P.J., Young J.C., Evert R.F., Sussman M.R. (2000). Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. USA 97: 13979–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Chino M. (1985). Nitrate and other anions in the rice phloem sap. Plant Cell Physiol. 26: 325–330 [DOI] [PubMed] [Google Scholar]
- Hayashi H., Chino M. (1986). Collection of pure phloem sap from wheat and its chemical composition. Plant Cell Physiol. 27: 1387–1393 [Google Scholar]
- Hu H.C., Wang Y.Y., Tsay Y.F. (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57: 264–278 [DOI] [PubMed] [Google Scholar]
- Huang N.C., Liu K.H., Lo H.J., Tsay Y.F. (1999). Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Suh S., Guan C., Tsay Y.F., Moran N., Oh C.J., An C.S., Demchenko K.N., Pawlowski K., Lee Y. (2004). A nodule-specific dicarboxylate transporter from alder is a member of the peptide transporter family. Plant Physiol. 134: 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke W.D., Pate J.S. (1991). Cation and chloride partitioning through xylem and phloem within the whole plant of Ricinus communis L. under conditions of salt stress. J. Exp. Bot. 42: 1105–1116 [Google Scholar]
- Knoblauch M., Peters W.S. (2010). Münch, morphology, microfluidics—our structural problem with the phloem. Plant Cell Environ. 33: 1439–1452 [DOI] [PubMed] [Google Scholar]
- Komarova N.Y., Thor K., Gubler A., Meier S., Dietrich D., Weichert A., Suter Grotemeyer M., Tegeder M., Rentsch D. (2008). AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiol. 148: 856–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Lalonde S., Tegeder M., Throne-Holst M., Frommer W.B., Patrick J.W. (2003). Phloem loading and unloading of sugars and amino acids. Plant Cell Environ. 26: 37–56 [Google Scholar]
- Lee K.C., Koh A., Loh C.S., Wong S.M. (2001). Cucurbit protoplast isolation for the study of plant virus replication. J. Virol. Methods 91: 21–27 [DOI] [PubMed] [Google Scholar]
- Lee R.B. (1980). Sources of reductant for nitrate assimilation in non-photosynthetic tissue: A review. Plant Cell Environ. 3: 65–90 [Google Scholar]
- Li J.Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wang Y., Okamoto M., Crawford N.M., Siddiqi M.Y., Glass A.D. (2007). Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman E.R., Tytgat J., Hess P. (1992). Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron 9: 861–871 [DOI] [PubMed] [Google Scholar]
- Lin S.H., Kuo H.F., Canivenc G., Lin C.S., Lepetit M., Hsu P.K., Tillard P., Lin H.L., Wang Y.Y., Tsai C.B., Gojon A., Tsay Y.F. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little D.Y., Rao H., Oliva S., Daniel-Vedele F., Krapp A., Malamy J.E. (2005). The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Natl. Acad. Sci. USA 102: 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.-Q., Allan D.L., Vance C.P. (2010). Systemic signaling and local sensing of phosphate in common bean: Cross-talk between photosynthate and microRNA399. Mol. Plant 3: 428–437 [DOI] [PubMed] [Google Scholar]
- Liu K.H., Huang C.Y., Tsay Y.F. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11: 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M., Chopin F., Leleu O., Smith S.J., Krapp A., Daniel-Vedele F., Miller A.J. (2006). Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol. 142: 1304–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate J.S. (1980). Transport and partitioning of nitrogenous solutes. Annu. Rev. Plant Physiol. 31: 313–340 [Google Scholar]
- Pate J.S. (1983). Patterns of nitrogen metabolism in higher plants and their ecological significance. In Nitrogen as an Ecological Factor, Lee J.A., McNeill S., Rorison I.H., eds (Oxford: Blackwell; ), pp. 225–255 [Google Scholar]
- Patrick J.W., Zhang W., Tyerman S.D., Offler C.E., Walker N.A. (2001). Role of membrane transport in phloem translocation of assimilates and water. Funct. Plant Biol. 28: 697–709 [Google Scholar]
- Redinbaugh M.G., Campbell W.H. (1991). Higher plant responses to environmental nitrate. Physiol. Plant. 82: 640–650 [Google Scholar]
- Rentsch D., Laloi M., Rouhara I., Schmelzer E., Delrot S., Frommer W.B. (1995). NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett. 370: 264–268 [DOI] [PubMed] [Google Scholar]
- Robert H.S., Friml J. (2009). Auxin and other signals on the move in plants. Nat. Chem. Biol. 5: 325–332 [DOI] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Rus A., Yokoi S., Sharkhuu A., Reddy M., Lee B.H., Matsumoto T.K., Koiwa H., Zhu J.K., Bressan R.A., Hasegawa P.M. (2001). AtHKT1 is a salt tolerance determinant that controls Na(+) entry into plant roots. Proc. Natl. Acad. Sci. USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H., Takei K., Hirose N. (2006). Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci. 11: 440–448 [DOI] [PubMed] [Google Scholar]
- Schneider A., Schatten T., Rennenberg H. (1994). Exchange between phloem and xylem during long distance transport of glutathione in spruce trees (Picea abies [Karst.] L.). J. Exp. Bot. 45: 457–462 [DOI] [PubMed] [Google Scholar]
- Scholl R.L., May S.T., Ware D.H. (2000). Seed and molecular resources for Arabidopsis. Plant Physiol. 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupp R., Schatten T., Willenbrink J., Rennenberg H. (1992). Long-distance transport of reduced sulphur in spruce (Picea abies L.). J. Exp. Bot. 43: 1243–1250 [Google Scholar]
- Sheen J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Shelp B.J. (1987). The composition of phloem exudate and xylem sap from broccoli (Brassica oleracea var. italica) supplied with NH4+, NO3- or NH4NO3. J. Exp. Bot. 38: 1619–1636 [Google Scholar]
- Smirnoff N., Stewart G.R. (1985). Nitrate assimilation and translocation by higher plants: Comparative physiology and ecological consequences. Physiol. Plant. 64: 133–140 [Google Scholar]
- Song J., Ding X., Feng G., Zhang F. (2006). Nutritional and osmotic roles of nitrate in a euhalophyte and a xerophyte in saline conditions. New Phytol. 171: 357–366 [DOI] [PubMed] [Google Scholar]
- Srivastava A.C., Ganesan S., Ismail I.O., Ayre B.G. (2008). Functional characterization of the Arabidopsis AtSUC2 sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol. 148: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Wallace I.S., Takano J., Roberts D.M., Fujiwara T. (2008). NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 20: 2860–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer J.R., Huffaker R.C. (1980). Determination of nitrate and nitrite by high-pressure liquid chromatography: Comparison with other methods for nitrate determination. Anal. Biochem. 102: 110–119 [DOI] [PubMed] [Google Scholar]
- Touraine B., Glass A.D. (1997). NO3- and ClO3- fluxes in the chl1-5 mutant of Arabidopsis thaliana. Does the CHL1-5 gene encode a low-affinity NO3- transporter? Plant Physiol. 114: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Sauer N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 196: 564–570 [DOI] [PubMed] [Google Scholar]
- Tsay Y.F., Chiu C.C., Tsai C.B., Ho C.H., Hsu P.K. (2007). Nitrate transporters and peptide transporters. FEBS Lett. 581: 2290–2300 [DOI] [PubMed] [Google Scholar]
- Tsay Y.F., Schroeder J.I., Feldmann K.A., Crawford N.M. (1993). The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Vidal E.A., Gutiérrez R.A. (2008). A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr. Opin. Plant Biol. 11: 521–529 [DOI] [PubMed] [Google Scholar]
- Vlot A.C., Klessig D.F., Park S.-W. (2008). Systemic acquired resistance: The elusive signal(s). Curr. Opin. Plant Biol. 11: 436–442 [DOI] [PubMed] [Google Scholar]
- Wallsgrove R.M., Keys A.J., Lea P.J., Miflin B.J. (1983). Photosynthesis, photorespiration and nitrogen metabolism. Plant Cell Environ. 6: 301–309 [Google Scholar]
- Wang R., Liu D., Crawford N.M. (1998). The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc. Natl. Acad. Sci. USA 95: 15134–15139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T., Shimada S., Guo W., Sato K., Kohmura E., Hayakawa T., Takagi T., Tohyama M. (1997). Cloning and functional expression of a brain peptide/histidine transporter. J. Biol. Chem. 272: 10205–10211 [DOI] [PubMed] [Google Scholar]
- Yoshimoto N., Inoue E., Saito K., Yamaya T., Takahashi H. (2003). Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 131: 1511–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Tan Q., Lee R., Trethewy A., Lee Y.-H., Tegeder M. (2010). Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 22: 3603–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.J., Theodoulou F.L., Muldin I., Ingemarsson B., Miller A.J. (1998). Cloning and functional characterization of a Brassica napus transporter that is able to transport nitrate and histidine. J. Biol. Chem. 273: 12017–12023 [DOI] [PubMed] [Google Scholar]