This work shows that gibberellin-regulated DELLA proteins regulate chlorophyll and carotenoid biosynthesis to stimulate cotyledon greening during deetiolation.
Abstract
In plants, light represents an important environmental signal that triggers the production of photosynthetically active chloroplasts. This developmental switch is critical for plant survival because chlorophyll precursors that accumulate in darkness can be extremely destructive when illuminated. Thus, plants have evolved mechanisms to adaptively control plastid development during the transition into light. Here, we report that the gibberellin (GA)-regulated DELLA proteins play a crucial role in the formation of functional chloroplasts during deetiolation. We show that Arabidopsis thaliana DELLAs accumulating in etiolated cotyledons derepress chlorophyll and carotenoid biosynthetic pathways in the dark by repressing the transcriptional activity of the phytochrome-interacting factor proteins. Accordingly, dark-grown GA-deficient ga1-3 mutants (that accumulate DELLAs) display a similar gene expression pattern to wild-type seedlings grown in the light. Consistent with this, ga1-3 seedlings accumulate higher amounts of protochlorophyllide (a phototoxic chlorophyll precursor) in darkness but, surprisingly, are substantially more resistant to photooxidative damage following transfer into light. This is due to the DELLA-dependent upregulation of the photoprotective enzyme protochlorophyllide oxidoreductase (POR) in the dark. Our results emphasize the role of DELLAs in regulating the levels of POR, protochlorophyllide, and carotenoids in the dark and in protecting etiolated seedlings against photooxidative damage during initial light exposure.
INTRODUCTION
Seedling development undergoes critical changes during the transition from life in the dark just after germination, toward life in a light environment when seedlings emerge through the soil surface (Von Arnim and Deng, 1996; Chen et al., 2004). In complete darkness, seedlings grow heterotrophically on seed reserves in the absence of chlorophyll and functional chloroplasts, a developmental program known as skotomorphogenesis. Once dark-grown seedlings break through the soil and reach the light, seedlings undergo photomorphogenic development, including cotyledon opening and the development of photosynthetically active chloroplasts, which enables autotrophic growth (Casal et al., 2004).
This delicate developmental switch from skotomorphogenesis to photomorphogenesis (called deetiolation) is tightly regulated and requires light. In preparation for this switch, dark-grown seedlings accumulate the chlorophyll precursor protochlorophyllide (Pchlide) to permit rapid assembly of functional photosynthetic machinery upon initial light irradiation (Reinbothe et al., 1996; Mochizuki et al., 2010). Once the seedlings are exposed to light, the light-dependent enzyme NADPH:protochlorophyllide oxidoreductase (POR) is photoactivated and catalyzes the conversion of Pchlide to chlorophyllide, which is subsequently esterified to give chlorophyll (Reinbothe et al., 1996; Oosawa et al., 2000; Su et al., 2001; Frick et al., 2003). Nevertheless, the amount of Pchlide must be stoichiometrically linked to the level of POR, as free Pchlide (not bound to POR) operates as a photosensitizer upon light exposure, producing reactive oxygen species (ROS) and thereby causing photooxidative damage (Sperling et al., 1997; op den Camp et al., 2003). Therefore, plants have evolved efficient mechanisms to carefully regulate the levels of Pchlide in the dark.
The phytochrome-interacting factors (PIFs) have been shown to play a critical role in the control of chlorophyll biosynthesis in the dark (Huq et al., 2004; Moon et al., 2008; Stephenson et al., 2009; Shin et al., 2009). PIFs (including PIF1, PIF3, PIF4, and PIF5) are a subset class of the basic helix-loop-helix family of transcriptional factors that function as negative regulators of phytochrome-mediated light responses (Leivar and Quail, 2011). PIFs act to repress photomorphogenic seedling development in darkness and light reverses this repression by stimulating the proteasome-mediated degradation of PIFs, thus releasing the repressive effect of PIFs on chlorophyll and photosynthetic gene expression (Shen et al., 2005; Al-Sady et al., 2006; Leivar et al., 2009; Shin et al., 2009). Accordingly, pif mutants display constitutive photomorphogenic phenotypes in darkness (short hypocotyls and open cotyledons) and accumulate higher amounts of Pchlide (Huq et al., 2004; Leivar et al., 2009; Shin et al., 2009; Stephenson et al., 2009). In consequence, cotyledons of etiolated pif mutant seedlings are severely bleached when transferred to light (Huq et al., 2004; Stephenson et al., 2009). Recently, the PIFs have also been shown to directly repress the phytoene synthase (PSY) gene expression (the main rate-determining enzyme of carotenoid biosynthesis) to downregulate the accumulation of carotenoids (Toledo-Ortiz et al., 2010). Carotenoid biosynthesis is strongly upregulated when seedlings that germinate in the dark emerge from the soil to protect the plastids against photooxidative damage by quenching excess excitation energy (Pogson and Rissler, 2000; Rodríguez-Villalón et al., 2009). It was proposed that the light-triggered degradation of PIFs results in a rapid production of carotenoids in coordination with chlorophyll biosynthesis to rapidly assemble a functional photosynthetic machinery (Toledo-Ortiz et al., 2010).
Seedling deetiolation is also subject to hormonal regulation (Vandenbussche et al., 2005; Alabadí et al., 2004, 2008; Zhong et al., 2009). For instance, mutants affecting the biosynthesis of the plant growth hormone gibberellin (GA) derepress photomorphogenesis in darkness, a phenotype that is reverted by lack of DELLA growth repressing proteins (DELLAs) (Alabadí et al., 2004, 2008; Achard et al., 2007). DELLAs (GA-INSENSITIVE [GAI], REPRESSOR OF GA1-3 [RGA], RGA-LIKE1 [RGL1], RGL2, and RGL3) are a subfamily of the GRAS family of transcriptional regulators that repress GA-mediated responses, and GA overcomes this DELLA-mediated restraint by stimulating the polyubiquitination of DELLAs by the specific SCFSLY E3 ubiquitin ligase, thus promoting their degradation by the 26S proteasome pathway (Peng et al., 1997; Silverstone et al., 1998; Cheng et al., 2004; Dill et al., 2004; Fu et al., 2004; Griffiths et al., 2006; Willige et al., 2007; Achard and Genschik, 2009). DELLAs were shown to integrate many other environmental signal inputs in addition to light and thus were proposed to provide a mechanism that enables plants to adapt their growth and development according to their surrounding environment (Achard et al., 2006).
Recent studies have reported that DELLAs exert their repressive function through their interaction with PIFs (at least with PIF3 and PIF4), which inhibits their ability to interact with their target gene promoters and thus blocks their ability to inhibit transcription (de Lucas et al., 2008; Feng et al., 2008). Consistent with this model, PIF3- and PIF4-mediated light regulation of hypocotyl elongation is abrogated in seedlings with low levels of GA and, thus, high levels of DELLA protein accumulation (de Lucas et al., 2008; Feng et al., 2008). In addition to their role in regulating hypocotyl elongation, the PIFs, as previously mentioned, are important in the regulation of chlorophyll biosynthesis in the dark, a process also known to be repressed by GA (Alabadí et al., 2004, 2008). However, the role of the DELLA-PIF interaction in chlorophyll biosynthesis and photobleaching resistance during the seedling deetiolation process is unclear. Here, we demonstrate that DELLAs are abundant in cotyledons of dark-grown seedlings and that they regulate chlorophyll and carotenoid biosynthesis in a PIF-dependent manner. We show that DELLA function derepresses the expression of chlorophyll and photosynthetic genes in the dark, which is reminiscent of multiple loss-of-function pif mutants. We further provide evidence that DELLAs are positive regulators of POR expression and limit the accumulation of ROS generation and photooxidative damage during seedling deetiolation. We propose that DELLAs play a prominent role in the regulation of chlorophyll biosynthesis in the dark to promote cotyledon greening during deetiolation.
RESULTS AND DISCUSSION
DELLAs Accumulate in Cotyledons of Dark-Grown Seedlings
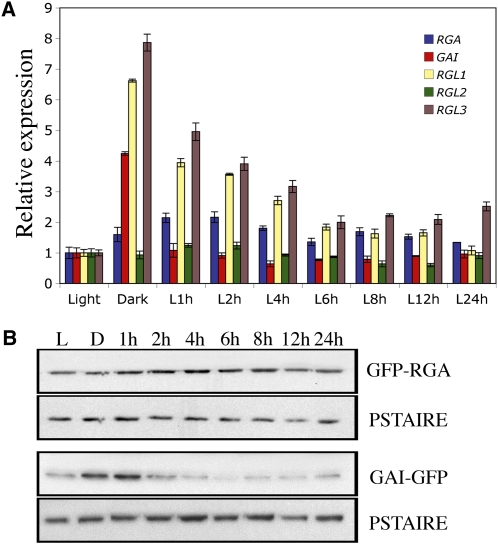
Recent genetic studies have revealed both distinct and overlapping functions of individual DELLAs in regulating GA responses (Cheng et al., 2004). However, their presence in cotyledons of dark-grown seedlings has not been established. To this end, we analyzed the expression pattern of the five Arabidopsis thaliana DELLA genes (GAI, RGA, RGL1, RGL2, and RGL3) in cotyledons of 5-d-old light- and dark-grown wild-type seedlings and of seedlings that have been transferred from dark to light conditions. Of the five DELLA genes, we found that transcript levels of GAI, RGL1, and RGL3 were higher by a factor of 4 to 8 in cotyledons of dark-grown seedlings compared with light-grown seedlings (Figure 1A). Moreover, higher levels of DELLA transcripts are directly linked to the absence of light and not the consequence of an altered developmental program, since the levels of GAI, RGL1, and RGL3 transcripts in etiolated seedlings decreased rapidly following transfer to white light, reaching the levels of light-grown seedlings within 6 h. Furthermore, although the expression of the DELLA genes RGA and RGL2 was not induced in darkness, their transcripts were detected in the dark (Figure 1A). Thus, the transcripts of all five DELLA genes accumulate in cotyledons of dark-grown seedlings.
Figure 1.
DELLAs Accumulate in Dark-Grown Cotyledons.
(A) Relative levels of RGA, GAI, RGL1, RGL2, and RGL3 gene transcripts (determined by quantitative RT-PCR) in cotyledons of 5-d-old seedlings grown in continuous white light (Light) or in the dark and transferred from dark to light for the time indicated (1 to 24 h). Data shown are the mean over three replicates and range of two biological repeats.
(B) Immunodetection of GFP-RGA and GAI-GFP in cotyledons of ProRGA:GFP-RGA and ProGAI:GAI-GFP seedlings grown in the light or in the dark and then transferred from dark to light; time as indicated (1 to 24 h). PSTAIRE serves as a sample-loading control. The blots shown are representative of three biological repeats.
[See online article for color version of this figure.]
DELLA proteins are subject to destabilization in the presence of GA (Cheng et al., 2004; Achard and Genschik, 2009). Thus, an increase in DELLA transcripts is not always accompanied by an increase in DELLA protein accumulation. Furthermore, previous studies have shown that light regulates GA levels in seeds and in seedling hypocotyls (Achard et al., 2007; Oh et al., 2007). To assess whether DELLAs accumulate in cotyledons of dark-grown seedlings, we used ProRGA:GFP-RGA and ProGAI:GAI-GFP transgenic lines expressing green fluorescent protein (GFP)-tagged versions of RGA and GAI that are detectable by protein gel blotting using an anti-GFP antibody (Achard et al., 2007). We detected the presence of both GFP-RGA and GAI-GFP in etiolated cotyledons (Figure 1B). GAI-GFP accumulated to even higher levels in etiolated cotyledons than in light-grown cotyledons and diminished rapidly following transfer of plants from dark to light. Thus, the amounts of GAI and RGA are directly proportional to theirs transcript levels, suggesting reduced bioactive GA levels in cotyledons. Overall, these results demonstrate that DELLAs are abundant in cotyledons of dark-grown seedlings and thus may cooperate with the PIFs in regulating chlorophyll biosynthesis in darkness (de Lucas et al., 2008; Feng et al., 2008; Leivar et al., 2009; Shin et al., 2009).
DELLA-PIF Complex Regulates Chlorophyll Biosynthesis in the Dark
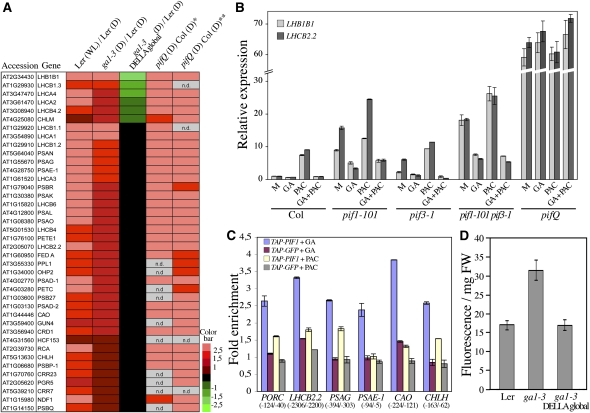
Previous studies reported that seedlings depleted in bioactive GA showed upregulation of light-regulated gene expression in darkness (Alabadí et al., 2004, 2008). To investigate the role of DELLAs at the genome level, we performed microarray analysis on 5-d-old etiolated wild-type (Landsberg erecta [Ler]), GA-deficient ga1-3, and ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1 rgl3-4 mutant seedlings (also called ga1-3 DELLA global-deficient mutant; Koini et al., 2009) (see Supplemental Data Set 1 online). Statistical analysis of the comparisons Ler white light versus Ler dark; ga1-3 dark versus Ler dark; and ga1-3 DELLA global dark versus Ler dark revealed 476 light-regulated genes displaying a DELLA-dependent change in the dark (see Supplemental Data Sets 1 to 3 online and Supplemental Figure 1A online; Bonferroni P value < 0.05). Half of these genes (238) were induced by light in the wild type and derepressed in a DELLA-dependent manner in an etiolated ga1-3 mutant (see Supplemental Data Set 2 online), while the other half was repressed by light in the wild type and repressed in a DELLA-dependent manner in etiolated ga1-3 (see Supplemental Data Set 3 online). Thus, DELLA function plays a critical role in the modulation of light-regulated genes in the dark. Next, to evaluate whether DELLAs regulate the expression pattern of the light-induced genes by modulating the transcriptional activity of PIFs, we compared the expression of the 238 light-induced DELLA-regulated genes (see Supplemental Data Set 2 online) with recent microarray data sets obtained from dark-grown quadruple pif mutants (pifQ; Leivar et al., 2009; Shin et al., 2009). Interestingly, as illustrated in Figure 2A, at least 40 chlorophyll biosynthesis and photosynthetic genes overlapped between ga1-3 and pifQ mutants, thus suggesting a tight regulation of these genes by the DELLA-PIF complex in the dark. To confirm the negative role of the DELLAs on PIF regulation of light-regulated genes, we measured the expression levels of three light-induced genes (LIGHT-HARVESTING CHLOROPHYLL-PROTEIN COMPLEX LHB1B1, LHCB2.2, and LHCB1.1, encoding chlorophyll binding proteins) that are regulated by both DELLAs and PIFs in etiolated wild-type seedlings and mutant seedlings lacking single or multiple PIFs treated with paclobutrazol (PAC; a GA biosynthesis inhibitor) and/or GA (Figure 2B; see Supplemental Figure 1B online). We found that PAC enhanced the expression levels of all three genes in the wild type and in single and double pif mutants, whereas GA abolished this upregulation (in seedlings treated with both PAC and GA). By contrast, LHB1B1, LHCB2.2, and LHCB1.1 expression was not affected by PAC or GA in the pifQ mutant, suggesting that DELLAs derepress light-induced gene expression in the dark by repressing the activity of the four PIF proteins absent in pifQ (PIF1, PIF3, PIF4, and PIF5). Thus, consistent with previous studies (Alabadí et al., 2008; de Lucas et al., 2008; Feng et al., 2008), our findings suggest that DELLAs positively regulate the expression of nuclear genes encoding chloroplast proteins in the dark by repressing the transcriptional activity of the PIFs. To provide direct evidence for this, we next investigated in vivo if the interaction of PIF1 with its targets was reduced in seedlings that accumulate DELLA proteins (PAC-treated seedlings). Given that the PIF proteins have been shown to bind to a G-box motif, CACGTG (Martínez-García et al., 2000; Moon et al., 2008), we first analyzed the upstream promoter region of the genes identified in the above transcriptome analysis for the presence of a G-box. We tested by chromatin immunoprecipitation (ChIP) assays using a transgenic line constitutively expressing a Myc-epitope-tagged PIF1 fusion protein in a pif1 background, whether LHCB2.2 was a direct target of PIF1. As shown in Figure 2C, PIF1 interacted with the LHCB2.2 promoter region containing a G-box, and this interaction was reduced when seedlings were treated with PAC. Two other photosynthetic genes, PHOTOSYSTEM 1 SUBUNIT G (PSAG) and PHOTOSYSTEM 1 SUBUNIT E-1 (PSAE-1), also interacted with PIF1 and showed similar regulation of this interaction. Chlorophyll biosynthesis genes were also identified as commonly regulated in ga1-3 and pifQ mutants (Figure 2A; see Figure 8 for pathway). We therefore tested whether PORC, CHLOROPHYLL A OXYGENASE (CAO), and CONDITIONAL CHLORINA (CHLH) were also direct targets of PIF1 (Figure 2C; Moon et al., 2008). Again, PIF1 was able to bind promoter regions containing a G-box of all three genes, and binding was reduced by PAC treatment. Thus, DELLA-PIF interactions directly affect the expression of genes encoding chlorophyll synthesis and other photosynthetic proteins in the dark.
Figure 2.
DELLAs Derepress Chlorophyll Biosynthesis in the Dark.
(A) Light-induced genes [up in Ler (white light, WL)/Ler (D)] showing similar upregulation to that seen for a DELLA-dependent increase in the dark for ga1-3 [up in ga1-3 (D)/Ler (D) and down or not differentially regulated in ga1-3 DELLA global (D)/Ler (D)] and to the quadruple pif mutant [up in pifQ (D)/Col (D)] as previously reported (*data analyzed by Shin et al. [2009]; **data analyzed by Leivar et al. [2009]). The color code (log2 ratios) depicts relative transcript levels. n.d., not determined.
(B) Relative transcript levels of LHB1B1 and LHCB2.2 in dark-grown wild type (Col), pif1-101, pif3-1, pif1-101 pif3-1, and pifQ mutants treated with PAC and/or GA (M, Mock). Data shown are the mean over three replicates and range of two biological repeats.
(C) DELLAs block the ability of PIF1 to bind promoters of nuclear genes encoding chloroplast proteins. Chromatin preparations from cotyledons of the 5-d-old dark-grown control line Pro35S:TAP-GFP or the Pro35S:TAP-PIF1 line treated with PAC or GA were subjected to ChIP followed by qPCR. Fold enrichment of each promoter region containing a G-box (PORC, LHCB2.2, PSAG, PSAE-1, CAO, and CHLH) was calculated by comparing the values with and without MYC antibody (as negative control), after normalizing to a control region (a sequence lacking a G-box). Data represent means ± sd of triplicate determinations from one ChIP experiment. Similar results were obtained in two independent biological experiments. The numbers below the gene names indicate base pairs upstream of the start site of transcription.
(D) Protochlorophyllide accumulation in wild-type (Ler), ga1-3, and ga1-3 DELLA global mutant seedlings in darkness. Data shown are the mean and range of two biological replicates each with four technical replicates.
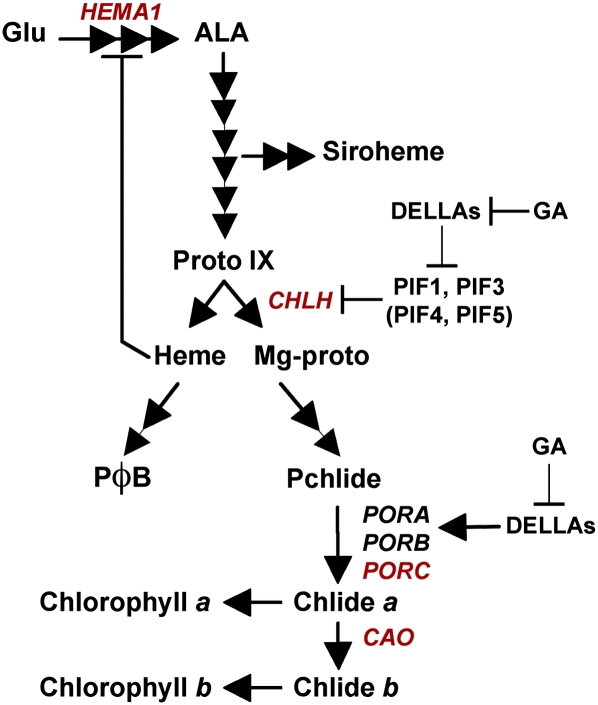
Figure 8.
Regulation of Chlorophyll Synthesis by the DELLA Family of Regulators.
A model for the action of DELLAs in regulating chlorophyll synthesis. Expression of CHLH is repressed by GA and promoted by DELLAs through the inactivation of PIF repressors. Other key light-induced chlorophyll biosynthesis genes (marked in red) that have been shown to be under PIF and DELLA regulation are proposed to use the same regulatory mechanism. Carotenoid synthesis through the induction of PSY is also promoted by DELLAs by a PIF-dependent mechanism. By contrast, the expression of light-repressed genes (PORA and PORB, in black) is independently promoted by DELLAs. Together, these regulatory responses promote plastid development while protecting etiolated seedlings against photooxidative damage during initial light exposure.
[See online article for color version of this figure.]
PIF factors are negative regulators of chlorophyll biosynthesis gene expression and in consequence repress Pchlide synthesis in the dark (Huq et al., 2004; Shin et al., 2009; Stephenson et al., 2009). We therefore assessed whether DELLAs also affect Pchlide accumulation in darkness. As shown in Figure 2D, we found that etiolated ga1-3 seedlings accumulated higher levels of Pchlide than the wild type. Moreover, the absence of DELLA proteins restored to wild-type values the level of Pchlide in ga1-3 DELLA global mutant seedlings.
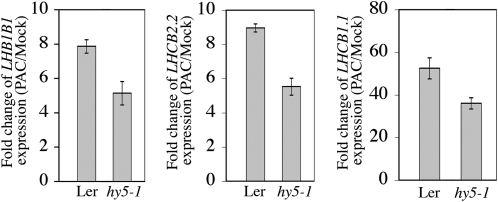
Previous work has shown that the GA pathway negatively regulates LONG HYPOCOTYL5 (HY5) activity to repress photomorphogenesis in darkness, and under these conditions, seedlings depleted in bioactive GA showed accumulation of HY5 (Alabadí et al., 2008). Consistent with an involvement of HY5 in GA-mediated repression of photomorphogenesis, the hy5 mutation reduced expression of LHB1B1, LHCB2.2, and LHCB1.1 in response to PAC in comparison to the wild type (Figure 3). However, the reduction was less than twofold, thus suggesting that the DELLA-PIF complex constitutes the main pathway by which GA modulates the expression of light-regulated genes in the dark.
Figure 3.
The hy5 Mutation Reduces Expression of LHB1B1, LHCB2.2, and LHCB1.1 in Response to PAC in Comparison to the Wild Type.
Relative transcript levels of LHB1B1, LHCB2.2, and LHCB1.1 in 5-d-old dark-grown wild type (Ler) and the hy5-1 mutant treated with PAC. Data (mean over three replicates) are represented as fold change of gene expression (PAC/Mock). Similar results were obtained in two independent experiments.
Overall, these observations confirm that DELLAs are important regulators of the chlorophyll biosynthetic pathway in the dark, as summarized in Figure 8.
The DELLA-PIF Complex Modulates PSY Expression to Regulate Carotenoid Accumulation
Carotenoids are essential photoprotective and antioxidant pigments synthesized by all photosynthetic organisms (Pogson and Rissler, 2000; Rodríguez-Villalón et al., 2009). Thus, carotenoid biosynthesis is strongly upregulated when dark-grown germinated seedlings first perceive light. In the dark, PIFs directly repress phytoene synthase (PSY) expression to downregulate the accumulation of the carotenoids, and light derepresses the PIF-mediated repression of carotenoid biosynthesis by stimulating the degradation of PIFs (Toledo-Ortiz et al., 2010). Previous work has shown that carotenoid biosynthesis is also induced when deetiolation is derepressed in the dark, including through the inhibition of GA biosynthesis (Rodríguez-Villalón et al., 2009; Toledo-Ortiz et al., 2010). We found that ga1-3 mutant seedlings accumulated higher levels of the four main species of carotenoids (lutein, β-carotene, violaxanthin, and neoxanthin) compared with wild-type seedlings (see Supplemental Figure 2A online). Conversely, lack of DELLA function (in ga1-3 DELLA global) abolished the increase in carotenoid accumulation observed in ga1-3 and resulted in carotenoid levels that were even lower than in wild-type seedlings (see Supplemental Figure 2A online). This result suggests that DELLAs derepress carotenoid biosynthesis in the dark. The rate-limiting step for the synthesis of carotenoids in the dark is the phytoene synthase encoded by PSY (Rodríguez-Villalón et al., 2009). We found a twofold increase in PSY transcript levels in etiolated ga1-3 seedlings relative to the wild type, and the absence of DELLA proteins suppressed this induction (see Supplemental Figure 2B online). Thus, the changes in PSY transcript levels closely correlate with changes in carotenoid levels. Finally, we assessed whether DELLAs act on PIF activity to regulate PSY expression. As was observed for the control of chlorophyll biosynthesis genes, we found that PAC treatment reduced PIF1 binding to the region encompassing the G-box motifs in the PSY promoter (see Supplemental Figure 2C online; Toledo-Ortiz et al., 2010). Altogether, these data indicate that the production of chlorophylls and carotenoids is coordinately regulated by DELLA-PIF complexes in the dark.
DELLA Function Prevents Photobleaching
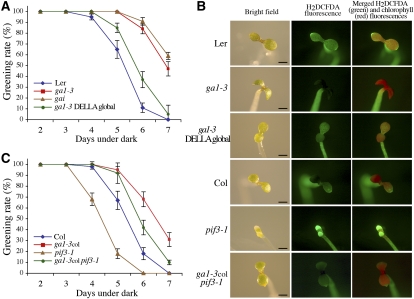
Etiolated seedlings that accumulate abnormally high levels of chlorophyll intermediates are bleached when transferred into the light (Sperling et al., 1997; op den Camp et al., 2003; Huq et al., 2004). Because etiolated ga1-3 mutant seedlings have high levels of Pchlide (Figure 2D), we assessed whether ga1-3 and various GA response mutants show a more severe bleaching phenotype when transferred into the light after extended periods in the dark. When grown for 6 d in the dark, ~90% of wild-type (Ler) seedlings were bleached after transfer to light (Figure 4A). Surprisingly, the ga1-3 and gai (a GA-insensitive mutant due to constitutive stability of the DELLA protein GAI; Peng et al., 1997) mutant seedlings were more resistant to photobleaching for the same period of dark treatment, despite a higher accumulation of Pchlide (as seen in ga1-3 in Figure 2D). Lack of DELLA function in ga1-3 DELLA global mutants partially suppressed the resistance conferred by ga1-3 (Figure 4A). Thus, unexpectedly and in contrast with pif mutants (Huq et al., 2004; Shin et al., 2009; Stephenson et al., 2009), mutants accumulating DELLAs prevent photobleaching.
Figure 4.
DELLAs Facilitate Greening of Etiolated Seedlings.
(A) and (C) Percentage of green cotyledons (greening rate) of etiolated seedlings (genotype indicated) grown in darkness before transfer to white light for 2 d. Data shown are the mean ± se of at least three independent experiments.
(B) Representative fluorescence microscopy images of ROS (H2DCFDA imaging) and chlorophyll fluorescence in cotyledons of seedlings grown for 4 d in the dark and transferred to white light for 2 d. Bars = 1 mm.
Previous studies suggest that failure of a seedling to green is attributable to photooxidative damage that is associated with increased accumulation of ROS in cotyledons (e.g., op den Camp et al., 2003). Consistent with the photobleaching phenotype, we found that the cotyledons of etiolated ga1-3 seedlings accumulated less ROS (as indicated by H2DCFDA fluorescence, a ROS-sensitive dye) and more chlorophyll (as indicated by autofluorescence) after transfer into light in comparison to wild-type seedlings (Figure 4B). Moreover, lack of DELLA function (in the ga1-3 DELLA global mutant) reduced chlorophyll accumulation and enhanced ROS accumulation of ga1-3 seedlings. Thus, DELLA function prevents photobleaching (and thus promotes the greening process) through the repression of ROS accumulation.
Previously, we have shown that DELLAs modulate the regulation of the chlorophyll biosynthetic pathway by PIFs (Figure 2). However, although the ga1-3 and pif mutants accumulate a high amount of Pchlide, these mutants behave oppositely during transfer into light. To assess the genetic interaction between the PIFs and the DELLAs, we crossed the ga1-3col (i.e., the ga1-3 allele in the Columbia [Col] ecotype) with the pif3-1 mutant. We found that accumulation of DELLAs (in ga1-3col pif3-1) substantially suppressed the photooxidative stress sensitivity conferred by pif3-1 (Figure 4C). Similarly, the increased ROS accumulation observed in cotyledons of pif3-1 seedlings after transfer from dark to light was significantly reduced in cotyledons of ga1-3col pif3-1 seedlings (Figure 4B). Thus, DELLAs, by a mechanism independent to PIF action, protect seedlings from photooxidative damage during deetiolation.
DELLAs Promote the Induction of POR Transcripts and Prolamellar Body Formation in the Dark
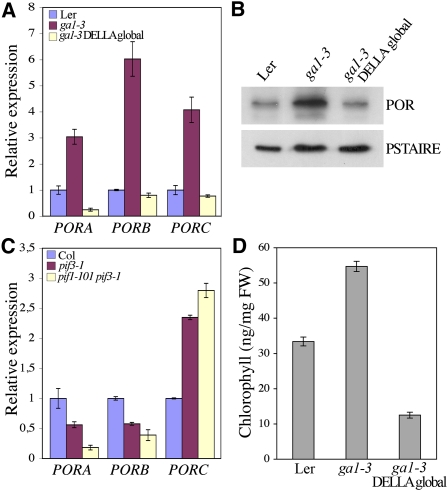
Coordinate regulation of Pchlide and POR levels is particularly crucial during deetiolation as it is the binding of Pchlide to POR that prevents it from causing photooxidative damage (Sperling et al., 1997). To investigate how DELLA function leads to increased resistance to photooxidative stress, we next determined whether DELLAs regulate the expression levels of the POR genes. In Arabidopsis, there are three closely related genes, PORA, PORB, and PORC, showing different expression patterns (Reinbothe et al., 1996; Oosawa et al., 2000; Su et al., 2001). While PORA and PORB are highly expressed in etiolated seedlings, PORC transcripts significantly accumulate only after the onset of illumination (Oosawa et al., 2000; Su et al., 2001; Matsumoto et al., 2004). We found that all three POR genes were induced in cotyledons of dark-grown ga1-3 seedlings compared with wild-type seedlings (Figure 5A). The induced abundance of POR transcripts was correlated with increased POR protein level as detected by immunoblotting using an anti-POR antibody that recognizes the three POR isoforms (Figure 5B). Moreover, the DELLA-dependent regulation of POR expression was maintained after the transfer of seedlings from dark to light, although the genes were still responsive to light (see Supplemental Figure 3 online). Consistent with our previous results, lack of DELLA proteins in ga1-3 DELLA global mutants suppressed the upregulation of the POR transcript and protein levels conferred by the ga1-3 mutation (Figures 5A and 5B). By contrast, analysis of POR expression in pif3-1 and the pif1-101 pif3-1 double mutant showed that expression was reduced for PORA and PORB, but higher for PORC, consistent with the expected light regulation of these genes (Figure 5C; Matsumoto et al., 2004). Previously, PORA and PORB were shown to be downregulated or not significantly different in pif mutants through microarray analysis (Leivar et al., 2009), while PORC was reported to be downregulated in the pif1-2 mutant (Moon et al., 2008), a result inconsistent with this study and its known light-signaling profile (Oosawa et al., 2000; Su et al., 2001; Matsumoto et al., 2004). Since PORC was shown to be a direct target of PIF1, this result was interpreted as PIF1 promoting PORC transcription (Moon et al., 2008), while our data are consistent with PIF1 repressing PORC expression as for other light-induced genes (Figure 2A). Taken together, our results suggest that DELLAs positively regulate (at least) PORA and PORB expression in the dark by a mechanism that is independent of regulation by PIFs. Accordingly, GA deficiency upregulated POR expression in the pif3-1 mutant (in ga1-3col pif3-1; see Supplemental Figure 4A online). The mechanism by which DELLAs regulate PORA and PORB expression is unclear but is likely to be indirect, as we were unable to obtain evidence for direct binding of GAI to the promoter of PORA and PORB by ChIP assays (see Supplemental Figure 4B online). The mechanism could eventually involve the regulation of HY5 activity (see Supplemental Figure 4C online). Nevertheless, DELLAs play a prominent role in the regulation of POR levels throughout the deetiolation process, providing a means of preventing ROS accumulation after illumination. Consistent with the POR levels, when grown in the dark for 5 d before transfer to white light, ga1-3 mutants accumulated higher levels of chlorophyll over the next 8 h than wild-type seedlings, whereas ga1-3 DELLA global mutants accumulated less chlorophyll (Figure 5D). Interestingly, the levels of the POR transcripts were also lower in the barley slender mutant, a loss-of-function mutant lacking the only DELLA protein (SLENDER) present in barley (Hordeum vulgare; Ougham et al., 2001). Therefore, this DELLA-dependent regulation of POR expression is evolutionarily conserved between monocots and dicots.
Figure 5.
DELLAs Activate POR Gene Expression.
(A) Relative levels of PORA, PORB, and PORC gene transcripts in cotyledons of 5-d-old dark-grown wild-type (Ler), ga1-3, and ga1-3 DELLA global mutant seedlings. Data shown are the mean over three replicates and range of two biological repeats.
(B) Immunodetection of POR proteins in cotyledons of 5-d-old dark-grown wild-type (Ler), ga1-3, and ga1-3 DELLA global mutant seedlings. The POR antibody recognizes the three POR isoforms.
(C) Relative levels of PORA, PORB, and PORC gene transcripts in 5-d-old dark-grown wild-type (Col), pif3-1, and pif1-101 pif3-1 mutant seedlings. Data shown are the mean over three replicates and range of two biological repeats.
(D) Chlorophyll levels in wild-type (Ler), ga1-3, and ga1-3 global DELLA mutant seedlings 8 h after transfer to white light following 5 d in the dark. Data shown are the mean over three replicates and range of two biological repeats.
[See online article for color version of this figure.]
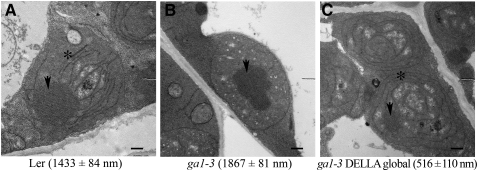
In etiolated angiosperms, POR is localized primarily with Pchlide and carotenoids in the prolamellar body (PLB), a lattice of tubular membranes that defines etioplasts (Park et al., 2002; Frick et al., 2003). It has been shown that a stoichiometric ratio between the POR, Pchlide, and carotenoids is required to form characteristic PLBs in etioplasts. Indeed, whereas the absence of any of these constituents results in a dramatic reduction in PLB size (or even their absence) in cotyledon etioplasts, overexpression of POR increases the size of PLBs (Sperling et al., 1997; Park et al., 2002; Frick et al., 2003). Accordingly, pif mutants also display etioplasts with reduced PLB size and increased prothylakoid membranes (Leivar et al., 2009; Stephenson et al., 2009). To determine whether DELLAs also regulate PLB size, we examined the ultrastructure of cotyledon etioplasts of 5-d-old wild-type, ga1-3, and ga1-3 DELLA global mutant seedlings. ga1-3 etioplasts generally showed an increased PLB size and decreased prothylakoid membranes compared with the wild type (Figures 6A and 6B). By contrast, ga1-3 DELLA global mutant etioplasts showed substantially smaller PLBs and increased membrane development (Figure 6C), although a PLB of similar size to that seen in wild-type seedlings was occasionally observed. Thus, DELLAs promote PLB formation, a result consistent with the increased POR levels in ga1-3 seedlings.
Figure 6.
DELLAs Regulate Prolamellar Body Formation.
Representative transmission electron microscopy images of cotyledon etioplasts from 5-d-old dark-grown wild-type (Ler [A]), ga1-3 (B), and ga1-3 DELLA global mutant (C) seedlings. The arrow indicates the PLB and the asterisk the prothylakoid membranes. Numbers represent cross section area (mean in nm ± se) for at least 50 PLBs per genotype. Bars = 500 nm.
Modulation of POR Levels Partially Suppresses the DELLA-Mediated Increased Resistance to Photobleaching
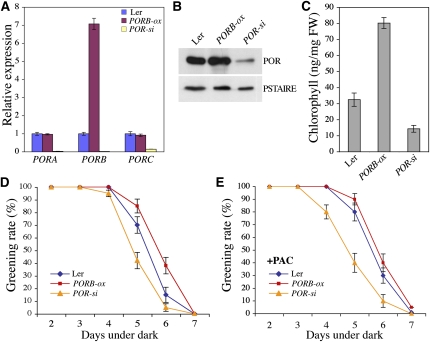
To examine the involvement of the POR proteins in the DELLA-mediated increased resistance to photobleaching, we created transgenic Arabidopsis plants that constitutively express PORB under the control of the 35S cauliflower mosaic virus promoter. Several T3 lines were identified, and two transgenic lines that moderately overexpress PORB (PORB-ox) and underexpress all three POR genes (hereafter called POR-si as PORA, PORB, and PORC are silenced) were selected for further studies (Figure 7A). Protein gel blot analysis confirmed that cotyledons of 5-d-old dark-grown PORB-ox and POR-si seedlings accumulated higher and lower amounts, respectively, of POR proteins in comparison to wild-type seedlings (Figure 7B). Consistent with previous studies (Frick et al., 2003), when grown in the dark for 5 d before transfer to white light, PORB-ox seedlings accumulated higher levels of chlorophyll over the next 8 h than wild-type seedlings, whereas POR-si accumulated less chlorophyll (Figure 7C). This result suggests that PORB-ox seedlings are greening faster and thus are able to reduce more rapidly the Pchlide into chlorophyllide than wild-type seedlings after illumination (Figure 8), while the converse is true with POR-si seedlings.
Figure 7.
Modulation of POR Levels Alters the Greening of Etiolated Seedlings.
(A) Relative levels of PORA, PORB, and PORC transcripts in cotyledons of 5-d-old dark-grown wild-type (Ler), PORB-ox, and POR-si seedlings. Data shown are the mean over three replicates and range of two biological repeats.
(B) Immunodetection of POR level in cotyledons of 5-d-old dark-grown wild-type (Ler), PORB-ox, and POR-si seedlings.
(C) Chlorophyll levels in wild-type (Ler), PORB-ox, and POR-si seedlings 8 h after transfer to white light following 5 d in the dark. Data shown are the mean over three replicates and range of two biological repeats.
(D) and (E) Percentage of green cotyledons (greening rate) of etiolated seedlings (genotype indicated) grown in darkness in absence (D) or in presence (E) of PAC before transfer to white light for 2 d. Data are the mean ± se of at least three independent experiments.
[See online article for color version of this figure.]
Because we presumed that DELLAs protect seedlings from photooxidation by inducing (at least in part) POR gene expression, we first investigated whether increased or decreased POR levels affect the greening rate of seedlings. When transferred into the light after extended periods in the dark, PORB-ox seedlings were substantially more resistant to photobleaching than POR-si seedlings (Figure 7D). Thus, as demonstrated previously (Sperling et al., 1997; Frick et al., 2003), PORs are critical in protecting seedlings from photooxidation. Then, we assessed the effect of PAC on the greening rate of PORB-ox and POR-si seedlings. Interestingly, while PAC treatment slightly increased resistance of wild-type seedlings to reach the rate of PORB-ox seedlings, reduced POR levels (in POR-si) prevent the positive effect of PAC in protecting seedlings from photobleaching (Figures 7D and 7E). Thus, reduced POR levels partially suppress the resistance conferred by increased accumulation of DELLAs.
Altogether, our results suggest that DELLAs play a prominent role, at least in part through the regulation of PIF activity, in coordinating the levels of POR, chlorophyll precursors, and carotenoids in cotyledons of dark-grown seedlings. Recently, two GATA-type transcription factors, GNC (GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM INVOLVED) and GNL (GNC-LIKE), were shown to repress GA signaling downstream from DELLAs and PIFs and to regulate POR levels and chlorophyll biosynthesis (Richter et al., 2010). While the loss of GNC and GNL impairs chlorophyll biosynthesis, their overexpression increases the amount of chlorophyll. Thus DELLAs may control chlorophyll biosynthesis at least in part by modulating the expression of these two GATA-type transcription factors. We propose a model (Figure 8) in which, in the dark, DELLAs interact with PIFs and repress their ability to bind to the promoters of light-regulated nuclear-encoded chloroplast genes (Figure 2C). As a consequence, DELLAs enhance the production of chlorophyll precursors and carotenoids in the dark (Figure 2D; see Supplemental Figure 2A online). Interestingly, PIF1 has been shown to regulate the transcription of GAI and RGA in germinating seeds, thus suggesting a feedback mechanism by which PIF1 regulates its activity (Oh et al., 2007). Our results also indicate that DELLAs regulate in a PIF-independent manner the expression levels of PORA and PORB encoding the light-dependent enzyme responsible for the conversion of phototoxic Pchlide to chlorophyllide. By doing this, DELLAs restrain ROS accumulation triggering photooxidative damage during initial light exposure, thereby promoting seedling greening. The promotion of PORA and PORB expression therefore acts as a safeguard mechanism for the induction of chlorophyll precursors in the absence of light. It is noteworthy that many signal inputs regulate the levels of POR transcripts (Kusnetsov et al., 1998; Zhong et al., 2009), thus suggesting that the regulation of the POR levels represents a critical regulatory step for the control of chlorophyll biosynthesis during seedling greening. However, because a reduction in POR levels (in POR-si) only prevents the positive effect of DELLA accumulation in protecting seedlings against photooxidative damage (and does not increase the sensitivity to PAC per se), it is likely that DELLAs act also through other mechanisms. Previously, it was reported that DELLAs promote survival of adversity by reducing the levels of ROS through the activation of ROS detoxification enzymes (Achard et al., 2008). In this study, we also found in the microarray data a large proportion of genes encoding antioxidant systems that were upregulated in dark-grown ga1-3 seedlings and thus could prevent ROS accumulation during seedling deetiolation (see Supplemental Data Set 2 online).
Finally, DELLAs have been proposed as integrators of different signal inputs to adapt plant growth and development to natural environments (Achard et al., 2006). We propose that DELLAs play also such a role during deetiolation, in adapting plastid development to cope with the delicate switch from heterotrophic to autotrophic growth.
METHODS
Arabidopsis thaliana Lines
Mutant and transgenic lines were derived from Ler (ga1-3; gai; ga1-3 DELLA global; hy5-1; ProRGA:GFP-RGA; ProGAI:GAI-GFP; and 35S:GAI-GFP) or Col (ga1-3col; pif3-1; pif1-101; pifQ; Pro35S:TAP-PIF1; and Pro35S:TAP-GFP) backgrounds as previously described (Tyler et al., 2004; Achard et al., 2006; Alabadí et al., 2008; Moon et al., 2008; Koini et al., 2009; Shin et al., 2009; Stephenson et al., 2009). The ga1-3col pif3-1 line was isolated from F3 progeny of the appropriate cross. Genomic PCR was used to confirm the genotype. PORB cDNA was amplified by RT-PCR and was cloned into the pGEM-T easy vector system (Promega). A BamHI-EcoRI fragment containing the PORB cDNA was inserted into pGreen0229 containing a 35S cauliflower mosaic virus cassette and Basta resistance marker (http://www.pgreen.ac.uk). The plasmid was introduced into Agrobacterium tumefaciens GV3101 by electroporation. Arabidopsis (Ler) plants were transformed by the floral dip method.
Growth Conditions
As ga1 mutants do not germinate without exogenous GA, all seeds including ga1-3 were pretreated at 4°C with 100 μM GA3 for 4 d to synchronize germination, washed thoroughly three times, and then surface sterilized and plated on 1× Murashige and Skoog (MS) 0.8% (w/v) agar plates containing 5 μM GA3 and/or 1 μM PAC when indicated. After germination was induced under white light (95 μmol m−2 s−1) for 6 h at 22°C, plates were placed in the dark or in continuous white light (95 μmol m−2 s−1) as indicated.
Gene Expression and Immunoblot Analysis
Seedlings were grown for 5 d in dark or white light (95 μmol m−2 s−1). Dark-grown seedlings were then placed under white light for the time indicated, and total RNA was purified from seedling cotyledons. Quantitative real-time RT-PCR analysis was performed using SYBR Green Master mix on a Lightcycler LC480 (Roche). AT4G34270 (TIP4-1 LIKE) and AT4G26410 genes were used as internal controls. The relative expression level of each gene was compared with that of wild-type cotyledon seedlings using GenEx Pro 4.3.5. software (MultiD Analyses) after normalization and averaging over three replicates. Quantitative RT-PCR analyses were performed on two biological repeats. PCR primers used are listed in Supplemental Table 1 online. Immunoblots were performed as previously described (Achard et al., 2007) with anti-GFP (Miltenyi Biotec) and anti-POR (Agrisera) antibodies.
ChIP Assays
ChIP assays were performed on cotyledons of 5-d-old dark-grown 35S:TAP-PIF1, 35S:TAP-GFP, and 35S:GAI-GFP seedlings. Cotyledons were vacuum-infiltrated with 1% formaldehyde for 10 min, and cross-linking was quenched by vacuum infiltration with 125 mM glycine for 5 min. Chromatin was sheared with a Bioruptor sonicator (Cosmo Bio, Tosho Denki) twice for 15 min with a 50% duty cycle and high power output to obtain 200- to 1000-bp DNA fragments. Chromatin was immunoprecipitated with specific antibodies (anti-MYC [Roche Diagnostics]; anti-GFP [Clontech]) together with protein A magnetic beads (Millipore). Following elution with Proteinase K (Invitrogen), DNA was recovered using Magna ChIP spin filters (Millipore). ChIP experiments using protein A magnetic beads without the addition of antibody were performed as negative controls. The resulting ChIP DNA was subjected to quantitative PCR (qPCR) analysis. PCR primers used are listed in Supplemental Table 1 online. Enrichment of promoter regions was determined using GenEx Pro 4.3.5 software (MultiD Analyses) after normalization and averaging over three replicates using control sequences lacking G-box (PSY −2571/−2466) or coding sequences (PORA +1025/+1146; PORB +1051/+1170) as references. ChIP-qPCR analyses were performed on two independent biological repeats.
Microarray Data Analysis
Microarray analysis on RNA from 5-d-old seedlings was performed on three independent biological replicates with the CATMA array (Unité de Recherche en Génomique Végétale). Microarray data from this article were deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; number GSE22681) and at CATdb (http://urgv.evry.inra.fr/CATdb/; Project RS2009-09 DELLAdark) according to the Minimum Information about a Microarray Experiment standards. RNA samples from three independent biological replicates were used. For each biological repeat, RNA samples for a given condition were obtained by pooling RNAs from more than 100 5-d-old seedlings. For each comparison, one technical replication with fluorochrome reversal was performed for each biological replicate (i.e., four hybridizations per comparison). The reverse transcription of RNA in the presence of Cy3-dUTP or Cy5-dUTP (Perkin-Elmer-NEN Life Science Products), the hybridization of labeled samples to the slides, and the scanning of the slides were performed as described previously (Lurin et al., 2004).
Data analysis was based on one dye swap per biological replicate (i.e., four arrays, each containing 24,576 GSTs and 384 controls) as described previously (Gagnot et al., 2008). Statistical methods were developed using the software R (R Development Core Team; http://www.R-project.org) in collaboration with the group Statistics and Genome at Unité Mixte de Recherche 518 Mathématique et Informatique Appliquées AgroParisTech/Institut National de la Recherche Agronomique and are available in the R package Anapuce at http://cran.r-project.org/web/packages/anapuce/index.html. For each CATMA array, the raw data include the logarithm of the median feature pixel intensity at wavelengths 635 nm (red) and 532 nm (green); no background is subtracted. Normalization per array was performed to remove systematic biases. First, spots that were considered to have badly formed features were excluded (spots with a flag value equal to –100). Then, a global intensity-dependent normalization was performed using the Lowess procedures (Yang and Thorne, 2003) to correct the dye bias. Finally, for each block, the log-ratio median calculated over the values for the entire block was subtracted from each individual log-ratio value to correct effects on each block, as well as print-tip, washing, and/or drying effects. At the end of the normalization step, a normalized log-ratio, which is equivalent to an expression difference (in log base 2) between the two samples cohybridized on the same array, is given for each spot. It is equal to the raw log-ratio minus the Lowess correction minus the block correction. Normalized logarithm intensity for each sample was also calculated. It was performed according to the within-array correction (Yang and Thorne, 2003), which is a redistribution of the correction calculated for the log2 ratio normalization on each channel.
To determine differentially expressed genes from a dye-swap, a paired t test was performed on the log2 ratios. The number of observations per spot is inadequate for calculating a gene-specific variance. For this reason, it is assumed that the variance of the log2 ratios is the same for all genes, and spots displaying extreme variances (too small or too large) were excluded. The raw P values were adjusted by the Bonferroni method (Ge et al., 2003), which controls the family-wise error rate and is the most stringent correction. When the Bonferroni P value is lower than 0.05, the gene is declared differentially expressed. Genes with a missing P value are genes with a too small or a too large variance or genes for which only one observation is available.
Protochlorophyllide Analysis
Seeds were stratified for 4 d at 4°C in 100 μM GA, washed, surface sterilized, and plated on 0.5× MS, 0.8% (w/v) agar plates, and germination was induced under white light (320 μmol m−2 s−1) for 6 h at 22°C. Twenty milligrams of 5-d-old dark-grown seedling material was extracted twice in 1.6 mL total of acetone:0.1 M NH4OH, 90:10 (v/v), centrifuging the samples at 13,000g for 10 min after each extraction. Pchlide was measured by relative fluorescence following excitation at 440 nm using a Hitachi fluorescence spectrophotometer F-2000.
Carotenoid and Chlorophyll Analysis
Arabidopsis pigments were extracted from 5-d-old seedling cotyledons and analyzed as previously described (Bouvier et al., 2006).
Photobleaching Assay
Seedlings were grown in the dark for 2 to 7 d on MS-agar Magenta boxes and moved into continuous white light (95 μmol m−2 s−1) for an additional 2 d. To determine the bleaching rate, the number of plants that were bleaching (yellowish or light green cotyledons) or greening (dark-green cotyledons) was counted. At least two independent biological repeats were performed for each series of analyses.
ROS Imaging
Seedlings were grown in the dark for 4 d and moved into continuous white light (95 μmol m−2 s−1) for an additional 2 d. Seedlings were incubated for 30 min at 4°C in 10 μM H2DCFDA (a ROS-sensitive dye) and then washed with 10 mM MES, 0.1 mM CaCl2, pH 6, for 60 min at 22°C. Dye excitation was at 480 nm; emitted light was detected at 535 to 550 nm with a Nikon SMZ1500 camera.
Electron Microscopy
Five-day-old dark-grown seedlings were fixed for 16 h in 4% (v/v) glutaraldehyde, 2 h in 2% (w/v) uranyl acetate and postfixed in 0.1% (v/v) osmium tetroxide in 150 mM sodium phosphate buffer, pH 7.2. Samples were dehydrated through an ethanol series and infiltrated with EPON 812 medium grade resin (Polysciences) and polymerized for 48 h at 60°C. Ultrathin sections (60 nm) were cut using an Ultracut E microtome (Reichert and Jung) and collected on grids coated with formvar (Electron Microscopy Sciences). Electron microscopy was performed with a Hitachi H-600 electron microscope at 75 kV. Images were captured with a CCD Advantage HR Hamamatsu camera and AMT software (Advanced Management Technology).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At2g01570 (RGA), At1g14920 (GAI), At1g66350 (RGL1), At3g03450 (RGL2), At5g17490 (RGL3), AT4g02780 (GA1), At2g20180 (PIF1), At1g09530 (PIF3), At2g43010 (PIF4), At3g59060 (PIF5), At5g11260 (HY5), At5g17230 (PSY), At5g54190 (PORA), At4g27440 (PORB), At1g03630 (PORC), At2g34430 (LHB1B1), At2g05070 (LHCB2.2), LHCB1.1 (At1g29920), At5g13630 (CHLH), At3g59400 (GUN4), At3g54890 (LHCA1), At1g44446 (CAO), At1g58290 (HEMA1), At1g55670 (PSAG), and At4g28750 (PSAE-1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Regulation of Chlorophyll Synthesis by the PIF and DELLA Families of Regulators.
Supplemental Figure 2. DELLAs Promote Carotenoid Biosynthesis in Darkness.
Supplemental Figure 3. DELLAs Upregulate POR Gene Expression.
Supplemental Figure 4. Role of PIF3 and HY5 in GA-Mediated Regulation of POR Gene Expression.
Supplemental Table 1. List of the Primers Used for Quantitative Real-Time PCR Analyses.
Supplemental Data Set 1. List of Differential Expressed Genes.
Supplemental Data Set 2. List of the 238 Light-Induced Genes Displaying a Similar Upregulation to That Seen for a DELLA-Dependent Increase in the Dark for ga1-3.
Supplemental Data Set 3. List of the 238 Light-Repressed Genes Displaying a Similar Downregulation to That Seen with ga1-3 in the Dark.
Acknowledgments
We thank N. Harberd for the ga1-3 DELLA global mutant and ProGAI:GAI-GFP and Pro35S:GAI-GFP lines, T.P. Sun for ga1-3col and ProRGA:GFP-RGA lines, C. Fankhauser for pif3-1 and pif1-101 pif3-1, G. Choi for pifQ, and E. Huq for Pro35S:TAP-PIF1 and Pro35S:TAP-GFP lines. This work was supported by Agence Nationale de la Recherche Grant 07-JCJC-0118.
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P., Genschik P. (2009). Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J. Exp. Bot. 60: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Achard P., Liao L., Jiang C., Desnos T., Bartlett J., Fu X., Harberd N.P. (2007). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143: 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Renou J.P., Berthomé R., Harberd N.P., Genschik P. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Alabadí D., Gallego-Bartolomé J., Orlando L., García-Cárcel L., Rubio V., Martínez C., Frigerio M., Iglesias-Pedraz J.M., Espinosa A., Deng X.W., Blázquez M.A. (2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53: 324–335 [DOI] [PubMed] [Google Scholar]
- Alabadí D., Gil J., Blázquez M.A., García-Martínez J.L. (2004). Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 134: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Bouvier F., Linka N., Isner J.C., Mutterer J., Weber A.P., Camara B. (2006). Arabidopsis SAMT1 defines a plastid transporter regulating plastid biogenesis and plant development. Plant Cell 18: 3088–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J.J., Fankhauser C., Coupland G., Blázquez M.A. (2004). Signalling for developmental plasticity. Trends Plant Sci. 9: 309–314 [DOI] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Cheng H., Qin L., Lee S., Fu X., Richards D.E., Cao D., Luo D., Harberd N.P., Peng J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064 [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A., Thomas S.G., Hu J., Steber C.M., Sun T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick G., Su Q., Apel K., Armstrong G.A. (2003). An Arabidopsis porB porC double mutant lacking light-dependent NADPH:protochlorophyllide oxidoreductases B and C is highly chlorophyll-deficient and developmentally arrested. Plant J. 35: 141–153 [DOI] [PubMed] [Google Scholar]
- Fu X., Richards D.E., Fleck B., Xie D., Burton N., Harberd N.P. (2004). The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnot S., Tamby J.P., Martin-Magniette M.L., Bitton F., Taconnat L., Balzergue S., Aubourg S., Renou J.P., Lecharny A., Brunaud V. (2008). CATdb: A public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 36(Database issue): D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Dudoit S., Speed T.P. (2003). Resampling-based multiple testing for microarray data analysis. Test 12: 1–77 [Google Scholar]
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z.L., Powers S.J., Gong F., Phillips A.L., Hedden P., Sun T.P., Thomas S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Koini M.A., Alvey L., Allen T., Tilley C.A., Harberd N.P., Whitelam G.C., Franklin K.A. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Kusnetsov V., Herrmann R.G., Kulaeva O.N., Oelmüller R. (1998). Cytokinin stimulates and abscisic acid inhibits greening of etiolated Lupinus luteus cotyledons by affecting the expression of the light-sensitive protochlorophyllide oxidoreductase. Mol. Gen. Genet. 259: 21–28 [DOI] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Huq E., Quail P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Matsumoto F., Obayashi T., Sasaki-Sekimoto Y., Ohta H., Takamiya K., Masuda T. (2004). Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol. 135: 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Tanaka R., Grimm B., Masuda T., Moulin M., Smith A.G., Tanaka A., Terry M.J. (2010). The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 15: 488–498 [DOI] [PubMed] [Google Scholar]
- Moon J., Zhu L., Shen H., Huq E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 9433–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H.S., Sun T.P., Kamiya Y., Choi G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa N., Masuda T., Awai K., Fusada N., Shimada H., Ohta H., Takamiya K. (2000). Identification and light-induced expression of a novel gene of NADPH-protochlorophyllide oxidoreductase isoform in Arabidopsis thaliana. FEBS Lett. 474: 133–136 [DOI] [PubMed] [Google Scholar]
- op den Camp R.G.L., Przybyla D., Ochsenbein C., Laloi C., Kim C., Danon A., Wagner D., Hideg E., Göbel C., Feussner I., Nater M., Apel K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ougham H.J., Thomas A.M., Thomas B.J., Frick G.A., Armstrong G.A. (2001). Both light-dependent protochlorophyllide oxidoreductase A and protochlorophyllide oxidoreductase B are down-regulated in the slender mutant of barley. J. Exp. Bot. 52: 1447–1454 [DOI] [PubMed] [Google Scholar]
- Park H., Kreunen S.S., Cuttriss A.J., DellaPenna D., Pogson B.J. (2002). Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B.J., Rissler H.M. (2000). Genetic manipulation of carotenoid biosynthesis and photoprotection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1395–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S., Reinbothe C., Lebedev N., Apel K. (1996). PORA and PORB, two light-dependent protochlorophyllide-reducing enzymes of angiosperm chlorophyll biosynthesis. Plant Cell 8: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R., Behringer C., Müller I.K., Schwechheimer C. (2010). The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Villalón A., Gas E., Rodríguez-Concepción M. (2009). Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 60: 424–435 [DOI] [PubMed] [Google Scholar]
- Shen H., Moon J., Huq E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Ciampaglio C.N., Sun T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling U., van Cleve B., Frick G., Apel K., Armstrong G.A. (1997). Overexpression of light-dependent PORA or PORB in plants depleted of endogenous POR by far-red light enhances seedling survival in white light and protects against photooxidative damage. Plant J. 12: 649–658 [DOI] [PubMed] [Google Scholar]
- Stephenson P.G., Fankhauser C., Terry M.J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q., Frick G., Armstrong G., Apel K. (2001). POR C of Arabidopsis thaliana: A third light- and NADPH-dependent protochlorophyllide oxidoreductase that is differentially regulated by light. Plant Mol. Biol. 47: 805–813 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Rodríguez-Concepción M. (2010). Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 107: 11626–11631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L., Thomas S.G., Hu J., Dill A., Alonso J.M., Ecker J.R., Sun T.P. (2004). Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., Verbelen J.P., Van Der Straeten D. (2005). Of light and length: Regulation of hypocotyl growth in Arabidopsis. Bioessays 27: 275–284 [DOI] [PubMed] [Google Scholar]
- Von Arnim A., Deng X.W. (1996). Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 215–243 [DOI] [PubMed] [Google Scholar]
- Willige B.C., Ghosh S., Nill C., Zourelidou M., Dohmann E.M., Maier A., Schwechheimer C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.H., Thorne N. (2003). Normalization for two-color cDNA microarray data. In Science and Statistics: A Festschrift for Terry Speed, IMS Lecture Notes, Monograph Series, Vol. 40, Goldstein D.R., ed, (Beachwood, OH: Institute of Mathematical Statistics; ), pp. 403–418 [Google Scholar]
- Zhong S., Zhao M., Shi T., Shi H., An F., Zhao Q., Guo H. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 106: 21431–21436 [DOI] [PMC free article] [PubMed] [Google Scholar]