This work describes the mechanism of statolith sedimentation during Arabidopsis gravity sensing. SGR9, a RING-type E3 ligase, is localized to statoliths, which interact with F-actin. SGR9 modulates the interaction between statoliths and F-actin and promotes the detachment of statoliths from F-actin, allowing the statoliths to sediment in the direction of gravity.
Abstract
Higher plants use the sedimentation of amyloplasts in statocytes as statolith to sense the direction of gravity during gravitropism. In Arabidopsis thaliana inflorescence stem statocyte, amyloplasts are in complex movement; some show jumping-like saltatory movement and some tend to sediment toward the gravity direction. Here, we report that a RING-type E3 ligase SHOOT GRAVITROPISM9 (SGR9) localized to amyloplasts modulates amyloplast dynamics. In the sgr9 mutant, which exhibits reduced gravitropism, amyloplasts did not sediment but exhibited increased saltatory movement. Amyloplasts sometimes formed a cluster that is abnormally entangled with actin filaments (AFs) in sgr9. By contrast, in the fiz1 mutant, an ACT8 semidominant mutant that induces fragmentation of AFs, amyloplasts, lost saltatory movement and sedimented with nearly statically. Both treatment with Latrunculin B, an inhibitor of AF polymerization, and the fiz1 mutation rescued the gravitropic defect of sgr9. In addition, fiz1 decreased saltatory movement and induced amyloplast sedimentation even in sgr9. Our results suggest that amyloplasts are in equilibrium between sedimentation and saltatory movement in wild-type endodermal cells. Furthermore, this equilibrium is the result of the interaction between amyloplasts and AFs modulated by the SGR9. SGR9 may promote detachment of amyloplasts from AFs, allowing the amyloplasts to sediment in the AFs-dependent equilibrium of amyloplast dynamics.
INTRODUCTION
Tropism in plants refers to the directional growth of organs in response to directional environmental stimuli, such as gravity, light, moisture, or touch, and is a consequence of the plants recognizing the directional information of the stimuli. In general, higher plant shoots show negative gravitropism (upward bending) and roots show positive gravitropism (downward bending), respectively. Since the direction and magnitude of gravity are almost constant across the surface of the earth, gravitropism can be regarded as a posture adjustment triggered by sensing the tilt of organs relative to the direction of gravity (Tasaka et al., 1999; Boonsirichai et al., 2002). Since gravity acts upon mass, a number of organisms use relatively heavy cellular components, called statoliths or otoliths, to sense the direction of gravity. Higher plants have unique plastids, termed amyloplasts, that can function as plant statoliths for gravity sensing. Amyloplasts are present within specific cells (statocytes); these plastids accumulate dense starch granules and sink in the direction of gravity (Morita and Tasaka, 2004). Based on this observed property of these plastids, the starch statolith hypothesis proposed that sedimentation of amyloplasts toward the gravity vector within specific cells (statocytes) is the trigger for the sensing of gravity, and this idea is now widely accepted (Sack, 1997; Morita and Tasaka, 2004).
Molecular and genetic studies using Arabidopsis thaliana provided evidence that the endodermal cells and the root cap columella cells, which contain sedimentable amyloplasts, act as statocytes in shoots and roots, respectively (Blancaflor et al., 1998; Fukaki et al., 1998). Studies on the phosphoglucomutase (pgm) mutant support the starch statolith hypothesis. The pgm mutant shows impaired starch synthesis in its plastids and exhibits a reduced gravitropic response both in shoots and roots (Caspar and Pickard, 1989; Kiss et al., 1989, 1997; Weise and Kiss, 1999). Amyloplasts do not sediment in the direction of gravity in the mutant statocytes (Caspar and Pickard, 1989; Kiss et al., 1989, 1997; Weise and Kiss, 1999). The extent of reduction in gravitropism is positively correlated with the reduction in starch content, suggesting that the mass of starch affects the magnitude of the gravitropic response (Caspar and Pickard, 1989; Kiss et al., 1989, 1997; Weise and Kiss, 1999).
Although the word statolith literally means stationary stone, the behavior of amyloplasts differs considerably from that of an ideal statolith, particularly in the shoot statocytes (Clifford and Barclay, 1980; Heathcote, 1981; Sack and Leopold, 1985; Saito et al., 2005). While most amyloplasts sediment to the bottom of the cells with Brownian movement, other amyloplasts exhibit dynamic saltatory movement, that is, directed (not necessarily toward gravity), non-Brownian movements constituting cytoplasmic streaming (Sack and Leopold, 1985). Both sedimentation and saltatory movements of amyloplasts have been observed before and after gravistimulation during live imaging of Arabidopsis stem endodermal cells using a vertical microscope (Saito et al., 2005). These observations demonstrate that endodermal amyloplasts are dynamic statoliths whose movements are probably influenced by intracellular components. Arabidopsis mutants with little or reduced shoot gravitropism (sgr mutants) indicate that the vacuole has a large effect on amyloplast dynamics for gravity perception in endodermal cells (Kato et al., 2002; Morita et al., 2002; Yano et al., 2003; Silady et al., 2004). A cellular defect common to sgr2, sgr3, zigzag (zig)/sgr4, and gravitropism defective2 (grv2)/sgr8/katamari2 (kam2) mutants is abnormal amyloplast localization within endodermal cells. Molecular cloning of SGR2, SGR3, ZIG/SGR4, and GRV2/SGR8/KAM2 indicated that vacuolar functions and/or vacuolar biogenesis supported by membrane trafficking in the endodermal cell are implicated in amyloplast behavior (Kato et al., 2002; Morita et al., 2002; Yano et al., 2003; Silady et al., 2004). In wild-type endodermal cells, the membrane of the large central vacuole exhibits dynamic and flexible properties, such as invagination and presence of transvacuolar strands, and amyloplasts are enclosed by the vacuolar membrane (Saito et al., 2005). By contrast, in the sgr mutants in which amyloplasts do not sediment in the direction of gravity, the vacuolar membranes lose their dynamic and flexible properties (Saito et al., 2005).
In addition to the vacuole, actin filaments (AFs) have been suggested to be involved in amyloplast movement on the basis of experiments using inhibitors of actin polymerization (Yamamoto and Kiss, 2002; Hou et al., 2003; Palmieri and Kiss, 2005; Saito et al., 2005), although some of the results are contradictory. Some studies suggest that the treatments have an inhibitory effect on amyloplast sedimentation (Palmieri and Kiss, 2005), whereas other found evidence of a promoting effect (Yamamoto and Kiss, 2002; Saito et al., 2005). AFs have been suggested to be involved not only in amyloplast dynamics but also in gravity signal transduction. Thus, it was assumed that AFs transmit the force generated by the movement of amyloplasts to a mechanoreceptor within the statocyte (Yoder et al., 2001; Perbal and Driss-Ecole, 2003). However, recent studies using inhibitors indicate that AFs are not required for gravity sensing and signal transduction (Yamamoto and Kiss, 2002; Saito et al., 2005). Consequently, the role of AFs in statocytes remains unclear.
Here, we show that Arabidopsis endodermal amyloplasts are in a dynamic equilibrium between sedimentation and saltatory movement and that this equilibrium is principally the result of interaction between the amyloplasts and AFs. Our results indicate a role for AFs in statocytes with regard to statolith movement, that is, AFs promote saltatory movement resulting in interference with amyloplast sedimentation. Moreover, we provide molecular genetic evidence that a RING-type E3 ligase, SGR9, which is localized to amyloplasts, not only modulates the interaction between the amyloplasts and AFs but also may promote detachment of the amyloplasts from AFs. As a result, SGR9 is an important component of the gravity perception mechanism that facilitates amyloplast sedimentation in the direction of gravity.
RESULTS
SGR9, a C3H2C3-type RING Finger Protein, Has E3 Ligase Activity
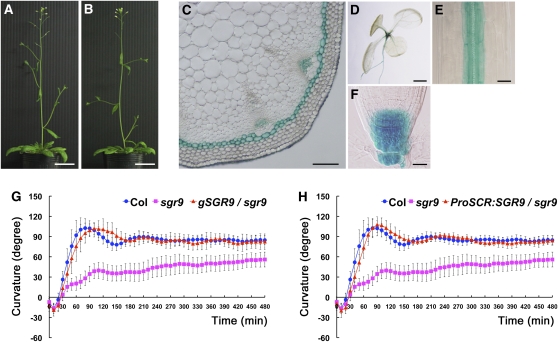
The recessive mutation sgr9 causes a weak gravitropic response in inflorescence stems; mutant plants have no obvious morphological phenotype except for lateral shoots growing horizontally (Figures 1A and 1B). When wild-type inflorescence stems were placed horizontally, they curved upward with a 90° curvature within 90 min (Figure 1G). By contrast, sgr9 stems exhibited only an ~50° curvature even after 8 h gravistimulation (Figure 1G). In addition, the sgr9 mutant displayed a slightly reduced gravitropic response in the hypocotyl and root (see Supplemental Figures 1A and 1B online). Because the gravitropic response requires organ growth, we examined the inflorescence stem of sgr9 for evidence of a growth defect. The growth rate of sgr9 during 5 d after bolting was comparable to that of the wild type, indicating that the gravitropic phenotype of sgr9 was not due to a defect in organ growth (see Supplemental Figure 1C online). It has been proposed that the gravitropic response shares a mechanism of differential organ growth with the phototropic response, which is regulated by the plant hormone auxin. We therefore compared light-oriented growth (phototropic responses) in the mutant and wild type. The sgr9 inflorescence stem showed a greater light-oriented growth response than the wild type (see Supplemental Figures 1D and 1E online), similar to reports for other gravitropic mutants (Fukaki et al., 1996b; Yamauchi et al., 1997). These results demonstrate that sgr9 does not affect the phototropic response in inflorescence stems.
Figure 1.
Characterization of the sgr9 Mutant and SGR9.
(A) and (B) Morphological phenotypes of 5-d-old plants after bolting. The wild type (A); sgr9 (B). Bars = 3 cm.
(C) to (F) GUS expression patterns under the control of the SGR9 promoter.
(C) Cross-section of the inflorescence stem at 2 to 3 cm below the apex from a 5-week-old plant. Bar = 100 μm.
(D) Seven-day-old seedling. Bar = 1 mm.
(E) Hypocotyl of a 3-d-old etiolated seedling. Bar = 50 μm.
(F) Root cap of a 7-d-old seedling. Bar = 25 μm.
(G) and (H) Time course of gravitropic responses of inflorescence stems.
(G) Gravitropic response of sgr9 and complementation analysis.
(H) Gravitropic response of ProSCR:SGR9/sgr9. SGR9 was expressed under the control of the SCR promoter in sgr9. Inflorescence stems were gravistimulated by being placed horizontally at 23°C under low nondirectional light. Error bars represent sd. A minimum of eight individuals was analyzed per sample point.
The SGR9 gene was mapped to chromosome 5 and cloned using its map position. A sequencing analysis identified a single G-to-A substitution that causes a nonsense mutation in At5g02750 (see Supplemental Figure 2A online). We performed a complementation analysis by inserting an ~2.5-kb wild-type genomic fragment containing the At5g02750 open reading frame and putative promoter region into sgr9 plants (gSGR9/sgr9). The inserted sequence rescued the gravitropic defect in transgenic sgr9 inflorescence stems, indicating that SGR9 is At5g02750 (Figure 1G). To obtain a null allele of sgr9, we analyzed the T-DNA insertion line SALK_070204. However, we could not detect the insertion for At5g02750.
The At5g02750 sequence encodes a C3H2C3-type RING finger protein. The SGR9 protein has a unique N-terminal half and a C3H2C3-type RING finger domain at the C-terminal region (see Supplemental Figure 2A online). A number of RING finger proteins are known to function as ubiquitin E3 ligases, and the RING finger domains are essential for this E3 activity (Stone et al., 2005). A computational classification analysis of the RING finger domains encoded by Arabidopsis genome indicated that SGR9 belonged to the same subclass as COP1-INTERACTING PROTEIN8 (CIP8) and ABI3-INTERACTING PROTEIN2 (AIP2) (Kosarev et al., 2002), which have been demonstrated to have ubiquitin E3 ligase activity (Hardtke et al., 2002; Zhang et al., 2005).
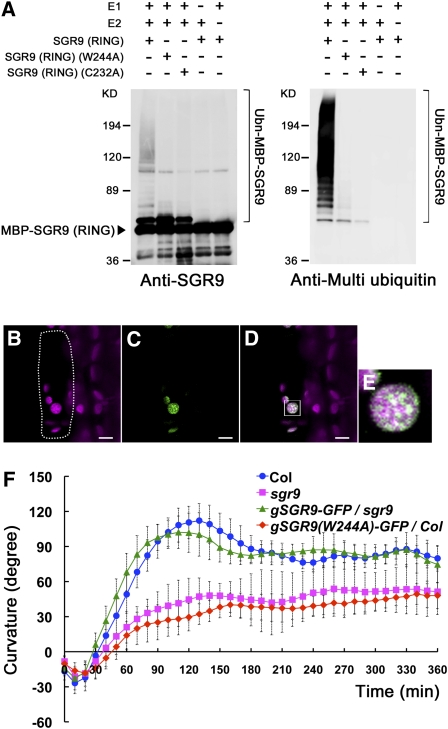
To determine whether SGR9 possesses E3 ligase activity, we constructed a recombinant protein, MBP-SGR9 (RING), in which maltose binding protein (MBP) was fused to the RING finger domain of SGR9 (Matsuda et al., 2001). This recombinant protein was then used in an in vitro ubiquitination assay. MBP-SGR9 (RING) exhibited E3 ligase activity in the presence of ubiquitin, ATP, E1, and E2 (Figure 2A). In the absence of E1 or E2, the recombinant protein did not exhibit any E3 ligase activity (Figure 2A). It has been reported that the activity of RING-type E3 ligases depends on the RING finger domain (Katoh et al., 2005). We therefore substituted Trp-244 with Ala (W244A) or Cys-232 with Ala (C232A) in the conserved amino acid sequence of the RING finger domain, with the expectation that each substitution would diminish E3 activity (see Supplemental Figure 2B online). In comparison to wild-type MBP-SGR9 (RING), MBP-SGR9 (RING) harboring W244A or C232A substitutions exhibited significantly reduced E3 ligase activity (Figure 2A). These results indicate that the RING finger domain of SGR9 has ubiquitin E3 ligase activity in vitro.
Figure 2.
Characterization of the SGR9 Protein.
(A) E3 ligase activity of the wild type and mutated SGR9 (RING). For the ubiquitination assay, MBP-SGR9 (RING), which is MBP fused to the RING finger domain of SGR9, was prepared, and amino acid substitutions W244A or C232A were introduced into the RING finger domain. An anti-SGR9 antibody was used to detect MBP-SGR9 (RING) or the mutated MBP-SGR9 (RING) (left panel), and an anti-multi ubiquitin antibody was used to detect ubiquitinated MBP-SGR9 (RING) or mutated MBP-SGR9 (RING) (right panel).
(B) to (E) Intracellular localization of mutated SGR9 protein in an endodermal cell of an inflorescence stem. Bars = 5 μm.
(B) Autofluorescence of a plastid chlorophyll. Dotted line indicates a single endodermal cell.
(C) SGR9(W244A)-GFP.
(D) Overlay of autofluorescence and GFP signal.
(E) Inset in (D). Note that mutated SGR9 protein is localized to the amyloplasts.
(F) Time course of gravitropic response of an inflorescence stem of gSGR9-GFP/sgr9 or gSGR9(W244A)-GFP/Col. Error bars represent sd. A minimum of six individuals was analyzed per sample point.
The SGR9 Gene Functions within Gravity-Sensing Tissue in Shoot Gravitropism
To analyze the expression pattern of SGR9, we expressed β-glucuronidase (GUS) under the control of the putative SGR9 promoter region used in the complementation analysis described above. Analysis of GUS activity showed that SGR9 was expressed in the seedling, hypocotyl, root, and stem (Figures 1C to 1F). Detailed analysis showed that SGR9 expression occurred in the hypocotyl endodermis (Figure 1E) and root cap columella (Figure 1F), tissues that act as statocytes. This indicates a good correlation with the gravitropic defect in the hypocotyl and root. Scrutiny of transverse sections of the inflorescence stem showed that strong GUS activity was present in the endodermis, the gravity-sensing tissue of the inflorescence stem, while weaker staining was present in vascular tissue (Figure 1C).
To confirm that SGR9 modulates shoot gravitropism in the endodermis, we transformed sgr9 plants with SGR9 driven by an endodermis-specific SCR promoter (ProSCR:SGR9/sgr9). The sgr9 transgenic plants with expression of SGR9 limited to the endodermis showed rescue of the gravitropic phenotype (Figure 1H). This result indicates that expression of SGR9 within the endodermis is sufficient for shoot gravitropism.
SGR9 Is Localized at Amyloplasts within Gravity-Sensing Cells
To investigate the subcellular localization of SGR9, we expressed an SGR9 protein fused with green fluorescent protein (GFP) at its C terminus (SGR9-GFP) under control of the SGR9 promoter region used for the complementation analysis (gSGR9-GFP/sgr9). The SGR9-GFP was able to rescue the gravitropic defect in sgr9 (Figure 2F), indicating that SGR9-GFP is a functional protein. However, the GFP fluorescence could not be detected (data not shown).
It has been reported that some RING-type ubiquitin E3 ligases are unstable due to their autoubiquitination activity (Jackson et al., 2000; Vaux and Silke, 2005). If SGR9 is also prone to autoubiquitination, then amino acid substitutions that diminish E3 activity should stabilize the protein. When SGR9(W244A)-GFP or SGR9(C232A)-GFP was expressed under control of the SGR9 promoter region in wild-type plants [gSGR9(W244A)-GFP)/Col or gSGR9(C232A)-GFP/Col], GFP fluorescence was detected. Interestingly, the fluorescent signals were localized to amyloplasts in the endodermal cell (Figures 2B to 2E; see Supplemental Figures 3A to 3D online). The localization of the mutated proteins, however, might not necessarily represent the normal localization of the wild-type SGR9 protein. The transgenic plants exhibited reduced gravitropic responses similar to those of sgr9 (Figure 2F; see Supplemental Figure 3E online), indicating that SGR9(W244A)-GFP or SGR9(C232A)-GFP caused a dominant-negative effect on the endogenous SGR9 protein. This result suggests that SGR9(W244A)-GFP and SGR9(C232A)-GFP acted at the site where endogenous SGR9 functions. Thus, our analyses suggest that SGR9 is likely to function in amyloplasts within the endodermis for shoot gravitropism.
SGR9 Is Involved in Amyloplast Sedimentation in Gravity-Sensing Cells
We previously reported that whereas most amyloplasts in endodermal cells sediment in the direction of gravity, a few show more dynamic movement. In addition, after gravistimulation by reorientation, amyloplasts relocate toward the new gravity vector within 3 min, which represents the minimum time to trigger gravity sensing (Fukaki et al., 1996a). The localization of SGR9(W244A)-GFP or SGR9(C232A)-GFP to amyloplasts prompted us to examine whether SGR9 is involved in amyloplast dynamics in the endodermal cell. Histological analysis of fixed stem tissue showed that almost all amyloplasts sediment in the direction of gravity in the wild type (see Supplemental Figure 1F online). By contrast, although the majority of amyloplasts sedimented as expected, some were located in the central region of the cell in sgr9 (see Supplemental Figure 1G online), suggesting that amyloplast sedimentation is abnormal in sgr9.
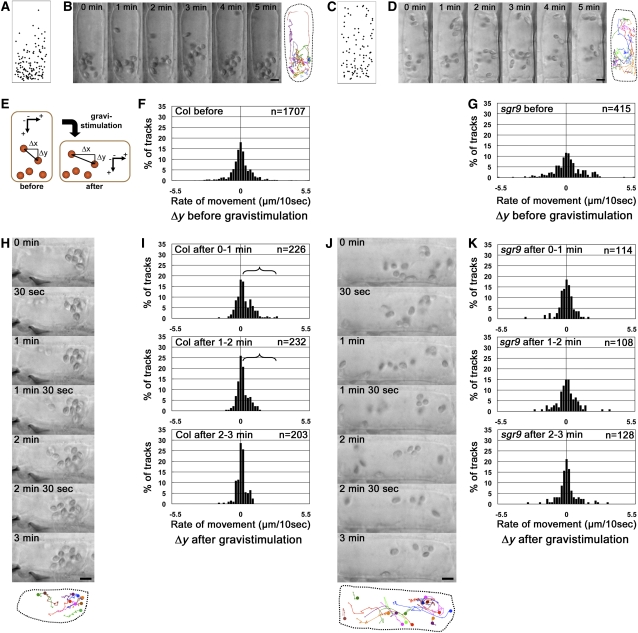
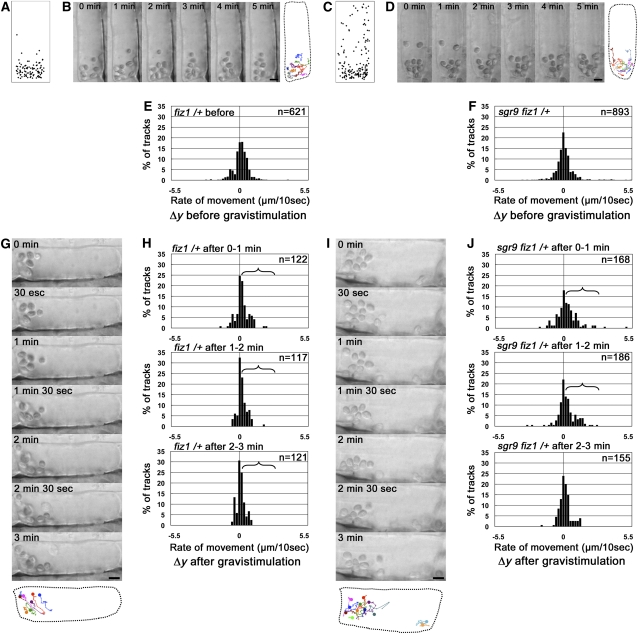
Then, we analyzed amyloplast movement in living endodermal cells held in a vertical orientation and subsequently subjected to gravistimulation by reorientation (Saito et al., 2005). First, we plotted amyloplast positions within endodermal cells to determine their normal pattern of distribution. In the wild type, amyloplasts showed an uneven distribution, with the majority located at the bottom of the cell (Figure 3A). By contrast, amyloplasts were almost evenly dispersed throughout sgr9 cells (Figure 3C).
Figure 3.
Amyloplast Dynamics before and after Gravistimulation of Endodermal Cells.
(A) and (C) Amyloplast distribution in endodermal cells. The wild type (A); sgr9 (C). The endodermal cell is shown as a simple rectangle, and the positions of amyloplasts from images at 0 min and after 5 min were plotted. Results from at least three individual cells were superimposed.
(B) and (D) Time-lapse images of amyloplasts at 1-min intervals before gravistimulation by reorientation. The wild type (B); sgr9 (D). The image on the right is a trace of amyloplast positions. Each color traces an individual amyloplast at 10-s intervals (marked by “X” on the lines). Dotted line indicates the outline of the endodermal cell analyzed. Bars = 5 μm.
(E) Definition of the axes for the quantitative analysis of amyloplast dynamics. The movement of an amyloplast at 10-s intervals was split into two components: a horizontal vector (Δx, μm/10 s) and a vertical vector (Δy, μm/10 s). A positive value for Δy indicates sedimentation toward the direction of gravity.
(F) and (G) Histogram showing the Δy component before gravistimulation. The wild type (F); sgr9 (G). Histograms show the rate of movement. The total number of tracks analyzed is shown by n. At least three individual cells were examined.
(H) and (J) Time-lapse images of amyloplasts at 1-min intervals after gravistimulation by reorientation. The wild type (H); sgr9 (J). At the bottom is a trace of individual amyloplasts. Each color traces an individual amyloplast at 10-s intervals. The circle indicates the start point. Dotted line indicates the outline of the endodermal cell analyzed. Bars = 5 μm.
(I) and (K) Histogram showing the Δy component after gravistimulation. The wild type (I); sgr9 (K). Histograms show the rate of movement. The brackets indicate statistically increased gravity oriented sedimentation of the amyloplasts (P value; Welch’s t test, one-tailed, P < 0.05, average of rate of movement after gravistimulation compared with that before stimulation). The total number of tracks analyzed is shown by n. At least three individual cells were examined.
Next, we examined the amyloplast movement using time-lapse imaging. In wild-type endodermal cells, some amyloplasts showed saltatory movement but most sedimented in the direction of gravity (Figure 3B; see Supplemental Movie 1 online). By contrast, in sgr9 endodermal cells, amyloplasts did not sediment but moved around randomly (Figure 3D; see Supplemental Movie 2 online). To quantify the amyloplast movement, each movement during a 10-s interval was divided into two components, a horizontal vector (Δx, μm/10 s) and a vertical vector (Δy, μm/10 s) (Figure 3E). The data are shown as histograms. Each histogram of Δx and Δy in the wild type was almost symmetric around the zero axis (Figure 3F; see Supplemental Figure 4A online). The population variances of Δx and Δy in sgr9 differed significantly from the wild type (F test, P < 0.05; Figures 3F and 3G; see Supplemental Figures 4A and 4B online). Although each histogram of Δx and Δy in sgr9 was also likely to be symmetric around the zero axis, the frequencies of Δx and Δy components with large absolute values were significantly higher than in the wild type (Figure 3G; see Supplemental Figure 4B online). Conversely, the frequencies of Δx and Δy with small absolute values, which indicate the incidence of nearly static amyloplasts, were lower in sgr9 than in the wild type. These results indicate that amyloplasts in sgr9 move more dynamically than those in the wild type before gravistimulation.
Amyloplasts were individually and continuously traced after gravistimulation as far as possible. In the wild type, the amyloplasts altered their positions toward the new bottom of the cell (Figure 3H; see Supplemental Movie 3 online). The amyloplast movement after gravistimulation was assessed quantitatively as described above, and the mean values of Δy before and after gravistimulation were compared (Saito et al., 2005). Positive values of Δy indicate movement toward the new gravity vector. To examine the time dependency of movement, the data were divided into three time intervals: 0 to 1 min, 1 to 2 min, and 2 to 3 min after initiation of gravistimulation by reorientation of the cell. In the wild type, the histograms show clearly asymmetric distributions of the Δy components at the 0 to 1 min and 1 to 2 min intervals toward the new gravity direction (Figure 3I). The mean value of Δy after gravistimulation increased significantly during the 0 to 1 min and 1 to 2 min intervals compared with prior to reorientation (Welch’s t test, one-tailed, P < 0.05) (Figure 3I, bracket), indicating that the amyloplasts changed their distribution toward the new gravity vector after gravistimulation.
By contrast, in sgr9, amyloplasts moved dynamically around the cell before and after gravistimulation, and the movement appeared to be random (Figure 3J; see Supplemental Movie 4 online). As expected from this, the histograms from sgr9 did not show an asymmetric distribution of the Δy components at any time interval (Figure 3K). In addition, the Δy component with the largest absolute value at the 0 to 1 min interval was slightly lower than prior to gravistimulation. Statistical analysis indicated that the mean value of Δy after gravistimulation did not significantly increase at any time interval compared with before gravistimulation (Figure 3K). These results suggest that the sedimentation of amyloplasts upon new gravistimulation is also defective in sgr9. Thus, the gravitropic defect of sgr9 is correlated with loss of significant amyloplast sedimentation.
Previously, we reported that amyloplast sedimentation in the direction of gravity was impaired in the mutants sgr2, sgr3, and zig/sgr4 and that this effect was probably due to a defect in vacuolar membrane dynamics in the endodermal cells of the inflorescence stem (Kato et al., 2002; Yano et al., 2003; Silady et al., 2004; Saito et al., 2005). To determine whether abnormal amyloplast dynamics in sgr9 might also be due to defects in the vacuolar membranes, we analyzed the dynamics of vacuolar membranes using a vertical microscope system. Vacuolar membranes in endodermal cells were visualized using GFP-Vam3 (Uemura et al., 2002) under the control of the SCR promoter. In wild-type endodermal cells, the vacuolar membranes underwent dynamic movements and transvacuolar strands were formed (see Supplemental Figure 5A online). By contrast, in sgr2, which is probably impaired in the biogenesis of vacuolar membrane, transvacuolar strand-like structures were not observed and abnormal vesicle-like structures accumulated (see Supplemental Figure 5C online). In sgr9, vacuolar membranes moved dynamically in a similar fashion as in the wild type (see Supplemental Figure 5B online), indicating that SGR9 is involved in the movement of amyloplasts in a manner that is independent of vacuolar function.
The sgr9 Phenotype Is Suppressed by the Disruption of AFs
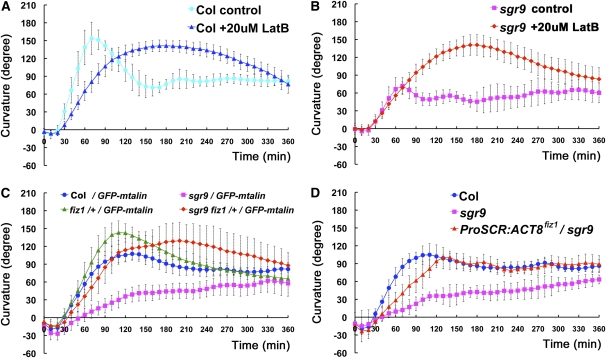
Our analyses showed that sgr9 had defective amyloplast sedimentation toward the direction of gravity and also had increased dynamic movements of its amyloplasts. We previously reported that the dynamic movement of the amyloplasts depends on AFs (Saito et al., 2005). Therefore, we sought to compare the relationship between AFs and amyloplast movement in the wild type and sgr9. First, we treated the cells with Latrunculin B (LatB), an inhibitor of AF polymerization. The LatB-treated wild-type inflorescence stem segments exhibited an obvious gravitropic response, although the kinetics of this response differed slightly from that of the untreated control (Figure 4A). Interestingly, the gravitropic response of LatB-treated sgr9 was comparable to that of the treated wild type (Figure 4B), suggesting that the gravitropic defect in sgr9 could be overcome by disruption of the AFs.
Figure 4.
The Effect of AFs Disruption on Gravitropism.
(A) and (B) Time course of gravitropic responses of inflorescence stem segments treated with 20 μM LatB. The wild type (A); sgr9 (B). The stem segments were treated with 20 μM for 12 to ~14 h, and then stems were gravistimulated by being placed horizontally at 23°C under low nondirectional light. Error bars represent sd. At least 10 individuals were examined per sample point.
(C) and (D) Time course of gravitropic responses of inflorescence stems.
(C) Gravitropic response of fiz1/+ and sgr9 fiz1/+. All plants using this analysis harbored the Pro35S:GFP-mTalin construct.
(D) Gravitropic response of ProSCR:ACT8 fiz1/sgr9. ACT8 fiz1 was expressed under the control of the SCR promoter in sgr9. Error bars represent sd. At least five individuals were examined per sample point.
To investigate this finding further, we made use of a semidominant mutant frizzy shoot1 (fiz1), which harbors an amino acid substitution in ACT8 that affects AFs formation. The fiz1 mutation has been shown to partially inhibit the formation of AFs in vegetative tissues (Kato et al., 2010). As homozygosity for fiz1 has a severely detrimental effect on organ growth, we used the mutation in the heterozygous state (fiz1/+) in our analysis. The inflorescence stems of fiz1/+ showed almost normal gravitropism, although their maximum curvature was greater than that of the wild type (Figure 4C). sgr9 fiz1/+ exhibited almost normal gravitropism (Figure 4C), indicating that the fiz1 mutation suppressed the sgr9 gravitropic phenotype in a similar manner to LatB treatment.
Next, we analyzed amyloplast movement in living endodermal cells. Almost all amyloplasts accumulated at the bottom of the cells in fiz1/+, and this biased distribution seemed to be even more striking than in the wild type (Figures 3A and 5A), indicating that amyloplast sedimentation toward the direction of gravity was facilitated by the fiz1 mutation. By contrast, the AF-dependent dynamic movements of amyloplasts were reduced in fiz1/+ compared with the wild type (Figures 3B, 3F, 5B, and 5E; see Supplemental Figures 4A and 4C and Supplemental Movie 5 online). Consistently, the population variances of Δy in fiz1/+ differed significantly from the wild type (F test, P < 0.05; Figures 3F and 5E). Interestingly, amyloplast distribution showed a greater shift toward the direction of gravity in sgr9 fiz1/+ compared with sgr9 (Figure 5C), indicating that amyloplast sedimentation was apparently restored by the fiz1 mutation. In contrast with the facilitation of amyloplast sedimentation, the dynamic movement of amyloplasts was significantly reduced in sgr9 fiz1/+ compared with sgr9 (Figures 5D and 5F; see Supplemental Figure 4D and Supplemental Movie 6 online).
Figure 5.
The Effect of AF Disruption on Amyloplast Dynamics.
(A) and (C) Amyloplast distribution in endodermal cells. fiz1/+ (A); sgr9 fiz1/+ (C). The endodermal cell is represented as a simple rectangle, and the positions of the amyloplasts from images at 0 min and after 5 min were plotted. Results from at least three individual cells were superimposed.
(B) and (D) Time-lapse images of amyloplasts at 1-min intervals before gravistimulation by reorientation. fiz1/+ (B); sgr9 fiz1/+ (D). The image on the right is a trace of amyloplast movements. Each color traces an individual amyloplast at 10-s intervals (marked by “X” on the lines). Dotted line indicates the outline of the endodermal cell analyzed. Bars = 5 μm.
(E) and (F) Histogram showing the Δy component before gravistimulation. fiz1/+ (E); sgr9 fiz1/+ (F). Histograms show the rate of movement as described in Figure 3. The total number of tracks analyzed is shown as n. At least three individual cells were examined.
(G) and (I) Time-lapse images of amyloplasts at 1-min intervals after gravistimulation by reorientation. fiz1/+ (G); sgr9 fiz1/+ (I). At the bottom is a trace of amyloplast positions. Each color traces an individual amyloplast at 10-s intervals. The circle indicates the start point. Dotted line indicates the outline of the endodermal cell analyzed. Bars = 5 μm.
(H) and (J) Histogram showing the Δy component after gravistimulation. fiz1/+ (H); sgr9 fiz1/+ (J). Histograms show the rate of movement. The brackets represent statistically increased gravity oriented sedimentation of the amyloplasts (P value; Welch’s t test, one-tailed, P < 0.05, average of rate of movement after gravistimulation compared with that before stimulation). The total number of tracks analyzed is shown as n. At least three individual cells were examined.
After reorientation of the cells to induce gravistimulation, the distribution patterns of the amyloplasts in fiz1/+ changed as they moved toward the new bottom of the cell, similar to the wild type (Figure 5G; see Supplemental Movie 7 online). The fiz1/+ cells showed an asymmetric distribution of the Δy components at all time intervals (Figure 5H). In addition, the mean value of Δy after gravistimulation increased in a statistically significant manner at all time intervals compared with prior to gravistimulation (Welch’s t test, one-tailed, P < 0.05) (Figure 5H, bracket). The amyloplasts also moved toward the new bottom of the cell in sgr9 fiz1/+ (Figure 5I; see Supplemental Movie 8 online). By contrast to sgr9, sgr9 fiz1/+ showed asymmetric distribution of Δy components, similar to fiz1/+, at the 0 to 1 min and 1 to 2 min intervals (Figure 5J). In addition, the mean value of Δy after gravistimulation showed a statistically significant increase at the 0 to 1 min and 1 to 2 min compared with before stimulation (Welch’s t test, one-tailed, P < 0.05) (Figure 5J, bracket). These results indicate that amyloplast sedimentation toward the new gravity vector after gravistimulation in sgr9 is restored by the fiz1 mutation.
The effects of LatB and the fiz1 mutation on AFs were displayed by tissues throughout the stem, including the epidermis, cortex, endodermis, and stele. Therefore, to eliminate the possible influence of fiz1 in tissues other than the endodermis, we used a limited expression of ACT8 fiz1. When ACT8 fiz1 is expressed in sgr9 under the control of the SCR promoter (ProSCR:ACT8 fiz1/sgr9), the transgenic plants show a partially restored gravitropic response (Figure 4D), although the extent of restoration is weaker than in sgr9 fiz1/+. This result suggests that the limited expression of ACT8 fiz1 is at least sufficient for partial restoration of the gravitropic phenotype in sgr9.
Jumping-Like Dynamic Movement of Amyloplasts Is Increased in sgr9 and decreased in fiz1/+
Our live-cell imaging and analyses of amyloplast movement imply that the increased dynamic amyloplast movement found in sgr9 is due to the AFs because such movement is greatly reduced by the fiz1 mutation (Figures 5D and 5F).
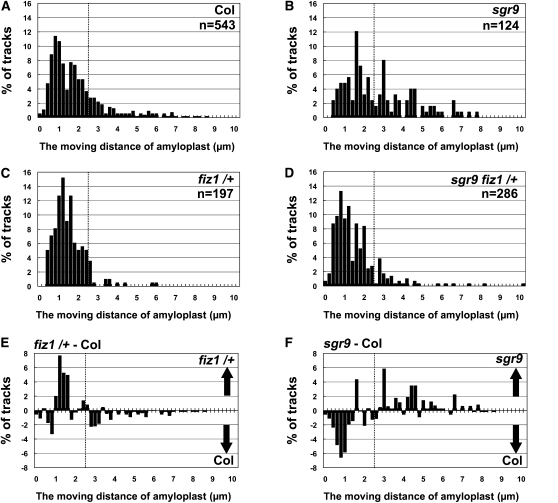
To describe the dynamic movement of amyloplasts, we analyzed amyloplast movement without regard to the direction of movement. The distance traveled by the amyloplast in 30-s windows was measured before gravistimulation. The data are shown as histograms (Figure 6). In wild-type cells, we frequently observed amyloplasts that traveled only short distances, that is, had a lower rate of movement. The distance such amyloplasts moved was less than their radius (≤ ~2.5 μm/30 s) (Figure 6A). We also observed a lower frequency of amyloplasts that moved relatively further, that is, had a higher rate of movement (>2.5 μm/30 s) (Figure 6A). As such amyloplasts tended to move out of the focal plane during the 30-s observation window, their frequency might have been underestimated. The population variances of the distances moved in fiz1/+ were significantly different to that of the wild type (F test, P < 0.05; Figures 6A and 6C). In addition, the difference of the frequency between the wild type and fiz1/+ clearly shows the increase of amyloplasts with lower a rate of movement and the decrease of those with a higher rate of movement in the fiz1/+ mutant as shown in Supplemental Movie 5 online (Figures 6C and 6E).
Figure 6.
The Moving Ddistance of Amyloplasts before Gravistimulation.
(A) to (D) Histograms showing the moving distance of amyloplasts for 30 s in the vertical vector (Δy component) before gravistimulation. The wild type (A), sgr9 (B), fiz1/+ (C), and sgr9 fiz1/+ (D). In contrast with Figures 3 and 5, percentages of tracks are shown without distinction of the direction of movement. n represents the total number of tracks subjected to this analysis. At least three individual cells were examined.
(E) Histogram showing the difference between the wild type and fiz1/+. The percentage of tracks of each compartment in the wild type was subtracted from that in fiz1/+. The positive and negative values mean that the tracks with indicated moving distance were counted more frequently in fiz1/+ and the wild type, respectively.
(F) Histogram showing the difference between the wild type and sgr9. The percentage of tracks of each compartment in the wild type was subtracted from that in sgr9. The positive and negative values mean that the tracks with indicated moving distance were counted more frequently in sgr9 and the wild type, respectively. Dotted lines indicate 2.5 μm/30 s.
In sgr9 cells, there was an increased frequency of amyloplasts with the higher rate of movement and a decreased frequency of those with the lower movement rate compared with the wild type (Figures 6B and 6F). The population variance for distance moved in sgr9 was significantly different from of the wild type (F test, P < 0.05; Figures 6A and 6B). Interestingly, the difference in frequency between the wild type and sgr9 appeared to be a mirror image of the difference between the wild type and fiz1/+ (Figures 6E and 6F). These results suggest that amyloplast movement shifted from a lower to a higher rate in sgr9, consistent with the predominance of dynamic amyloplast movement in sgr9 as shown in Figures 3D and 3G. In sgr9 fiz1/+ cells, the frequency of amyloplasts with the higher rate of movement apparently decreased, while those with the lower rate apparently increased compared with sgr9 (Figure 6D). This suggests that the shift in amyloplast movement to a higher rate in sgr9 was due to AFs.
Interaction between Amyloplasts and AFs Is Abnormal in sgr9 Endodermal Cells
Based on the above results, we posited that there might be defects in the interaction between AFs and amyloplasts in sgr9. We therefore used time-lapse imaging to observe the dynamics of the interaction between amyloplasts and AFs within endodermal cells positioned vertically.
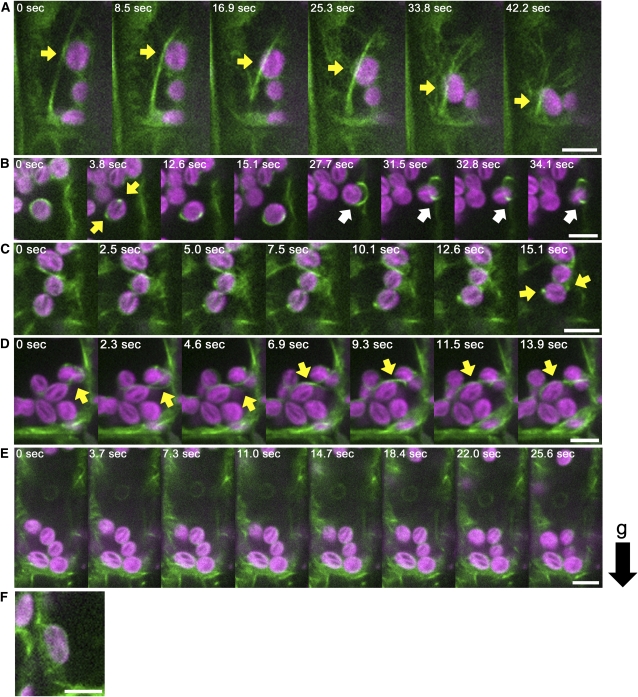
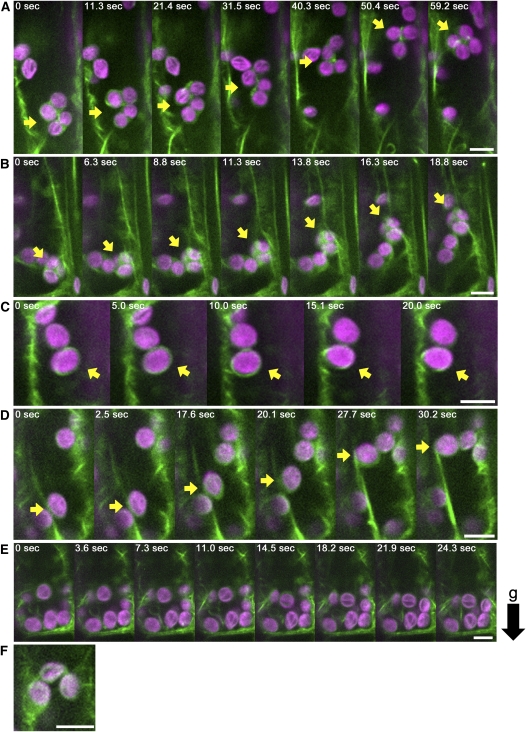
In wild-type endodermal cells (see Supplemental Movie 9 online), four types of AFs that interact with amyloplasts were observed: actin meshwork that enwrapped amyloplasts (Figure 7F; see Supplemental Figure 6F online), linear actin cables (Figure 7A; see Supplemental Figure 6A online), curving actin bundles attached to amyloplasts (Figure 7D; see Supplemental Figure 6D online), and ring-like actin bundles surrounding amyloplasts (Figure 7B; see Supplemental Figure 6B online). Amyloplasts enwrapped by an AF meshwork generally did not change their location (Figure 7F; see Supplemental Figure 6F online). Amyloplasts attached to linear actin cables occasionally moved a long distance along the cable (Figure 7A; see Supplemental Figure 6A and Supplemental Movie 10 online). This type of movement may be identical to the actin-dependent dynamic movements of amyloplasts described above. Amyloplasts attached to curving actin bundles only moved short distances, and this type of actin bundle slid smoothly across the surfaces of the amyloplasts (Figure 7D; see Supplemental Figure 6D and Supplemental Movie 11 online). The ring-like actin bundle produced a distinctive double peak of GFP signals on the amyloplast (Figure 7B; see Supplemental Figure 6B online). Smertenko et al. (2010) described ring-like structures, analogous to these ring-like actin bundles, in Arabidopsis root hairs (Smertenko et al., 2010). Amyloplasts associated with ring-like actin bundles moved only short distances and also rotated (Figure 7B; see Supplemental Figure 6B and Supplemental Movie 12 online). In the time-lapse imaging analysis, ring-like actin bundles were observed to appear and disappear. For example, a ring-like structure appeared gradually on an amyloplast enwrapped by a meshwork, consolidated itself at the equatorial position (Figure 7C; see Supplemental Figure 6C and Supplemental Movie 13 online), remained in association with the amyloplast for a time (Figure 7B; see Supplemental Figure 6B and Supplemental Movie 12 online), and, thereafter, gradually disengaged from the amyloplast (Figure 7B; see Supplemental Figure 6B and Supplemental Movie 12 online). It seemed that the interactions of the amyloplast with each type of AFs was not constant but could vary with time (see Supplemental Movie 9 online)
Figure 7.
Dynamics of Amyloplasts and AFs in Wild Type and fiz1/+ Endodermal Cells.
(A) to (D) Time-lapse images of AFs (green) and amyloplasts (magenta) in wild-type cells.
(A) Linear actin cable and amyloplast. The arrow indicates an amyloplast that has moved a long distance along a linear actin cable.
(B) Ring-like actin bundle and amyloplast. The yellow arrow indicates a ring-like actin bundle that can be recognized by the two peaks of GFP signal on either side of the amyloplast. The white arrow indicates an amyloplast that has disengaged from a ring-like actin bundle.
(C) Appearance of a ring-like actin bundle. Arrow represents the ring-like actin bundle recognized by two peaks of GFP signal at the sides of the amyloplast.
(D) Curving actin bundle and amyloplasts. Arrow represents a curving actin bundle.
(E) Time-lapse images of AFs (green) and amyloplasts (magenta) in fiz1/+.
(F) Meshwork actin (green) and amyloplasts (magenta) in the wild type.
AFs were visualized by GFP-mTalin under the control of the 35S promoter. The black arrow to the right represents the direction of gravity (g). Bars = 5 μm.
The most prominent difference between sgr9 and wild-type endodermal cells was the relatively frequent tethering by AFs of a cluster of several amyloplasts in sgr9 cells (Figure 8A; see Supplemental Figure 7A and Supplemental Movie 14 online). This type of interaction was observed in sgr9 endodermal cells but rarely seen in wild-type endodermal cells. Each amyloplast frequently changed position in the cluster. At the same time, the pattern of GFP-mTalin surrounding the amyloplasts changed continually. In addition, the cluster gradually moved in a random fashion within the cell. Thus, the clustered amyloplasts found in sgr9 seemed to be entangled with AFs.
Figure 8.
Dynamics of Amyloplasts and AFs in sgr9 and sgr9 fiz1/+ Endodermal Cells.
(A) to (D) Time-lapse images of AFs (green) and amyloplasts (magenta) in sgr9 cells.
(A) Clustered amyloplasts entangled with AFs. The arrow indicates a cluster of several amyloplasts tethered by AFs.
(B) A few amyloplasts entangled with AFs. The arrow indicates a cluster containing a few amyloplasts that have moved a long distance along a linear actin cable.
(C) Ring-like actin bundle and amyloplast. Arrow points to the ring-like actin bundle.
(D) Linear actin cable and amyloplast. Arrow shows that the amyloplast has moved a long distance along the linear actin cable.
(E) Time-lapse images of AFs (green) and amyloplasts (magenta) in sgr9 fiz1/+.
(F) Meshwork actin (green) and amyloplast (magenta) in sgr9.
AFs are visualized by GFP-mTalin under the control of 35S promoter. The black arrow to the right represents the direction of gravity (g). Bars = 5 μm.
Three of the four types of AFs interaction with amyloplasts found in the wild type were observed in sgr9: actin meshworks (Figure 8F; see Supplemental Figure 7F online), actin cable (Figure 8D; see Supplemental Figure 7D and Supplemental Movie 15 online), and ring-like actin structure (Figure 8C; see Supplemental Figure 7C and Supplemental Movie 16 online). However, curving actin bundles that slide on amyloplasts were not observed in sgr9. We also noted that clusters containing a few amyloplasts occasionally moved long distances along the linear actin cables in sgr9 (Figure 8B; see Supplemental Figure 7B and Supplemental Movie 17 online). Since it was easy to identify amyloplasts with a ring-like structure in both the wild type and sgr9, we analyzed the frequency of this type of interaction. We found that 25% of amyloplasts in wild-type cells and 10.5% in sgr9 cells had ring-like structures (Table 1). The reduction in rate of amyloplasts with a ring-like structure in sgr9 may be inversely correlated with the appearance of trapped clusters of amyloplasts (Figures 8A and 8B; see Supplemental Figures 7A and 7B online).
Table 1.
The Incidence of Ring-Like Actin Bundle
| Genotype | Cell No. | Total Amyloplast No. | Incidence (%) |
| Col/GFP-mTalin | n = 7 | n = 56 | 25% |
| sgr9/GFP-mTalin | n = 9 | n = 86 | 10.5% |
The incidence of ring-like actin bundles was calculated from the ratio between the number of amyloplasts surrounded by ring-like actin bundle structures and the total number of amyloplasts within about 1 min from time-lapse images.
To determine whether the fiz1 mutation affected interaction between AFs and amyloplasts, we performed similar experiments as above with fiz1/+ and sgr9 fiz1/+. Linear actin cables, curving actin bundles, and ring-like actin structures were not detected in fiz1/+ (Figure 7E; see Supplemental Figure 6E and Supplemental Movie18 online) or sgr9 fiz1/+ cells (Figure 8E; see Supplemental Figure 7E and Supplemental Movie 19 online), despite the presence of AFs at the periphery of the endodermal cells. Moreover, abnormal AFs tethering amyloplast clusters, as found in sgr9, were not observed in sgr9 fiz1/+ cells (Figure 8E; see Supplemental Figure 7E and Supplemental Movie 19 online). These results suggest that the restoration of amyloplast sedimentation by the fiz1 mutation is due to loss of the entangled AFs that surround amyloplasts in sgr9. Overall, these results indicate that SGR9 might be involved in modulating the interaction between amyloplasts and AFs. Furthermore, the modulation conferred by SGR9 function might be important for amyloplast sedimentation toward the direction of gravity.
DISCUSSION
SGR9 Is Involved in Amyloplast Dynamics
Here, we isolated a gene encoding a novel RING finger protein and showed that this protein is responsible for the reduced shoot gravitropism of the Arabidopsis sgr9 mutant (Figure 1G). SGR9 is mainly expressed in the endodermis, the gravity-sensing tissue that is essential for gravitropism (Figure 1C). The gravitropic phenotype of sgr9 was fully complemented by endodermis-specific expression of SGR9 using the SCR promoter; thus, SGR9 function is required within the endodermis for shoot gravitropism (Figure 1H). Consistent with this conclusion, endodermal amyloplasts displayed abnormal dynamics in sgr9 (Figures 3C, 3D, 3G, 3J, and 3K). A previous study demonstrated that vacuolar integrity, such as vacuolar formation and/or dynamic features of the vacuolar membrane, in the endodermal cells is important for normal amyloplast dynamics (Kato et al., 2002; Yano et al., 2003; Saito et al., 2005). However, vacuolar formation and dynamics appeared to be normal in sgr9 endodermal cells (see Supplemental Figure 5B online), indicating that SGR9 is involved in amyloplast dynamics by a route other than interference with vacuolar integrity.
Role of AFs in Amyloplast Dynamics
In wild-type endodermal cells, amyloplasts showed a complicated pattern of movement even before gravistimulation (Figure 3B; see Supplemental Movie 1 online). Based on the analysis shown in Figure 6, the movement of amyloplasts can be kinetically separated into two categories: a higher rate of movement and a lower rate of movement. The movement associated with the higher rate was very similar to saltatory movement, which has been described previously (Saito et al., 2005). The disruption of AFs simultaneously reduced the frequency of amyloplasts along with the higher rate of movement (Figure 6C) and eliminated saltatory movement by the amyloplasts (Figure 5B). Amyloplasts undertaking saltatory movement appeared to attach to and move along long linear actin cables (Figure 7A). These long actin cables were absent from fiz1/+ mutant cells (Figure 7E). Thus, the saltatory movement of amyloplasts is likely to depend on long linear actin cables.
By contrast, amyloplast movement at the lower rate is not dependent on long actin cables. Instead, disruption of AFs increased the frequency of amyloplasts moving at the lower rate (Figure 6C). Additionally, AFs disruption enhanced amyloplast sedimentation as most amyloplasts sedimented in the direction of gravity (Figures 5A and 5B; Saito et al., 2005); this indicates that the increased frequency of amyloplasts moving at the lower rate largely contribute to sedimentation. Thus, we define this movement as sedimentable movement, which is independent of long actin cables.
Together, our observations and quantitative analyses suggest that there may be some form of dynamic equilibrium between the saltatory movement state and the sedimentable movement state that is mediated by AFs (long actin cables, more specifically) associated with the endodermal amyloplasts. In wild-type cells, only a small proportion of the amyloplasts participate in saltatory movement; rather, most amyloplasts sediment to the bottom of the cell via sedimentable movement. Possibly, some of the amyloplasts involved in sedimentable movement might attach to linear actin cables and, in this way, alter their status from the sedimentable movement state to the rapid saltatory movement state.
SGR9 May Modulate the Dynamic Equilibrium of Amyloplasts
We found here that most amyloplasts in endodermal cells of the sgr9 mutant moved around dynamically and were not located toward the bottom of the cells either before or after gravistimulation by reorientation (Figures 3D and 3J). This observation was confirmed by our quantitative analysis of amyloplast movement (Figures 3G and 3K). In the endodermal cells of sgr9, rapid saltatory movement was increased and sedimentable movement was decreased compared with wild-type cells (Figures 6B and 6F). Moreover, the saltatory movements in endodermal cells of sgr9 were blocked by the fiz1 mutation (Figure 6D), while the gravitropic phenotype of sgr9 was suppressed by LatB treatment and by the fiz1 mutation (Figure 4). These results indicate that SGR9 has a role in switching the dynamic equilibrium of amyloplast movement from the saltatory state to the sedimentable mode. Consistently, the interaction between AFs and amyloplasts was aberrant in the sgr9 endodermis. The appearance of clusters of amyloplasts enmeshed in AFs was the most remarkable abnormality observed in sgr9 cells (Figure 8A). Such clusters, which were never seen in wild-type cells, exhibited relatively rapid movements of the amyloplast (Figures 3D, 3G, 3J, and 3K). In addition, in sgr9 cells, amyloplasts attached to and moved along a long actin cable; these amyloplasts appeared to be interconnected by a fine actin mesh (Figure 8B). Only a few amyloplasts were observed to be located at the bottom of endodermal cells in sgr9, consistent with a reduction in the sedimentable movement rate in the mutant (Figure 6B). These results imply that once the amyloplasts attach to AFs, they rarely detach in the absence of SGR9, resulting in the predominance of amyloplasts with saltatory movements.
It is possible that the ring-like actin surrounding amyloplasts might be a means for detaching AFs from amyloplasts in wild-type cells. This ring-like structure gradually develops around amyloplasts surrounded by actin mesh (Figure 7C), becomes consolidated, and is then released from the amyloplasts in wild-type cells (Figure 7B). Although the ring-like structure is also found in sgr9 cells, its incidence is less than half that of wild-type cells (Table 1), possibly as a result of an impaired modulation of the interaction between amyloplasts and AFs. The idea that SGR9 might modulate this interaction is consistent with the localization of SGR9 to amyloplasts (Figures 2B to 2E; see Supplemental Figures 3A to 3D online). Although we used mutated SGR9-GFP proteins to determine localization, the fact that they showed a dominant-negative effect on endogenous SGR9 indicates that the mutated SGR9-GFP proteins were located at the site of action of endogenous SGR9 (Figure 2F; see Supplemental Figure 3E online).
Recently, it has been reported that some components of the Translocon of Outer Membrane of Chloroplasts (TOC) complex are involved in root gravitropism (Stanga et al., 2009). The modifier of arg1 (mar1) and mar2 mutants, which affect TOC75-III and TOC132, respectively, enhance the gravitropic defect of altered response to gravity1 (arg1). In both mar1 arg1 and mar2 arg1, amyloplasts show normal sedimentation behavior in columella cells upon gravistimulation, suggesting a role for the TOC components in gravity signal transduction following amyloplasts sedimentation. By contrast, sgr9 has a defect in amyloplast sedimentation within the endodermal cells (Figure 3). Thus, SGR9 probably acts in a different process in gravitropism from the TOC components, despite a similarity in their sites of action.
Role of SGR9 in Gravity Sensing
Based on our findings here, we propose that sedimentation of amyloplasts, which is an important element of gravity sensing in shoot gravitropism, largely depends on the status of the dynamic equilibrium mediated by AFs. The fact that LatB treatment and the fiz1 mutation did not inhibit gravitropism indicates that AF-dependent saltatory movement of amyloplasts is not necessary for gravity sensing (Figures 4A to 4C). When amyloplasts are freed from AFs, they sediment in the direction of gravity. This is probably due to their mass, as they contain dense starch granules. Possibly, amyloplasts in the sedimentable mode might be caught by actin cables and start to move in the saltatory mode. SGR9 may have a role for releasing amyloplasts caught by AFs. Alternatively, SGR9 may prevent amyloplasts from interacting with AFs, but once an interaction exceeds a certain threshold, amyloplasts become caught by the AFs. Only amyloplasts that are freed from AFs by the function of SGR9 are able to enter the sedimentable mode at the bottom of cell and also to sediment toward a new gravity vector in response to a sudden gravistimulation. In this way, SGR9 might contribute to the sensing of directional changes.
Most chloroplasts are anchored to the plasma membrane by AFs in plant cells. It has been reported that some proteins residing on the outer membrane of plastids can bind to actin (Oikawa et al., 2003, 2008; Higaki et al., 2007; Schmidt von Braun and Schleiff, 2008; Jouhet and Gray, 2009). One such protein is CHLOROPLAST UNUSUAL POSITIONING1 (CHUP1), which is well characterized as having a role in chloroplast photorelocation. The relocation of chloroplasts in mesophyll cells in response to light stimulation is mediated by short AFs (Kadota et al., 2009). CHUP1 may be required for the regulation of the interaction of short AFs with chloroplasts during this process of movement (Kadota et al., 2009). In contrast with plastid (chloroplast) movement based on short AFs, plastids (amyloplasts) that are free of AFs appear to be essential for gravity sensing. This implies that this plastid characteristic, which may depend on SGR9 function, is crucial for gravity-sensing cells. It is also consistent with the specific expression of the gene in the endodermis of the stem and hypocotyl and in the columella cells in the root cap. The extent of contribution of SGR9 to gravity sensing might differ between organs possibly due to variations in the intracellular characteristics around the amyloplasts.
Molecular Function of SGR9
The Arabidopsis genome contains ~480 genes that encode RING finger proteins. Computational analysis based on the amino acid sequence of the RING domain in Arabidopsis indicated that SGR9 belongs to a cluster that includes AIP2 and CIP8 (Kosarev, et al., 2002). CIP8 and AIP2 proteins exhibit polyubiquitination activity in photomorphogenesis and abscisic acid hormone signaling, respectively (Hardtke et al., 2002; Zhang et al., 2005). In addition, AIP2 is involved in ABA-INSENSITIVE3 protein degradation in a polyubiquitination manner (Zhang et al., 2005). Similarly, the SGR9 RING finger domain possessed polyubiquitination activity in an E1 and E2 conjugating enzyme-dependent manner (Figure 2A), indicating that SGR9 also functions as a ubiquitin E3 ligase. The amino acid substitution in the SGR9 RING finger domain markedly decreased E3 ligase activity of MBP-SGR9(RING) (Figure 2A), and an SGR9-GFP that harbored the amino acid substitution did not rescue the gravitropic defect of sgr9 (data not shown). This indicates that the SGR9 E3 ligase activity that is dependent on the RING finger domain is important for gravity sensing. The amino acid sequence of the N-terminal region of SGR9 is unique within the Arabidopsis genome. As the RING finger domain functions as a binding domain with an E2 conjugating enzyme, the unique N-terminal region may bind to a unique substrate. The substrate of SGR9 might be either actin-associated proteins or the adopter proteins between amyloplasts and AFs. The ubiquitination of these substrates might lead to protein degradation or changes in protein–protein interaction, which might modulate the interaction between amyloplasts and AFs. Identification of the substrate of SGR9 would clarify how the protein modulates the interaction between AFs and amyloplasts.
In addition, mutations in the RING finger domain of SGR9 that diminish E3 ligase activity appeared to stabilize SGR9 (Figures 2B to 2E; see Supplemental Figures 3A to 3D online). This implies that SGR9 may also have autoubiquitination activity. Thus, how the activity of SGR9 is regulated is the question that needs to be addressed.
METHODS
Plant Materials and Growth Conditions
The Columbia (Col) accession of Arabidopsis thaliana was used as the wild type. The sgr9 mutant was isolated from ethyl methanesulfonate–mutagenized M2 seeds (Col) provided by Lehle Seeds. Plants carrying the Pro35S:GFP-mTalin construct or the fiz1 mutation were also derived from the Col accession and were described in our previous study (Kato et al., 2010). Seedlings were germinated on Murashige and Skoog (MS) plates under constant white light at 23°C for 7 to 10 d. Plants were transplanted and grown on soil under constant white light at 23°C.
Isolation and Mapping of SGR9
A Landsberg erecta (Ler) wild-type plant was crossed to the sgr9 mutant plant to generate a mapping population. The SGR9 locus was mapped toward the top of chromosome 5. For fine-scale mapping, DNA was prepared from ~200 F2 progeny. We generated cleaved-amplified polymorphic sequence markers to identify polymorphisms between Col and Ler based on information provided by The Arabidopsis Information Resource.
Gravitropism Analysis
Primary inflorescence stems 4 to 8 cm in length were used for shoot gravitropism analyses. They were transferred to nondirectional dim light (1 μmol m−2 s−1) at 23°C, and then the plants were placed horizontally after preincubation for 1 h. Photographs were taken automatically every 10 min by digital camera (Canon) under the control of ZoomBrowser EX software (Canon). The stem curvature was defined as the angle formed between the growing direction of the apex and the horizontal base line and was measured on the digital images using Image J software (http://rsb.info.nih.gov/ij/).
DNA Manipulation and Plant Transformation
For complementation analysis, the genomic fragment of SGR9 including the putative promoter region and 3′ downstream region was amplified from plant genomic DNA by PCR (primer set: SGR9_gF, SGR9_gR; see Supplemental Table 1 online for oligonucleotide sequences), and then the PCR products were subcloned using a Zero Blunt TOPO PCR cloning kit (Invitrogen) (pTOPO-gSGR9). The genomic fragment of SGR9 was cloned into binary vector pBIN19. Total RNA was isolated from inflorescence stems using the RNeasy Plant Mini Kit (Qiagen). cDNAs were synthesized using SuperScript II reverse transcriptase (Invitrogen). The SGR9 cDNA was amplified by PCR (primer set: SGR9_cDNA-B-K_F and SGR9_cDNA-B-K_R) and then subcloned using the Zero Blunt TOPO PCR cloning kit (pTOPO-cSGR9). The SGR9 cDNA was inserted into downstream of the SCR promoter on the pBI binary vector (ProSCR:SGR9) (Morita et al., 2002). The ACT8 cDNA was amplified by PCR (primer set: ACT8_cDNA_F and ACT8_cDNA_R) and then subcloned using the Zero Blunt TOPO PCR cloning kit. The fiz1 type ACT8 cDNA was produced by PCR using mutagenic primer (primer set: ACT8_fiz1_F and ACT8_fiz1_R). The fiz1-type ACT8 cDNA was inserted into downstream of the SCR promoter on the pBI binary vector (ProSCR:ACT8 fiz1) (Morita et al., 2002). The SGR9 promoter region was amplified from plant genomic DNA by PCR [primer set: SGR9_gF and pSGR9(Bam)_R], and then the PCR products were subcloned using the Zero Blunt TOPO PCR cloning kit. The SGR9 promoter fragment was inserted into upstream of the uidA gene in the pBI101 (ProSGR9:GUS) (Morita et al., 2002). To make translational fusion constructs, the SmaI site was inserted into immediately upstream of stop codon in SGR9 genomic fragment (pTOPO-gSGR9) by PCR (primer set: SGR9_CPF_F and SGR9_CPF_R) (pTOPO-gSGR9-SmaI). The gSGR9-SmaI fragment was cloned into binary vector pBIN19. GFP fragment (Morita et al., 2002) was inserted into SGR9 genomic fragment using SmaI. To obtain mutated SGR9 fused with GFP [gSGR9-GFP (W244A) and gSGR9-GFP (C232A)], pTOPO-gSGR9-SmaI was mutagenized by PCR using mutagenic primer [primer set (W244A): SGR9_W244A_F and SGR9_W244A_R; primer set (C232A): SGR9_C232A_F and SGR9_ZnR2). The SGR9 genomic fragment containing the mutation was cloned into binary vector pBIN19. The GFP-encoding DNA fragment was inserted into the SmaI site. The DNA sequence of all constructs was confirmed by sequencing. The resulting constructs were transformed into Agrobacterium tumefaciens strain GV3101 (MP90) and introduced into wild-type (Col) or sgr9 plants by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected based on resistance to kanamycin.
Histochemical Analysis of GUS Activity
For GUS staining of transgenic plants seedling, hypocotyl, flower, and inflorescence stem, samples were first immersed in 90% (v/v) ice-cold acetone for 15 to 30 min and then incubated in GUS staining solution (100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 5 mM ferricyanide, 5 mM ferrocyanide, 0.1% [v/v] Triton X-100, and 0.52 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) at 37°C. After being cleared in 70% (v/v) ethanol/chloral hydrate solution (8 g chloral hydrate, 1 mL glycerol, and 2 mL water), tissues were observed with a light microscope. For histological sections, GUS-stained inflorescence stems were dehydrated using a 70 to 100% (v/v) ethanol series and then embedded in Technovit 7100 (Heraeus Kulzer) and sectioned with a microtome.
Protein Expression and Purification for in Vitro Assay
To obtain the DNA fragment encoding the SGR9 RING finger domain, the SGR9 cDNA (pTOPO-cSGR9) was truncated by PCR (primer set: SGR9_RING_F and SGR9_RING_R) (pTOPO-SGR9-RING). The SGR9-RING fragment was cloned into pMALp2 (New England Biolabs). To obtain mutated SGR9-RING constructs [cSGR9-RING (C232A) and cSGR9-RING (W244A)], pTOPO-cSGR9-RING was mutagenized by PCR using mutagenic primer [primer set (W244A): SGR9_W244A_F and SGR9_W244A_R; primer set (C232A): SGR9_C232A_F and SGR9_ZnR2]. Each fragment was cloned into pMALp2 (New England Biolabs). For preparation of recombinant proteins, the resulting pMALp2 plasmids were transformed into Escherichia coli BL21. Cells were grown at 37°C for 2 h to an OD600 of 0.4 to 0.6 in medium (1% [w/v] tryptone, 0.5% [w/v] yeast extract, 0.5% [w/v] NaCl, 0.2% [w/v] Glc, 10 μM ZnSO4, and 100 μg/mL ampicillin). Isopropyl β-d-thiogalactopyranoside (1 mM) was added and incubated for 6 h at 37°C. Cells were harvested by centrifugation and lysed in lysis buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 100 μM ZnSO4, and 1 mM DTT) and then sonicated. The lysates were centrifuged at 4°C and 9000g for 30 min, and the supernatants were applied to amylose resin affinity column (New England Biolabs) at 4°C. MBP-fused proteins were eluted with buffer containing 10 mM maltose.
In Vitro Ubiquitination Assay and Immunoblot Analysis
Ubiquitination assays were performed using a ubiqutinylation kit (BIOMOL) and performed according to the manufacturer's protocol. For the SGR9 ubiquitination assay, each reaction contained at least 700 ng purified MBP-SGR9 (RING), MBP-SGR9 (RING) (W244A), or MBP-SGR9 (RING) (C232A), human E1, human E2 (UbcH5c), and ATP. After incubation at 30°C for 8 h, the reaction was stopped with 2× SDS-PAGE sample buffer at 95°C for 5 min. Proteins were separated on a 7.5% SDS-polyacrylamide gel and then blotted to polyvinylidene difluoride membrane. For detection of ubiquitinated proteins, the membranes were treated with anti-SGR9 (1:5000 dilution) or anti-Multiubiquitin (7:10,000 dilution) (Medical & Biological Laboratories Co.) for 1 h. After washing, the membranes were incubated with anti-rabbit IgG (1:10,000 dilution) or anti-mouse IgG (1:10,000 dilution), respectively. Immunoreactive signals were detected with the ECL detection system (Amersham Bioscience).
Preparation of SGR9 Antibody
To obtain recombinant SGR9 protein, the full-length SGR9 cDNA was cloned into pCold II (Takara). The plasmid was transformed into E. coli BL21. Cells were grown at 37°C for 3 h to an OD600 of 0.4 to 0.6 in Luria-Bertani medium (1% [w/v] tryptone, 0.5% [w/v] yeast extract, and 1% [w/v] NaCl) containing 100 μg/mL ampicillin. The culture was placed at 15°C for 30 min, and then isopropyl β-d-thiogalactopyranoside (1 mM) was added and incubated for 24 h at 15°C. Cells were harvested by centrifugation at 4°C and 4500g for 15 min. Recombinant SGR9 proteins were purified and used as antigen in the preparation of specific antibodies.
Confocal Microscopy
Fluorescence of SGR9(W244A)-GFP and SGR9(C232A)-GFP was imaged by confocal laser scanning microscopy (FV1000; Olympus). GFP fluorescence (green) and chlorophyll autofluorescence (magenta) were detected with 500- to 510-nm and 640- to 700-nm spectral settings, respectively, with 488-nm excitation.
Sample Preparation for the Vertical Stage Microscope
Before cutting the inflorescence stem segment, the slide glass was prepared (see Supplemental Figure 8 online). First, double-sided adhesive tape was prepared using two sheets of an adhesive tape stuck together by waterproof adhesive agent (Aron Alpha; Toagosei). The double-sided adhesive tape was cut and stuck to the slide glass (0.9- to ~1.2-mm thick; Matsunami). The silicon isolator (FastWell FW20; Grace Bio-Labs) was applied to the slide glass. The inflorescence stem segment (<1 cm in length, the region of 1 to 2 cm from the top of the inflorescence stem) was excised. To split the stem segment longitudinally, we prepared a hand-built instrument. The stem segment was set on a hand-built instrument and then cut longitudinally using a razor blade, maintaining the direction relative to gravity. Immediately after cutting, a small amount of GM liquid medium (1× MS salt mixture and 1% [w/v] Suc) with 0.1% (w/v) agar was added. The segment was stuck to double-sided adhesive tape on the slide glass maintaining the direction relative to gravity. To add the GM liquid medium with 0.1% (w/v) agar to isolated area, the slide glass was inclined. After adding the medium, the isolated area was immediately covered with a cover slip. The slide glass was reoriented vertically.
Live-Cell Imaging Using a Vertical Stage Microscope
Imaging of endodermal cells was performed with a vertical stage microscope (model BX50; Olympus) (Saito et al., 2005) equipped with a dry (air-immersion) objective (UPlanApo ×40, 0.85 numerical aperture; Olympus) and an oil-immersion objective (UPlanApo ×100, 1.35 numerical aperture; Olympus). Longitudinally sectioned inflorescence stems were mounted on the rotatable vertical stage with 45° click stop function (model U-SRP; Olympus) and turned to 90° while monitoring the endodermal cells. Bright-field images were acquired with a cooled charge-coupled device camera (model CoolSNAP cf; Photometrics, Nippon Roper) and processed by IPLab image analysis software (Scanalytics). Fluorescence images were acquired with a spinning-disk confocal scanning unit (model CSU10; Yokogawa) equipped with a fiber-coupled diode laser (488 nm) (model HPU50101-PFS; FITEL, Furukawa Electric Co.), an emission filter wheel (model 99A351; Ludl Electronic Products), and a back-illuminated electron multiplying–charge-coupled device camera (model iXON+ DU897E-CS0-#BV; Andor Technology) and processed by iQ image analysis software (Andor Technology). GFP fluorescence and autofluorescence of amyloplasts after excitation at 488 nm were detected at 503 to 552 nm (model Em01-R488-25; Semrock) and 665 to 705 nm (model FF01-685/40-25; Semrock), respectively. The band-pass emission filters were changed using a filter wheel controller system (model MAC 6000; Ludl Electronic Products).
Plots of the Amyloplast Distribution
The time-lapse images from several independent cells that were vertically settled were used to plots analyses of amyloplast distribution. The first image (0 min) and an image taken after 5 min from one cell were used for the analysis. x and y coordinates of each amyloplast were defined by the center position of amyloplast and then coordinates were recorded. To superpose the results from several independent cells, the endodermal cell was simplified as a rectangle (longitudinal axis is 2; horizontal axis is 1), and then each coordinate of amyloplast was related to rectangle. The position of amyloplasts was plotted on this rectangle.
Quantitative and Statistical Analysis of the Amyloplast Movement
Quantitative and statistical analyses were performed as described previously (Saito et al., 2005). The movement of amyloplasts was subjected to quantitative analysis using time-lapse images. Amyloplasts that could be traced as much as possible were selected. Each coordinate of the amyloplast in the image frame was recorded, and Δx and Δy was calculated at 10-s intervals. The x axis indicates a horizontal line, and the y axis indicates a vertical line. Pseudomovement caused by the directional and negligible stage drifting was corrected using the coordinate of an immobile marker (for example, the corner of the cell) in the first and last frames. In this study, the frequency of Δy at each 10-s interval was presented in a histogram. Variances of the populations of Δy from the wild type and those from mutants before gravistimulation were compared using F tests. For statistical analysis of amyloplast sedimentation after gravistimulation, the mean value of Δy after gravistimulation was compared with the mean value of Δy before gravistimulation (Welch’s t test, one-tailed, P < 0.05).
Trace of Amyloplast Movement
The time-lapse images of endodermal cells were subjected to trace analysis of amyloplasts. Since some amyloplasts move out of the focal plane, amyloplasts traced as long as possible were indicated. Each coordinate of amyloplast in the image frame was defined by the center position of amyloplast, and each coordinate of the amyloplast at the 10-s intervals was recorded. The positions of each amyloplast were connected with lines.
Quantitative Analysis of the Moving Distance of Amyloplast for 30 s
To analyze the distribution of rates of movement of amyloplasts at steady state, distances moved in 30 s were recounted based on the data sets of Δy component (μm/10 s) before gravistimulation from quantitative analysis of the amyloplast movements (shown in Figures 3 and 5). To eliminate the directional element, the absolute values of Δy components were used. The value of Δy component (μm/10 s) was summed within serial 30 s. Variances of the populations of Δy for 30 s from the wild type and those from mutants were compared using F tests.
Treatment of the Influorescence Stem with LatB
A 20 mM stock solution of LatB (Calbiochem) was prepared in dimethyl sulfoxide. The stem segments of the primary inflorescence stem were used for gravitropic assays. The stem segments were cut from the primary inflorescence stems with a razor blade, and their basal ends were immersed in a 200 μL tube containing liquid medium (0.5× MS salt mixture and 1% [w/v] Suc). The stem segments were preincubated for 12 to ~14 h with DMSO or 20 μM LatB at 23°C under constant light condition. After incubation, stems were gravistimulated by being placed horizontally at 23°C under dim nondirectional light. Gravitropism analysis is described above.
Gravitropism Analysis in the Hypocotyl
For hypocotyl gravitropism analysis, seedlings were etiolated at 23°C for 3 d in darkness (vertical position). Before gravistimulation, photographs were taken by digital camera. Then, they were rotated through 90° and incubated for 12 h in darkness under the same condition (horizontal position). After incubation, photographs were taken by digital camera. The curvature degree was determined by the angle between the growing direction of the vertical position and the horizontal position, measured from digital images using Image J software, and then presented in histogram.
Gravitropism Analysis in Root
For root gravitropism analysis, seedlings were grown at 23°C for 4 d in darkness (vertical position). Before gravistimulation, photographs were taken by digital camera. Then, they were rotated through 90° and incubated for 18 h in darkness under the same condition (horizontal position). After incubation, photographs were taken by digital camera. The curvature degree was determined by the angle between the growing direction of the vertical position and the horizontal position, measured from digital images using Image J software, and then presented in histogram.
Histological Analysis
For observation of amyloplast sedimentation, inflorescence stem segments were fixed, embedded in Technovit 7100 (Heraeus Kulzer), and sectioned as described previously (Morita et al., 2002).
Construction of the GFP-Vam3 Marker Line
The GFP with AtVam3 construct (GFP-Vam3) was provided by Masahiko Sato (Uemura et al., 2002). GFP-Vam3 fragment was inserted downstream of the SCR promoter on the pBI binary vector (Morita et al., 2002). The resulting constructs were transformed into Agrobacterium strain GV3101 (MP90) and introduced into wild-type (Col) plants by the floral dip method. Transgenic plants were selected by resistance to kanamycin and then used as the endodermal vacuolar membrane marker.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At5g02750 (SGR9), At1g31480 (SGR2), At5g46860 (SGR3), At5g39510 (ZIG/SGR4), At5g64920 (CIP8), and At5g20910 (AIP2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of the sgr9 Mutant.
Supplemental Figure 2. Molecular Cloning of SGR9 and the SGR9 Protein Structure.
Supplemental Figure 3. Intracellular Localization of SGR9(C232A)-GFP and Gravitropic Response of gSGR9(C232A)-GFP/Col.
Supplemental Figure 4. Amyloplast Dynamics before Gravistimulation.
Supplemental Figure 5. Vacuolar Membrane Dynamics in Endodermal Cell.
Supplemental Figure 6. Dynamics of AFs in Wild-Type and fiz1/+ Endodermal Cells.
Supplemental Figure 7. Dynamics of AFs in sgr9 and sgr9 fiz1/+ Endodermal Cells.
Supplemental Figure 8. Sample Preparation for the Vertical Stage Microscope.
Supplemental Table 1. Oligonucleotides Used in DNA Manipulation.
Supplemental Movie 1. Amyloplast Dynamics in the Wild Type before Gravistimulation, Related to Figure 3B.
Supplemental Movie 2. Amyloplast Dynamics in sgr9 before Gravistimulation, Related to Figure 3D.
Supplemental Movie 3. Amyloplast Dynamics in the Wild Type after Gravistimulation by Reorientation, Related to Figure 3H.
Supplemental Movie 4. Amyloplast Dynamics in sgr9 after Gravistimulation by Reorientation, Related to Figure 3J.
Supplemental Movie 5. Amyloplast Dynamics in fiz1/+ before Gravistimulation, Related to Figure 5B.
Supplemental Movie 6. Amyloplast Dynamics in sgr9 fiz1/+ before Gravistimulation, Related to Figure 5D.
Supplemental Movie 7. Amyloplast Dynamics in fiz1/+ after Gravistimulation by Reorientation, Related to Figure 5G.
Supplemental Movie 8. Amyloplast Dynamics in sgr9 fiz1/+ after Gravistimulation by Reorientation, Related to Figure 5I.
Supplemental Movie 9. Dynamics of AFs and Amyloplasts in Wild-Type Whole Endodermal Cell.
Supplemental Movie 10. Dynamics of Linear Actin Cable and Amyloplasts in the Wild Type, Related to Figure 7A.
Supplemental Movie 11. Dynamics of Curving Actin Bundle and Amyloplasts in the Wild Type, Related to Figure 7D.
Supplemental Movie 12. Dynamics of Ring-Like Actin Bundle and Amyloplasts in the Wild Type, Related to Figure 7B.
Supplemental Movie 13. Appearance of Ring-Like Actin Bundle and Amyloplasts in the Wild Type, Related to Figure 7C.
Supplemental Movie 14. Dynamics of Clustered Amyloplasts and AFs in sgr9, Related to Figure 8A.
Supplemental Movie 15. Dynamics of Linear Actin Cable and Amyloplasts in sgr9, Related to Figure 8D.
Supplemental Movie 16. Dynamics of Ring-Like Actin Bundle and Amyloplasts in sgr9, Related to Figure 8C.
Supplemental Movie 17. Dynamics of a Few Amyloplasts and AFs in sgr9, Related to Figure 8B.
Supplemental Movie 18. Dynamics of AFs and Amyloplasts in fiz1/+, Related to Figure 7E.
Supplemental Movie 19. Dynamics of AFs and Amyloplasts in sgr9 fiz1/+, Related to Figure 8E.
Acknowledgments
We thank Takehide Kato for providing Pro35S:GFP-mTalin transgenic plants and the fiz1 mutant and Nam-Hai Chua for providing the Pro35S:GFP-mTalin vector. We also thank Noriyuki Matsuda for helpful discussion with ubiquitination assay, Chieko Saito for helpful discussion with analysis of amyloplast dynamics, and Nauko Inui, Seiko Ishihara, and Kaori Kaminoyama for technical assistance. The financial support of a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan (16085205 to M.T.M.) and a grant from PRESTO project, Japan Science and Technology Agency (to M.T.M.) are gratefully acknowledged.
References
- Blancaflor E.B., Fasano J.M., Gilroy S. (1998). Mapping the functional roles of cap cells in the response of Arabidopsis primary roots to gravity. Plant Physiol. 116: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonsirichai K., Guan C., Chen R., Masson P.H. (2002). Root gravitropism: An experimental tool to investigate basic cellular and molecular processes underlying mechanosensing and signal transmission in plants. Annu. Rev. Plant Biol. 53: 421–447 [DOI] [PubMed] [Google Scholar]
- Caspar T., Pickard B.G. (1989). Gravitropism in a starchless mutant of Arabidopsis: Implications for the starch-statolith theory of gravity sensing. Planta 177: 185–197 [PubMed] [Google Scholar]
- Clifford P.E., Barclay G.F. (1980). The sedimentation of amyloplasts in living statocytes of the dandelion flower stalk. Plant Cell Environ. 3: 381–386 [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Fukaki H., Fujisawa H., Tasaka M. (1996a). Gravitropic response of inflorescence stems in Arabidopsis thaliana. Plant Physiol. 110: 933–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H., Fujisawa H., Tasaka M. (1996b). SGR1, SGR2, and SGR3: Novel genetic loci involved in shoot gravitropism in Arabidopsis thaliana. Plant Physiol. 110: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H., Wysocka-Diller J., Kato T., Fujisawa H., Benfey P.N., Tasaka M. (1998). Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 14: 425–430 [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Okamoto H., Stoop-Myer C., Deng X.W. (2002). Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J. 30: 385–394 [DOI] [PubMed] [Google Scholar]
- Heathcote D. (1981). The geotropic reaction and statolith movements following geostimulation of mung bean hypocotyl. Plant Cell Environ. 4: 131–140 [Google Scholar]
- Higaki T., Sano T., Hasezawa S. (2007). Actin microfilament dynamics and actin side-binding proteins in plants. Curr. Opin. Plant Biol. 10: 549–556 [DOI] [PubMed] [Google Scholar]
- Hou G., Mohamalawari D.R., Blancaflor E.B. (2003). Enhanced gravitropism of roots with a disrupted cap actin cytoskeleton. Plant Physiol. 131: 1360–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P.K., Eldridge A.G., Freed E., Furstenthal L., Hsu J.Y., Kaiser B.K., Reimann J.D. (2000). The lore of the RINGs: Substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10: 429–439 [DOI] [PubMed] [Google Scholar]
- Jouhet J., Gray J.C. (2009). Interaction of actin and the chloroplast protein import apparatus. J. Biol. Chem. 284: 19132–19141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota A., Yamada N., Suetsugu N., Hirose M., Saito C., Shoda K., Ichikawa S., Kagawa T., Nakano A., Wada M. (2009). Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 13106–13111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Morita M.T., Fukaki H., Yamauchi Y., Uehara M., Niihama M., Tasaka M. (2002). SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14: 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Morita M.T., Tasaka M. (2010). Defects in dynamics and functions of actin filament in Arabidopsis caused by the dominant-negative actin fiz1-induced fragmentation of actin filament. Plant Cell Physiol. 51: 333–338 [DOI] [PubMed] [Google Scholar]
- Katoh S., Tsunoda Y., Murata K., Minami E., Katoh E. (2005). Active site residues and amino acid specificity of the ubiquitin carrier protein-binding RING-H2 finger domain. J. Biol. Chem. 280: 41015–41024 [DOI] [PubMed] [Google Scholar]
- Kiss J.Z., Guisinger M.M., Miller A.J., Stackhouse K.S. (1997). Reduced gravitropism in hypocotyls of starch-deficient mutants of Arabidopsis. Plant Cell Physiol. 38: 518–525 [DOI] [PubMed] [Google Scholar]
- Kiss J.Z., Hertel R., Sack F.D. (1989). Amyloplasts are necessary for full gravitropic sensitivity in roots of Arabidopsis thaliana. Planta 177: 198–206 [PubMed] [Google Scholar]
- Kosarev P., Mayer K.F., Hardtke C.S. (2002). Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 3: research 0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N., Suzuki T., Tanaka K., Nakano A. (2001). Rma1, a novel type of RING finger protein conserved from Arabidopsis to human, is a membrane-bound ubiquitin ligase. J. Cell Sci. 114: 1949–1957 [DOI] [PubMed] [Google Scholar]
- Morita M.T., Kato T., Nagafusa K., Saito C., Ueda T., Nakano A., Tasaka M. (2002). Involvement of the vacuoles of the endodermis in the early process of shoot gravitropism in Arabidopsis. Plant Cell 14: 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M.T., Tasaka M. (2004). Gravity sensing and signaling. Curr. Opin. Plant Biol. 7: 712–718 [DOI] [PubMed] [Google Scholar]
- Oikawa K., Kasahara M., Kiyosue T., Kagawa T., Suetsugu N., Takahashi F., Kanegae T., Niwa Y., Kadota A., Wada M. (2003). Chloroplast unusual positioning1 is essential for proper chloroplast positioning. Plant Cell 15: 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K., Yamasato A., Kong S.G., Kasahara M., Nakai M., Takahashi F., Ogura Y., Kagawa T., Wada M. (2008). Chloroplast outer envelope protein CHUP1 is essential for chloroplast anchorage to the plasma membrane and chloroplast movement. Plant Physiol. 148: 829–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M., Kiss J.Z. (2005). Disruption of the F-actin cytoskeleton limits statolith movement in Arabidopsis hypocotyls. J. Exp. Bot. 56: 2539–2550 [DOI] [PubMed] [Google Scholar]
- Perbal G., Driss-Ecole D. (2003). Mechanotransduction in gravisensing cells. Trends Plant Sci. 8: 498–504 [DOI] [PubMed] [Google Scholar]
- Sack F.D. (1997). Plastids and gravitropic sensing. Planta 203(Suppl. 1): S63–S68 [DOI] [PubMed] [Google Scholar]
- Sack F.D., Leopold A.C. (1985). Cytoplasmic streaming affects gravity-induced amyloplast sedimentation in maize coleoptiles. Planta 164: 56–62 [PubMed] [Google Scholar]
- Saito C., Morita M.T., Kato T., Tasaka M. (2005). Amyloplasts and vacuolar membrane dynamics in the living graviperceptive cell of the Arabidopsis inflorescence stem. Plant Cell 17: 548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt von Braun S., Schleiff E. (2008). The chloroplast outer membrane protein CHUP1 interacts with actin and profilin. Planta 227: 1151–1159 [DOI] [PubMed] [Google Scholar]
- Silady R.A., Kato T., Lukowitz W., Sieber P., Tasaka M., Somerville C.R. (2004). The gravitropism defective 2 mutants of Arabidopsis are deficient in a protein implicated in endocytosis in Caenorhabditis elegans. Plant Physiol. 136: 3095–3103, discussion 3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A.P., Deeks M.J., Hussey P.J. (2010). Strategies of actin reorganisation in plant cells. J. Cell Sci. 123: 3019–3028 [DOI] [PubMed] [Google Scholar]
- Stanga J.P., Boonsirichai K., Sedbrook J.C., Otegui M.S., Masson P.H. (2009). A role for the TOC complex in Arabidopsis root gravitropism. Plant Physiol. 149: 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Hauksdóttir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaka M., Kato T., Fukaki H. (1999). The endodermis and shoot gravitropism. Trends Plant Sci. 4: 103–107 [DOI] [PubMed] [Google Scholar]
- Uemura T., Yoshimura S.H., Takeyasu K., Sato M.H. (2002). Vacuolar membrane dynamics revealed by GFP-AtVam3 fusion protein. Genes Cells 7: 743–753 [DOI] [PubMed] [Google Scholar]
- Vaux D.L., Silke J. (2005). IAPs, RINGs and ubiquitylation. Nat. Rev. Mol. Cell Biol. 6: 287–297 [DOI] [PubMed] [Google Scholar]
- Weise S.E., Kiss J.Z. (1999). Gravitropism of inflorescence stems in starch-deficient mutants of Arabidopsis. Int. J. Plant Sci. 160: 521–527 [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Kiss J.Z. (2002). Disruption of the actin cytoskeleton results in the promotion of gravitropism in inflorescence stems and hypocotyls of Arabidopsis. Plant Physiol. 128: 669–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Fukaki H., Fujisawa H., Tasaka M. (1997). Mutations in the SGR4, SGR5 and SGR6 loci of Arabidopsis thaliana alter the shoot gravitropism. Plant Cell Physiol. 38: 530–535 [DOI] [PubMed] [Google Scholar]
- Yano D., Sato M., Saito C., Sato M.H., Morita M.T., Tasaka M. (2003). A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc. Natl. Acad. Sci. USA 100: 8589–8594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder T.L., Zheng H.Q., Todd P., Staehelin L.A. (2001). Amyloplast sedimentation dynamics in maize columella cells support a new model for the gravity-sensing apparatus of roots. Plant Physiol. 125: 1045–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Garreton V., Chua N.H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]