This work examines the molecular mechanism of jasmonate regulation of anthocyanin biosynthesis and trichome initiation. It identifies three bHLH transcription factors and two MYB transcription factors as new targets of JAZ proteins, showing that JAZ proteins attenuate the transcriptional function of WD-repeat/bHLH/MYB complexes to regulate anthocyanin accumulation and trichome.
Abstract
Jasmonates (JAs) mediate plant responses to insect attack, wounding, pathogen infection, stress, and UV damage and regulate plant fertility, anthocyanin accumulation, trichome formation, and many other plant developmental processes. Arabidopsis thaliana Jasmonate ZIM-domain (JAZ) proteins, substrates of the CORONATINE INSENSITIVE1 (COI1)–based SCFCOI1 complex, negatively regulate these plant responses. Little is known about the molecular mechanism for JA regulation of anthocyanin accumulation and trichome initiation. In this study, we revealed that JAZ proteins interact with bHLH (Transparent Testa8, Glabra3 [GL3], and Enhancer of Glabra3 [EGL3]) and R2R3 MYB transcription factors (MYB75 and Glabra1), essential components of WD-repeat/bHLH/MYB transcriptional complexes, to repress JA-regulated anthocyanin accumulation and trichome initiation. Genetic and physiological evidence showed that JA regulates WD-repeat/bHLH/MYB complex-mediated anthocyanin accumulation and trichome initiation in a COI1-dependent manner. Overexpression of the MYB transcription factor MYB75 and bHLH factors (GL3 and EGL3) restored anthocyanin accumulation and trichome initiation in the coi1 mutant, respectively. We speculate that the JA-induced degradation of JAZ proteins abolishes the interactions of JAZ proteins with bHLH and MYB factors, allowing the transcriptional function of WD-repeat/bHLH/MYB complexes, which subsequently activate respective downstream signal cascades to modulate anthocyanin accumulation and trichome initiation.
INTRODUCTION
Anthocyanins, a class of flavonoids widely found in plants, are responsible for plant coloration (Springob et al., 2003; Lepiniec et al., 2006). They allure insects to pollinate flowers (Vogt et al., 1994), participate in pollen–pistil interactions (Mo et al., 1992), protect plants from UV radiation (Harborne and Williams, 2000; Solovchenko and Schmitz-Eiberger, 2003) and stress damage (Winkel-Shirley, 2002), and function as antimicrobial agents against insect attack and pathogen infection (Harborne and Williams, 2000; Winkel-Shirley, 2001; Gould, 2004).
Anthocyanin accumulation is stimulated by a series of endogenous developmental signals, such as Suc (Teng et al., 2005; Solfanelli et al., 2006) and plant hormones (Deikman and Hammer, 1995; Loreti et al., 2008; Shan et al., 2009), and by various environmental stresses, including UV irradiation, pathogen infection, insect attack, drought, wounding, and nutrient depletion (Harborne and Williams, 2000; Cominelli et al., 2008; Gonzalez et al., 2008; Lillo et al., 2008; Gonzalez, 2009).
The WD-repeat/bHLH/MYB complex acts as an important regulatory machinery to modulate anthocyanin accumulation in plant species (Payne et al., 2000; Broun, 2005; Ramsay and Glover, 2005). In Arabidopsis thaliana, the WD-repeat protein Transparent Testa Glabra1 (TTG1) (Walker et al., 1999) recruits basic-helix-loop-helix (bHLH) transcription factors (Toledo-Ortiz et al., 2003), such as Glabra3 (GL3) (Payne et al., 2000), Transparent Testa8 (TT8) (Nesi et al., 2000; Baudry et al., 2006), or Enhancer of Glabra3 (EGL3) (Zhang et al., 2003), and R2R3 MYB transcriptional factors (MYB75, MYB90, MYB113, or MYB114) (Borevitz et al., 2000; Stracke et al., 2001; Zimmermann et al., 2004; Stracke et al., 2007; Allan et al., 2008; Gonzalez et al., 2008; Rowan et al., 2009) to form the WD-repeat/bHLH/MYB complex, which mediates anthocyanin biosynthesis mainly by upregulating the expression of late anthocyanin biosynthetic genes, including NADPH-dependent dihydroflavonol reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), and UDP-Glc:flavonoid 3-Oglucosyltransferase (UF3GT) (Dooner et al., 1991; Kubasek et al., 1992; Shirley et al., 1995; Gonzalez et al., 2008).
The WD-repeat/bHLH/MYB complex, consisting of TTG1 (Walker et al., 1999), the bHLH protein GL3 (Payne et al., 2000), or EGL3 (Bernhardt et al., 2003) and the R2R3 MYB protein GL1 or MYB23 (Oppenheimer et al., 1991; Kirik et al., 2005; Morohashi and Grotewold, 2009), also mediates trichome development (Payne et al., 2000; Zhao et al., 2008; Pesch and Hülskamp, 2009). Trichomes are fine structures differentiated from epidermal cells in the aerial part of plant (Ishida et al., 2008), which assist seed dispersal (Wagner et al., 2004) and function as barriers to protect plants against herbivores, insects, abiotic damage, UV irradiation, and excessive transpiration (Wagner et al., 2004; Ishida et al., 2008). Trichome formation is initiated by various environmental cues, such as wounding and insect attack (Larkin et al., 1996; Serna and Martin, 2006; Yoshida et al., 2009), and by different endogenous developmental signals, including phytohormones, such as gibberellin (Perazza et al., 1998) and jasmonate (JA; Traw and Bergelson, 2003; Li et al., 2004; Yoshida et al., 2009).
JA, including jasmonic acid and its oxylipin derivatives, is an important plant hormone (Creelman and Mullet, 1997; Feussner and Wasternack, 2002) that functions as a regulatory molecule to mediate multiple plant developmental processes (Staswick et al., 1992; Feys et al., 1994; McConn and Browse, 1996; Stintzi and Browse, 2000; Wang et al., 2005; Schommer et al., 2008; Cheng et al., 2009; Ren et al., 2009; Sun et al., 2009; Shan et al., 2011) and also acts as a defense signal to mediate plant resistance against insect attack, UV damage, pathogen infection, wounding, and many other abiotic stresses (Howe et al., 1996; McConn et al., 1997; Vijayan et al., 1998; Rao et al., 2000; Reymond et al., 2000; Farmer, 2001; Nibbe et al., 2002; Xiao et al., 2004; Schilmiller and Howe, 2005; Wasternack, 2007; Kim et al., 2009). The F-box protein Coronatine Insensitive1 (COI1) associates with ASK1/ASK2, Cullin1, and Rbx1 to form the SCFCOI1 complex that mediates the diverse JA responses (Xie et al., 1998; Xu et al., 2002; Liu et al., 2004; Ren et al., 2005). The JA-ZIM-domain (JAZ) proteins, substrates of the SCFCOI1 complex, contain 12 members and function as negative regulators to repress diverse JA responses, probably by directly inhibiting various transcriptional regulators (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Fonseca et al., 2009a; Seo et al., 2011; Song et al., 2011). Upon perception of a JA signal, COI1 recruits JAZ proteins for ubiquitination and subsequent degradation through the 26S proteasome to activate downstream signal cascades that modulate JA-mediated plant responses (Chini et al., 2007; Thines et al., 2007; Melotto et al., 2008; Fonseca et al., 2009b; Yan et al., 2009; Sheard et al., 2010).
Previous studies showed that JA can induce anthocyanin accumulation (Franceschi and Grimes, 1991; Tamari et al., 1995) and trichome initiation (Traw and Bergelson, 2003; Li et al., 2004; Maes et al., 2008). JA upregulates several anthocyanin biosynthetic genes that are essential for anthocyanin accumulation (Loreti et al., 2008; Shan et al., 2009) and enhances expression of GL3, which is required for trichome formation (Maes et al., 2008; Yoshida et al., 2009). However, the molecular mechanism of how JA regulates anthocyanin biosynthesis and trichome initiation is largely unknown. In this study, we demonstrate that JAZ proteins directly interact with bHLH members (GL3, EGL3, and TT8) and MYB factors (MYB75 and GL1) of WD-repeat/bHLH/MYB complexes, attenuate their transcriptional function, and subsequently repress anthocyanin biosynthesis and trichome initiation. We speculate that, upon degradation of JAZ proteins in response to a JA signal, the WD-repeat/bHLH/MYB complexes are released to activate downstream signal cascades and mediate JA-induced anthocyanin biosynthesis and trichome initiation.
RESULTS
JAZ Proteins Interact with the bHLH Transcription Factors GL3, EGL3, and TT8 and the MYB Factors MYB75 and GL1
We fused JAZ1 with the LexA DNA binding domain (BD) to screen an Arabidopsis cDNA library for identification of JAZ-interacting proteins in the yeast two-hybrid (Y2H) system. Sequence analysis of putative colonies showed that a bHLH transcription factor, GL3, and a MYB transcription factor, MYB75, interacted with JAZ1 in yeast.
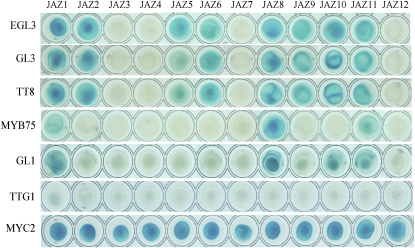
Previous studies demonstrated that the bHLH factors, including GL3 and its closely related members EGL3 and TT8 (Pires and Dolan, 2010), the R2R3 MYB factors, including MYB75 and its closely related members MYB90, MYB113, MYB114, and GL1 (Stracke et al., 2001), and a WD-repeat protein, TTG1, form WD-repeat/bHLH/MYB complexes to mediate diverse plant responses (Zimmermann et al., 2004; Gonzalez et al., 2008; Zhao et al., 2008). We used Y2H to investigate interactions of the 12 JAZ proteins with the major members of the WD-repeat/bHLH/MYB complexes, including three bHLH factors (TT8, GL3, and EGL3), two representative MYB factors (MYB75 and GL1), and the WD-repeat protein TTG1. Positive interactions of MYC2 (Lorenzo et al., 2004; Dombrecht et al., 2007) with 12 JAZ proteins (Figure 1) (Chini et al., 2009) and immunoblot analysis of the yeast strains (see Supplemental Figure 1 online) confirmed that all 12 JAZ fusion proteins were expressed in yeasts. As shown in Figure 1, the three bHLH transcription factors EGL3, GL3, and TT8 strongly interacted with eight JAZ proteins (JAZ1, JAZ2, JAZ5, JAZ6, JAZ8, JAZ9, JAZ10, and JAZ11) but not with the remaining four JAZ proteins (JAZ3, JAZ4, JAZ7, and JAZ12). The two MYB factors MYB75 and GL1 exhibited interactions with JAZ1, JAZ8, and JAZ11. GL1 exhibited additional interaction with JAZ10. However, the WD-repeat protein TTG1 did not interact with any JAZ protein in yeast. In conclusion, these results imply that several JAZ proteins may physically interact with the bHLH and MYB members but not with the WD-repeat protein from WD-repeat/bHLH/MYB complexes.
Figure 1.
Arabidopsis JAZ Proteins Interact with EGL3, GL3, TT8, MYB75, and GL1 in Yeast.
Y2H assays to test the interactions of the 12 JAZ proteins with EGL3, GL3, TT8, MYB75, GL1, TTG1, and MYC2. The 12 JAZ proteins were fused with the LexA DNA binding domain. EGL3, GL3, TT8, MYB75, GL1, TTG1, and MYC2 were fused with activation domain. MYC2 was used as a positive control (Chini et al., 2009). Protein–protein interactions were assessed on SD/Gal/Raf/X-gal (-Ura/-His/-Trp/-Leu) medium in 96-well plates.
[See online article for color version of this figure.]
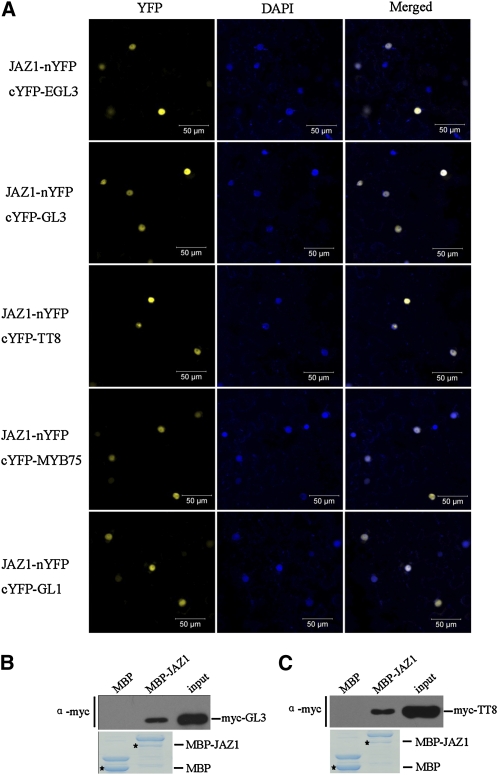
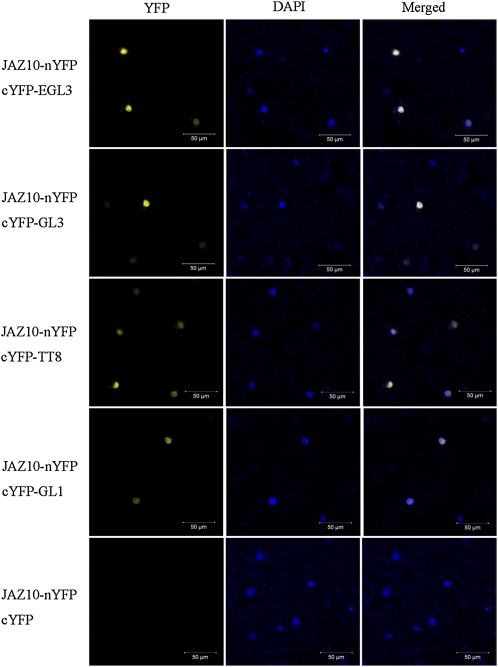
We next adopted bimolecular fluorescence complementation (BiFC) assays (Weinthal and Tzfira, 2009) to verify interactions of JAZ proteins with the bHLH (GL3, EGL3, and TT8) and MYB members (MYB75 and GL1) in planta. The N-terminal fragment of yellow fluorescent protein (nYFP) was ligated with JAZ1 and JAZ10 to produce JAZ1-nYFP and JAZ10-nYFP. The bHLH and MYB members were individually fused with the C-terminal fragment of YFP (cYFP). JAZ1-nYFP was transiently coexpressed with each bHLH or MYB member in Nicotiana benthamiana leaves. We found that coexpression of JAZ1-nYFP with cYFP-EGL3 resulted in strong YFP fluorescence in the nucleus of epidermal cells in N. benthamiana leaves (Figure 2A), whereas no YFP fluorescence was detected in negative controls (JAZ1-nYFP coexpressed with cYFP or nYFP coexpressed with cYFP-EGL3) (see Supplemental Figure 2 online). Similar results were observed for coexpression of JAZ1-nYFP with GL3, TT8, MYB75, and GL1 cYFP fusions (Figure 2A; see Supplemental Figure 2 online) and for coexpression of JAZ10-nYFP with EGL3, GL3, TT8, and GL1 cYFP fusions (Figure 3).
Figure 2.
Arabidopsis JAZ1 Interacts with EGL3, GL3, TT8, MYB75, and GL1.
(A) BiFC assay to detect the interactions of JAZ1 (fused with N-terminal fragment of YFP) with EGL3, GL3, TT8, MYB75, and GL1 (fused with C-terminal fragment of YFP). Construct pairs indicated on the left were coexpressed in leaves of N. benthamiana. YFP fluorescence was detected 50 h after infiltration. The nuclei are indicated by DAPI staining.
(B) In vitro pull-down assay to verify the interaction of GL3 with JAZ1. Purified MBP and the MBP-JAZ1 fusion protein were incubated with the myc-GL3 expressed in leaves of N. benthamiana. Bound proteins were washed, separated on SDS-PAGE, and immunoblotted with the anti-c-myc antibody (α-myc, top panel). The input lane shows the expression level of myc-GL3 expressed in leaves of N. benthamiana. The positions of purified MBP and MBP-JAZ1 separated on SDS-PAGE are indicated with an asterisk (bottom panel; stained by Coomassie blue).
(C) In vitro pull-down assay to verify the interaction of TT8 with JAZ1.The operational procedure was the same as (B), except that myc-TT8 was used instead of myc-GL3.
Figure 3.
Arabidopsis JAZ10 Interacts with EGL3, GL3, TT8, and GL1.
BiFC assay to detect the interactions of JAZ10 (fused with nYFP) with EGL3, GL3, TT8, and GL1 (fused with cYFP). Construct pairs indicated on the left were coexpressed in leaves of N. benthamiana. YFP fluorescence was detected 50 h after infiltration. The nuclei are indicated by DAPI staining.
We further applied pull-down assays to verify the JAZ interactions with these transcription factors in vitro. Maltose binding protein (MBP) and MBP-fused JAZ1 (MBP-JAZ1) were expressed in Escherichia coli and purified using amylose resin. Two transcription factors, GL3 and TT8, were fused with six myc tags (myc-GL3 and myc-TT8) and expressed in N. benthamiana leaves. The MBP-JAZ1 resin was incubated with the N. benthamiana leaves extracts expressing myc-GL3 (Figure 2B) or myc-TT8 (Figure 2C) and then separated on SDS-PAGE for immunoblotting with anti-c-myc antibody. As shown in Figure 2, the negative control (MBP resin) was unable to pull down myc-GL3 (Figure 2B, top panel) or myc-TT8 (Figure 2C, top panel), whereas the myc-TT8 and myc-GL3 were efficiently retained by the MBP-JAZ1 resin, suggesting that JAZ1 physically interacts with GL3 (Figure 2B, top panel) and TT8 (Figure 2C, top panel) in vitro.
Taken together, the Y2H assays, BiFC assays, and pull-down assays consistently demonstrate that the bHLH (GL3, EGL3, and TT8) and MYB members (MYB75 and GL1), which belong to different transcription factor families, interact with JAZ proteins, implying that these transcription factors may function as direct targets of JAZ proteins.
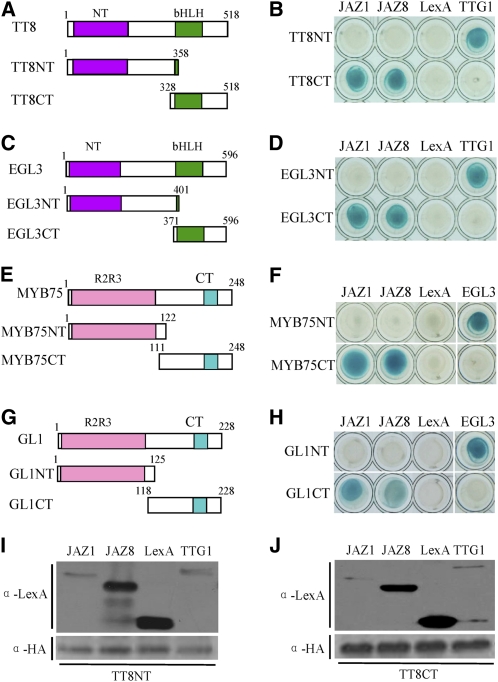
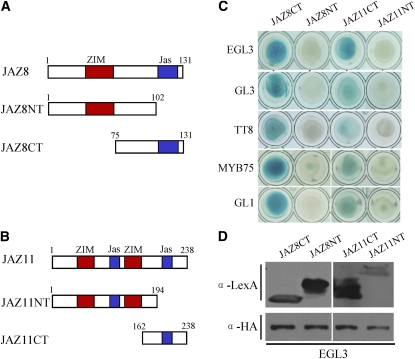
C-Terminal Domains of EGL3, TT8, MYB75, and GL1 Are Involved in Interactions with JAZ1 and JAZ8
Previous studies showed that the N-terminal domain of the bHLH factor MYC2 interacts with JAZ proteins (Chini et al., 2007). We examined which domains of the bHLH and MYB members in the WD-repeat/bHLH/MYB complex were responsible for interaction with JAZ proteins. The bHLH members (TT8 and EGL3) were divided into N-terminal fragments (TT8NT and EGL3NT) and C-terminal fragments (TT8CT and EGL3CT) containing the bHLH DNA binding domain (Figures 4A and 4C). The MYB members (MYB75 and GL1) were similarly divided into N-terminal fragments, containing the R2R3 DNA binding domain, and C-terminal fragments to produce MYB75NT, GL1NT, MYB75CT, and GL1CT, respectively (Figures 4E and 4G).
Figure 4.
C-Terminal Domains in TT8, EGL3, MYB75, and GL1 Are Involved in the Interactions with JAZ1 and JAZ8.
(A) and (C) Schematic diagrams show domain constructs of TT8 (A) and EGL3 (C). The diagrams display the conserved NT domain and bHLH domain. The numbers indicate positions of the first and the last amino acid of the domain constructs.
(E) and (G) Schematic diagrams show domain constructs of MYB75 (E) and GL1 (G). The diagrams display the conserved R2R3 domain and CT domain.
(B) and (D) Y2H assays to detect the interactions of JAZ1 and JAZ8 with different domains of TT8 (TT8NT and TT8CT fused with AD domain) (B) and EGL3 (EGL3NT and EGL3CT fused with AD domain) (D). pLexA vector (LexA) was used as a negative control. TTG1, which can interact with the N terminus of these bHLH factors (Payne et al., 2000), was used as a positive control.
(F) and (H) Y2H assays to test the interactions of JAZ1 and JAZ8 with different domains of MYB75 (MYB75NT and MYB75CT fused with AD domain) (F) and GL1 (GL1NT and GL1CT fused with AD domain) (H). pLexA vector (LexA) was used as a negative control. EGL3, which can interact with these MYB factors (Zimmermann et al., 2004), was used as a positive control.
(I) Expression of JAZ1, JAZ8, TTG1, and TT8NT was detected in yeast strains used for Y2H assays shown in the top panel of (B). JAZ1, JAZ8, TTG1, and LexA were detected with anti-LexA antibody (top). TT8NT was detected with anti-HA antibody (bottom).
(J) Expression of JAZ1, JAZ8, TTG1, and TT8CT was detected in yeast strains used for Y2H assays shown in the bottom panel of (B). JAZ1, JAZ8, TTG1, and LexA were detected with anti-LexA antibody (top). TT8CT was detected with anti-HA antibody (bottom).
[See online article for color version of this figure.]
As shown in Figure 4, all the C-terminal fragments (TT8CT, EGL3CT, MYB75CT, and GL1CT), but no N-terminal fragments (TT8NT, EGL3NT, MYB75NT, and GL1NT) in these transcription factors, exhibited interactions with JAZ1 and JAZ8 in yeast. These results suggest that the C-terminal domains of EGL3, TT8, MYB75, and GL1 are responsible for interaction with JAZ proteins. This is different from the interaction of JAZ proteins with N-terminal domain of MYC2, which belongs to different bHLH subfamily (Toledo-Ortiz et al., 2003).
The C-Terminal Jas Domain in JAZ8 and JAZ11 Is Responsible for Interactions with TT8, GL3, EGL3, MYB75, and GL1
To investigate which domain of JAZ proteins was responsible for interactions with the bHLH and MYB members, we divided JAZ8 and JAZ11 into N-terminal (JAZ8NT and JAZ11NT) and C-terminal fragments (JAZ8CT and JAZ11CT). All the C-terminal fragments contained a Jas domain. JAZ8NT contained a ZIM domain, whereas JAZ11NT harbored two ZIM domains and one Jas domain (Figures 5A and 5B) (Chini et al., 2007; Thines et al., 2007; Katsir et al., 2008a). We found that GL3, EGL3, TT8, MYB75, and GL1 interacted with C-terminal fragments but not with N-terminal fragments of JAZ8 and JAZ11 in yeast (Figure 5). Consistent with previous data (Chini et al., 2007), these results indicate that the Jas domain in JAZ8 and JAZ11 is responsible for protein–protein interactions between JAZs and their downstream transcription factors. Our results further indicate that, in JAZ11, the Jas domain nearer to the N terminus is not involved in the interactions with EGL3, GL3, TT8, MYB75, and GL1.
Figure 5.
The C-Terminal Regions in JAZ8 and JAZ11 Are Responsible for Interactions with EGL3, GL3, TT8, MYB75, and GL1.
(A) and (B) Schematic diagrams show domain constructs of JAZ8 and JAZ11. The diagrams display the conserved ZIM domain and Jas domain.
(C) Y2H assays to assess the interactions of EGL3, GL3, TT8, MYB75, and GL1 with different domains of JAZ8 and JAZ11 (JAZ8CT, JAZ8NT, JAZ11CT, and JAZ11NT individually fused with the LexA BD domain).
(D) Expression of JAZ8CT, JAZ8NT, JAZ11CT, JAZ11NT, and EGL3 was detected in yeast strains used for Y2H assays shown in the top panel of (C). JAZ8CT, JAZ8NT, JAZ11CT, and JAZ11NT were detected with anti-LexA antibody (top). EGL3 was detected with anti-HA antibody (bottom).
[See online article for color version of this figure.]
JAZ1 Affects the Interactions between bHLH and MYB Proteins
Having demonstrated the interaction of JAZ proteins with the bHLH and MYB transcription factors (Figures 1 to 5), we further examined the effect of JAZ proteins on the interaction of the bHLH proteins (GL3 or TT8) with the MYB proteins (GL1 or MYB75) in the BiFC assay.
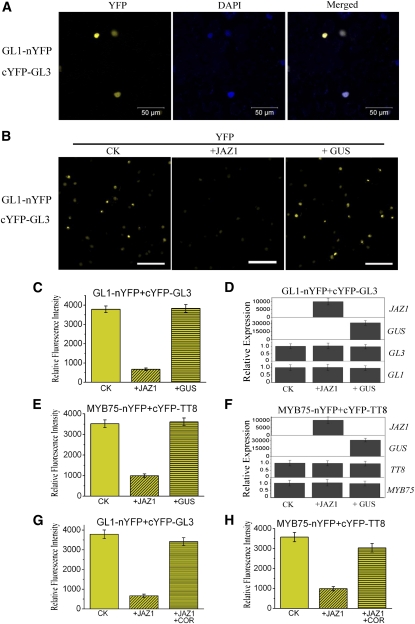
When GL1-nYFP and cYFP-GL3 were transiently coexpressed in N. benthamiana leaves, we detected strong YFP fluorescence in the nucleus of epidermal cells in N. benthamiana leaves (Figures 6A and 6B), consistent with the previous observation that GL1 interacts with GL3 (Payne et al., 2000). When JAZ1 was coexpressed with GL1-nYFP and cYFP-GL3, the fluorescence signal was obviously reduced (Figures 6B to 6D), whereas coexpression of β-glucuronidase (GUS) with GL1-nYFP/cYFP-GL3 did not reduce the fluorescence intensity (Figures 6B to 6D). These results indicate that JAZ1 can affect the interaction, through repression and/or structural modification, between the MYB GL1 and the bHLH GL3, essential components of the GL1/GL3-based WD-repeat/bHLH/MYB complex required for trichome formation.
Figure 6.
Arabidopsis JAZ1 Affects the Interaction of GL3 with GL1 and the Interaction of TT8 with MYB75.
(A) BiFC assays to detect the interaction of GL3 (fused with cYFP) with GL1 (fused with nYFP). cYFP-GL3 and GL1-nYFP were coexpressed in leaves of N. benthamiana. YFP fluorescence was detected 50 h after infiltration. DAPI staining indicates the nuclei.
(B) BiFC assays showing that JAZ1 attenuates the interaction of GL3 with GL1. YFP fluorescence was detected 50 h after coexpression of GL1-nYFP/cYFP-GL3 (CK), JAZ1/GL1-nYFP/cYFP-GL3 (+JAZ1), or GUS/GL1-nYFP/cYFP-GL3 (+GUS). The identical amounts of Agrobacterium tumefaciens strains containing GL1-nYFP and cYFP-GL3 were used in these three combinations (CK, +JAZ1, and +GUS). The three combinations were infiltrated into the same leaf of N. benthamiana and detected under the microscope with identical gain settings (lasers, 514 nm, 8.0%; pinhole, 34 μm; master gain, 732). The experiment was repeated in more than 10 independent biological replicates with similar results. Bar = 100 μm.
(C) Quantitative analysis of YFP fluorescence intensity in (B). Fifty independent fluorescent spots were assessed for fluorescence intensity. Error bars represent se.
(D) Real-time PCR analysis of JAZ1, GUS, cYFP-GL3 (GL3), and GL1-nYFP (GL1) expression in (B). Total RNAs were extracted from leaves of N. benthamiana coinfiltrated with the combinations of various constructs in (B). The primers specific for amplification of GUS, Arabidopsis JAZ1, cYFP-GL3 (GL3), and GL1-nYFP (GL1) are shown in Supplemental Table 1 online. Nicotiana Actin was used as the internal control. Expression levels of genes in the CK treatment were standardized as one unit in the real-time PCR system. Error bars represent se.
(E) JAZ1 attenuates the interaction of TT8 with MYB75. YFP fluorescence was detected 50 h after coexpression of MYB75-nYFP/cYFP-TT8 (CK), JAZ1/MYB75-nYFP/cYFP-TT8 (+JAZ1), or GUS/MYB75-nYFP/cYFP-TT8 (+GUS) in the same leaf of N. benthamiana.
(F) Real-time PCR analysis of JAZ1, GUS, cYFP-TT8 (TT8), and MYB75-nYFP (MYB75) expression in (E). Error bars represent se.
(G) COR attenuates JAZ1 inhibition of the interaction between GL3 and GL1. N. benthamiana leaves were coinfiltrated with Agrobacterium strains carrying the indicated constructs. CK, treatment with buffer on coexpression of GL1-nYFP/cYFP-GL3; +JAZ1, treatment with buffer on coexpression of JAZ1/GL1-nYFP/cYFP-GL3; +JAZ1+COR, treatment with 5 μM COR on coexpression of JAZ1/GL1-nYFP/cYFP-GL3. Fifty independent fluorescent spots were assessed for fluorescence intensity to generate the quantitative data. Error bars represent se.
(H) COR attenuates JAZ1 inhibition of the interaction between MYB75 and TT8. CK, treatment with buffer of coexpression of MYB75-nYFP/cYFP-TT8; +JAZ1, treatment with buffer of coexpression of JAZ1/MYB75-nYFP/cYFP-TT8; +JAZ1+COR, treatment with 5 μM COR of coexpression of JAZ1/MYB75-nYFP/cYFP-TT8.
Similar results were observed for coexpression of JAZ1 with MYB75-nYFP and cYFP-TT8 in N. benthamiana leaves (Figures 6E and 6F; see Supplemental Figure 3 online). The fluorescence signal was obviously reduced by JAZ1, but not by GUS, in BiFC assays (Figures 6E and 6F), which suggests that JAZ1 affects the interaction, through repression and/or structural modification, between MYB75 and TT8, essential components of the MYB75/TT8-based WD-repeat/bHLH/MYB complex required for anthocyanin accumulation.
Having shown that JAZ1 affected the interactions between the bHLH and MYB proteins in the BiFC assays (Figures 6B, 6C, and 6E), we investigated whether exogenous JA treatment could attenuate the JAZ1 repression of those interactions. Consistent with the data shown in Figures 6B and 6C, YFP fluorescence signal was obviously reduced when JAZ1 was coexpressed with GL1-nYFP and cYFP-GL3 (Figure 6G). Furthermore, we found that exogenous application of coronatine, a JA mimic molecule (Katsir et al., 2008b; Fonseca et al., 2009b; Sheard et al., 2010), restored the YFP fluorescence obviously (Figure 6G). Similarly, we observed that coronatine also rescued the JAZ1-repressed YFP fluorescence signal of MYB75-nYFP/cYFP-TT8 (Figure 6H). These results suggest that JA can reverse the JAZ1-mediated interference with the interaction between the bHLH and MYB proteins in these BiFC assays, probably by inducing the depletion of the transiently expressed Arabidopsis JAZ1 via the COI1 homolog and 26S proteasome of N. benthamiana.
JAZ1 Represses the Transcriptional Function of the bHLH Factor TT8 and MYB Member MYB75
DFR, the anthocyanin biosynthetic gene, is a direct target of the WD-repeat/bHLH/MYB complex (Nesi et al., 2000; Zhang et al., 2003; Gonzalez et al., 2008; Matsui et al., 2008). We further verified effect of JAZ1 on the transcriptional function of TT8 and MYB75 using the DFR promoter-driven luciferase reporter in a well-established transient expression assay.
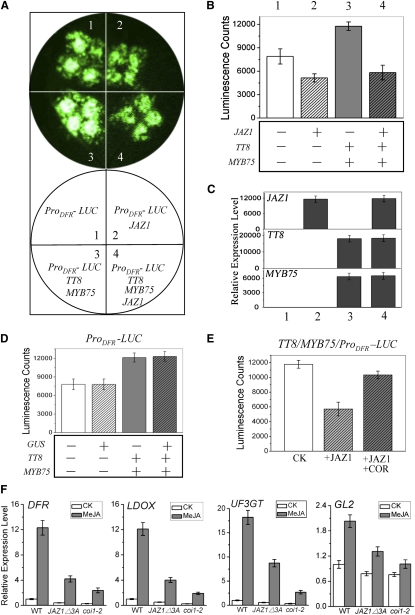
When the promoter of DFR was fused with luciferase (LUC) reporter gene (ProDFR-LUC) and infiltrated into N. benthamiana leaves, we observed a certain amount of LUC activity that was reflected by luminescence intensity (Figure 7A), indicating that endogenous bHLH/MYB proteins of N. benthamiana may activate expression of ProDFR-LUC. Coexpression of ProDFR-LUC with TT8 and MYB75 generated much stronger luminescence intensity in the N. benthamiana leaves (Figures 7A to 7C). These results indicate that TT8 and MYB75 positively regulate the expression of DFR, consistent with previous research (Zimmermann et al., 2004; Matsui et al., 2008).
Figure 7.
Arabidopsis JAZ1 Represses the Transcriptional Function of TT8 and MYB75.
(A) Transient expression assays show that JAZ1 represses the transcriptional function of TT8 and MYB75. Luminescence imaging of N. benthamiana leaves is shown 50 h after coinfiltration with the constructs indicated in the bottom circle.
(B) Quantitative analysis of luminescence intensity in (A). Five independent determinations were assessed. Error bars represent se.
(C) Real-time PCR analysis of JAZ1, TT8, and MYB75 in (A). Total RNAs were extracted from leaves of N. benthamiana coinfiltrated with the constructs in (A). The primers specific for amplification of the Arabidopsis JAZ1, TT8, and MYB75 are shown in Supplemental Table 1 online. Nicotiana Actin was used as the internal control. Error bars represent se.
(D) GUS has no effect on the transcriptional function of TT8 and MYB75. LUC activity was detected 50 h after coinfiltration of constructs as indicated. Quantitative analysis of luminescence intensity is presented. Error bars represent se.
(E) COR attenuates JAZ1 inhibition of the transcriptional function of TT8 and MYB75. N. benthamiana leaves were coinfiltrated with Agrobacterium strains carrying the indicated constructs. CK, buffer treatment of coinfiltration of TT8/MYB75/ProDFR–LUC; +JAZ1, buffer treatment of coinfiltration of JAZ1/TT8/MYB75/ProDFR–LUC; +JAZ1+COR, treatment with 5 μM COR of coinfiltration of JAZ1/TT8/MYB75/ProDFR–LUC. Quantitative analysis of luminescence intensity is presented. Error bars represent se.
(F) Real-time PCR analysis for DFR, LDOX, UF3GT, and GL2 in Arabidopsis wild-type (WT), JAZ1△3A, and coi1-2 plants treated without (CK) or with 100 μM MeJA for 8 h. ACTIN8 was used as the internal control. Error bars represent se.
[See online article for color version of this figure.]
When ProDFR-LUC was coinfiltrated in N. benthamiana leaves with JAZ1 driven by the 35S cauliflower mosaic virus (CaMV35S) promoter, the LUC activity was obviously reduced in comparison with infiltration of ProDFR-LUC alone or coinfiltration of ProDFR-LUC and the GUS gene driven by the CaMV35S promoter (Figures 7A to 7D), indicating that JAZ1 may repress the transcriptional function of endogenous N. benthamiana bHLH/MYB proteins. Furthermore, coexpression of JAZ1 with ProDFR-LUC, TT8, and MYB75 dramatically reduced the luminescence intensity compared with the coexpression of ProDFR-LUC, TT8, and MYB75 without JAZ1 or with GUS (Figures 7A to 7D; see Supplemental Figure 4 online), demonstrating that JAZ1 represses the transcriptional function of TT8 and MYB75. In addition, we applied coronatine, the JA mimic molecule, onto the N. benthamiana leaf area coexpressing JAZ1/TT8/MYB75/ProDFR-LUC to investigate whether JA could attenuate the JAZ1 repression and restore the expression level of ProDFR-LUC. As shown in Figure 7E, treatment with coronatine (COR) obviously increased the luminescence intensity, suggesting that JA can reverse the JAZ1-mediated interference with the transcriptional function of TT8 and MYB75. Taken together, these results demonstrate that JAZ1 represses the transcriptional function of the bHLH factor TT8 and MYB member MYB75, thereby attenuating expression of ProDFR-LUC in the transient assay.
To examine further whether accumulation of endogenous JAZ proteins could repress the expression of endogenous DFR in planta, we detected the expression levels of DFR and other target genes of the TT8/MYB75-based complex in the Arabidopsis dominant-negative transgenic plant JAZ1△3A and the coi1-2 mutant, both of which highly accumulate JAZ proteins (Thines et al., 2007; Zhang and Turner, 2008; Chini et al., 2009; Chung and Howe, 2009). The results in Figure 7F showed that the JAZ1△3A transgenic plants and the coi1-2 mutant exhibited obvious repression of DFR, LDOX, and UF3GT, the anthocyanin biosynthetic genes that are direct targets of the TT8/MYB75-based WD-repeat/bHLH/MYB complex (Zhang et al., 2003; Tohge et al., 2005; Gonzalez et al., 2008). In addition, we found that JAZ1△3A and coi1-2 also exhibited obvious repression of the trichome initiation factor GL2 (Figure 7F), a target gene of the GL1/GL3-based WD-repeat/bHLH/MYB complex (Zhang et al., 2003; Morohashi and Grotewold, 2009). These results suggest accumulation of endogenous JAZ proteins represses the respective target genes of MYB75/TT8- and GL1/GL3-based WD-repeat/bHLH/MYB complexes in planta.
The WD-Repeat/bHLH/MYB Complex Is Involved in JA-Regulated Anthocyanin Accumulation and Trichome Initiation
It has been reported that, in Arabidopsis, bHLH factors (GL3, EGL3, and TT8), MYB factors, including MYB75, and the WD-repeat protein TTG1 form the WD-repeat/bHLH/MYB complex to mediate anthocyanin accumulation (Zhang et al., 2003; Baudry et al., 2006). The bHLH factors (GL3 and EGL3) and TTG1 also form an alternative WD-repeat/bHLH/MYB complex with the MYB member GL1 to mediate trichome initiation (Payne et al., 2000; Bernhardt et al., 2003). We further investigated the role of these WD-repeat/bHLH/MYB complexes in JA-regulated anthocyanin accumulation and trichome initiation.
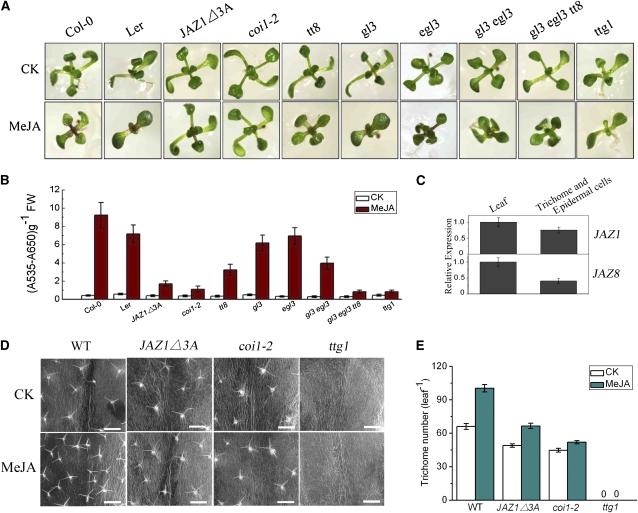
The Arabidopsis WD-repeat/bHLH/MYB mutants, the coi1-2 mutant (Xu et al., 2002), and the JAZ1△3A transgenic line (Thines et al., 2007) were used for measurement of JA-induced anthocyanin accumulation and trichome initiation. As shown in Figure 8, the anthocyanin content in wild-type seedlings was obviously upregulated when they were treated with JA. However, JA-induced anthocyanin accumulation decreased in the WD-repeat/bHLH/MYB mutants compared with that in the wild type (Figures 8A and 8B). When treated with methyl jasmonate (MeJA), the gl3 egl3 double mutant exhibited an obvious reduction in anthocyanin accumulation compared with the gl3 or egl3 single mutants (Figures 8A and 8B); the gl3 egl3 tt8 triple mutant displayed much lower anthocyanin contents compared with the gl3 egl3 double mutant (Figures 8A and 8B), indicating that GL3, EGL3, and TT8 function redundantly in mediating JA-induced anthocyanin accumulation. In the ttg1 mutant, JA-induced anthocyanin accumulation was severely disrupted (Figures 8A and 8B), indicating that the single WD-repeat protein TTG1 is indispensable and essential for the integrity of the WD-repeat/bHLH/MYB complex (Zhao et al., 2008). These results suggest that the WD-repeat/bHLH/MYB complex is involved in JA-regulated anthocyanin accumulation.
Figure 8.
WD-Repeat/bHLH/MYB Complexes Are Required for JA-Regulated Anthocyanin Accumulation and Trichome Initiation.
(A) Eleven-day-old Arabidopsis seedlings of wild-type (Col-0 and Landsberg erecta), JAZ1△3A, coi1-2, tt8, gl3, egl3, gl3 egl3, gl3 egl3 tt8, and ttg1 grown on MS medium supplied without (CK) or with 25 μM MeJA.
(B) Anthocyanin contents of the seedlings in (A). FW, fresh weight. Error bars represent se (n = 3).
(C) Real-time PCR analysis of JAZ1 and JAZ8 expression in trichomes and epidermal cells. The trichomes and epidermal cells were isolated from Arabidopsis leaves using sharp forceps under a microscope. ACTIN8 was used as the internal control. Expression levels of the genes in leaves were standardized as one unit in the real-time PCR analysis. Error bars represent se.
(D) Images of trichome in wild-type Col-0 (WT), JAZ1△3A, coi1-2, and ttg1 plants treated without (CK) or with MeJA. The fifth true leaves were chosen to observe trichomes using an environmental scanning electron microscope. No trichomes were observed in ttg1. Bars = 500 μm.
(E) Total trichome number in the fifth true leaves described in (D). Error bars represent se (n ≥ 12).
We further measured total trichome number per leaf in ttg1, gl1, and the gl3 egl3 double mutant in response to JA induction. We found that JA-induced trichome formation was abolished in ttg1 (Figures 8D and 8E), whereas an obvious increase of trichome numbers was detected in JA-treated wild-type (Traw and Bergelson, 2003; Maes et al., 2008; Yoshida et al., 2009). Similar to previous observations (Maes et al., 2008; Yoshida et al., 2009), we also noticed that the JA-induced trichome initiation was disrupted in gl1 and the gl3 egl3 double mutant. These results suggest that the WD-repeat/bHLH/MYB complex is also involved in JA-regulated trichome initiation.
Consistent with our previous observation (Shan et al., 2009), we found that the coi1-2 mutant failed to accumulate anthocyanin in response to JA treatment (Figures 8A and 8B). Compared with the wild type, the trichome number in the coi1-2 mutant was also reduced obviously. In response to JA induction, trichome formation was dramatically attenuated in the coi1-2 mutant (Figures 8D and 8E) (Yoshida et al., 2009). These results indicate that JA regulates anthocyanin accumulation and trichome initiation in a COI1-dependent manner.
In the JAZ1△3A transgenic plants, JA-induced anthocyanin accumulation was largely decreased in comparison with that in the wild type (Figures 8A and 8B). Similarly, trichome formation, when treated with or without JA, was clearly reduced in the JAZ1△3A transgenic plants (Figures 8D and 8E). These results suggest that JAZ1△3A dominantly represses JA-regulated anthocyanin accumulation and trichome initiation. Consistent with the role for JAZ in repressing anthocyanin accumulation, expression of some JAZ genes, including JAZ1 and JAZ8, was detected in the leaves by quantitative real-time PCR (Figure 8C). We also used sharp forceps, under a microscope, to harvest trichomes and the leaf epidermal cells where trichomes are differentiated for quantitative real-time PCR analysis: the expression of JAZ1 and JAZ8 was also detected, consistent with a role for JAZ in affecting trichome initiation (Figure 8C). As the level of JAZ transcripts is not necessarily correlated with the level of JAZ proteins (Thines et al., 2007), the expression levels of JAZ proteins in trichomes and epidermal cells remain to be determined.
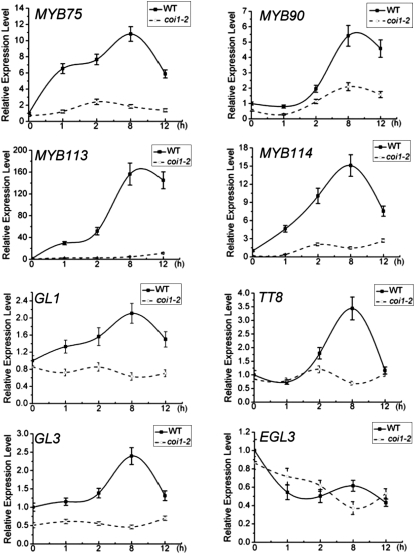
Previous studies showed that MYC2 (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007), a target gene of JAZ proteins (Chini et al., 2007), is rapidly induced upon JA treatment (Chung et al., 2008; Pauwels et al., 2008). We found that, among the WD-repeat/bHLH/MYB components, MYB113, MYB75, and MYB114 were rapidly upregulated to ~29.6-, 6.5-, and 4.6-fold in response to JA induction within 60 min (Figure 9); TT8, GL3, MYB90, and GL1 were gradually induced by JA within 8 h (Figure 9); however, EGL3 was not obviously upregulated by JA treatment (Figure 9). We further found that JA-induced expression of the bHLH factors TT8 and GL3 and all the MYB members (MYB113, MYB75, MYB114, MYB90, and GL1) was severely attenuated in coi1-2 (Figure 9), suggesting that expression of these WD-repeat/bHLH/MYB components is regulated by JA in a COI1-dependent manner.
Figure 9.
Real-Time PCR Analysis for Gene Expression of Transcription Factors in the WD-Repeat/bHLH/MYB Complex.
Real-time PCR analysis of MYB75, MYB90, MYB113, MYB114, GL1, TT8, GL3, and EGL3 in the wild type (WT) and coi1-2 treated with 100 μM MeJA for 0, 1, 2, 8, and 12 h. ACTIN8 was used as the internal control. Error bars represent se.
Overexpression of MYB75 Rescues Anthocyanin Accumulation in the coi1 Mutant
The JAZ proteins, which accumulate to high levels in the coi1 mutant (Thines et al., 2007), interact with their downstream transcription regulators, including MYC2 (Chini et al., 2007), to repress JA responses (Katsir et al., 2008a; Fonseca et al., 2009a). The accumulated JAZ proteins in the coi1 mutant may attenuate transcriptional function of bHLH and MYB factors, including MYB75, TT8, GL3, and GL1 (Figures 6 and 7), essential components of the WD-repeat/bHLH/MYB complexes required for anthocyanin accumulation and trichome formation. We examined whether overexpression of MYB75 could rescue anthocyanin accumulation in the coi1 mutant.
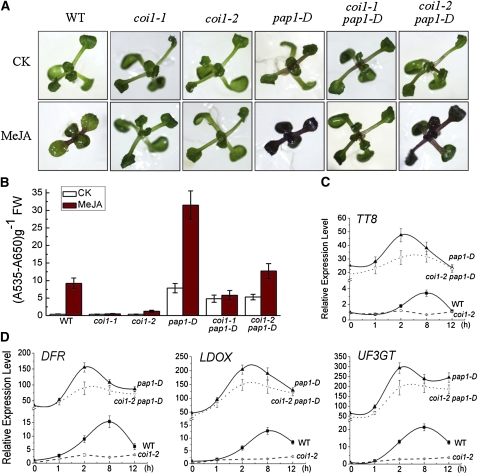
The transgenic line pap1-D overexpressing MYB75 (alternatively named as PAP1) (Borevitz et al., 2000) was crossed with coi1-1 and coi1-2 to generate the coi1-1 pap1-D and coi1-2 pap1-D plants with high expression of MYB75. As expected, the anthocyanin content levels were low in untreated wild-type seedlings and in the coi1 seedlings with or without JA induction but high in the JA-treated wild-type seedlings (Figures 10A and 10B). The transgenic pap1-D seedlings accumulated anthocyanin at high levels even without JA induction and at very high levels in response to JA induction (Figures 10A and 10B).
Figure 10.
Overexpression of MYB75 Rescues Anthocyanin Accumulation in coi1 Mutants.
(A) Eleven-day-old seedlings of the wild type (WT), coi1-1, coi1-2, pap1-D, coi1-1 pap1-D, and coi1-2 pap1-D grown on MS medium supplied without (CK) or with 25 μM MeJA. pap1-D (Borevitz et al., 2000) is a T-DNA activation line with overexpression of MYB75 driven by four CaMV35S promoters.
(B) Anthocyanin contents of seedlings in (A). FW, fresh weight. Error bars represent se (n = 3).
(C) Real-time PCR analysis for TT8 in the wild type, coi1-2, pap1-D, and coi1-2 pap1-D treated with 100 μM MeJA for 0, 1, 2, 8, and 12 h. ACTIN8 was used as the internal control. Error bars represent se.
(D) Real-time PCR analysis for DFR, LDOX, and UF3GT in the wild type, coi1-2, pap1-D, and coi1-2 pap1-D treated with 100 μM MeJA for 0, 1, 2, 8, and 12 h. ACTIN8 was used as the internal control. Error bars represent se.
As shown in Figure 10, overexpression of MYB75 in the coi1 background largely rescued anthocyanin accumulation. coi1-1 pap1-D and coi1-2 pap1-D constitutively accumulated anthocyanin at high levels. We noticed that coi1-2 pap1-D showed increased anthocyanin accumulation in response to JA treatment, although the level remained much lower than that in JA-treated pap1-D seedlings. The responsiveness to JA signal in coi1-2 pap1-D probably resulted from the leakiness of coi1-2, a weak allele of COI1 (Xu et al., 2002). In conclusion, these results demonstrate that overexpression of MYB75 suppresses the defect of anthocyanin accumulation in the coi1 mutants.
Consistent with the anthocyanin accumulation results, the anthocyanin biosynthetic genes DFR, LDOX, and UF3GT in coi1-2 pap1-D were upregulated to high levels (Figure 10D). In coi1-2 pap1-D, but not in coi1-2, a high expression level was detected for TT8 (Figure 10C), a bHLH member of the WD-repeat/bHLH/MYB complex that regulates expression of several anthocyanin biosynthetic genes (Nesi et al., 2000; Baudry et al., 2004). These data showed that overexpression of MYB75 obviously suppressed the defect in JA-induced expression of the bHLH factor TT8 and the anthocyanin biosynthetic genes (DFR, LDOX, and UF3GT) in coi1-2.
Taken together, our results suggest that MYB75 acts downstream of COI1 to regulate JA-induced anthocyanin accumulation positively and that overexpression of MYB75 restores anthocyanin accumulation in the coi1 mutants.
Overexpression of EGL3 or GL3 Rescues Trichome Initiation in coi1-2
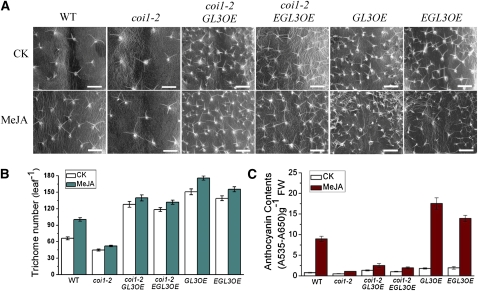
GL3 and EGL3 play a dual role in regulation of JA-induced trichome initiation and anthocyanin accumulation (Zhang et al., 2003; Yoshida et al., 2009). We induced overexpression of GL3 and EGL3 in the coi1-2 background to examine the effects of elevated GL3 and EGL3 on trichome initiation and anthocyanin accumulation.
As shown in Figures 11A and 11B, the trichome number in coi1-2 was reduced compared with that in the wild type, and the JA induction of trichomes was severely attenuated in coi1-2. The transgenic lines coi1-2 GL3OE and coi1-2 EGL3OE that overexpress GL3 or EGL3 (see Supplemental Figure 5 online) obviously activated trichome initiation even without JA induction (Figures 11A and 11B). Overexpression of GL3 or EGL3 in the wild type resulted in a dramatic increase in trichome formation under all circumstances without (Morohashi et al., 2007) or with JA induction (Figures 11A and 11B). These results suggest that GL3 and EGL3 act downstream of COI1 to regulate JA-induced trichome initiation positively and that overexpression of GL3 or EGL3 restores trichome initiation in coi1-2.
Figure 11.
Overexpression of GL3 and EGL3 Rescues Trichome Initiation in coi1-2.
(A) Images of trichomes in the wild type (WT), coi1-2, coi1-2 GL3OE (overexpression of GL3 in coi1-2), coi1-2 EGL3OE (overexpression of EGL3 in coi1-2), GL3OE (overexpression of GL3 in wild-type), and EGL3OE (overexpression of EGL3 in wild-type) treated without (CK) or with 150 nM MeJA. The fifth true leaves were observed using environmental scanning electron microscopy. Bars = 500 μm.
(B) Total trichome number in the fifth true leaves described in (A). Error bars represent se (n ≥ 12).
(C) Anthocyanin contents of 11-d-old seedlings of the wild type, coi1-2, coi1-2 GL3OE, coi1-2 EGL3OE, GL3OE, and EGL3OE grown on MS medium supplied without (CK) or with 25 μM MeJA. FW, fresh weight. Error bars represent se (n = 3).
[See online article for color version of this figure.]
As the transcription factors GL3 and EGL3 were found to regulate both trichome initiation and anthocyanin accumulation (Zhang et al., 2003; Broun, 2005), we further investigated whether overexpression of GL3 or EGL3 could rescue anthocyanin accumulation in the coi1-2 mutant. As expected, an obvious increase in anthocyanin accumulation, in response to JA induction, was detected in the wild type but not in coi1-2 (Figures 10 and 11C). The transgenic lines coi1-2 GL3OE and coi1-2 EGL3OE exhibited a mild increase in anthocyanin compared with coi1-2 (Figure 11C), indicating that overexpression of GL3 or EGL3 could slightly restore anthocyanin accumulation in coi1-2. However, no obvious increase in anthocyanin was detected in coi1-2 GL3OE and coi1-2 EGL3OE compared with the obvious restoration caused by the MYB75 overexpression in coi1-2 pap1-D (Figures 10 and 11C). Similar data were observed for overexpression of GL3 or EGL3 in the wild type: the transgenic lines GL3OE and EGL3OE exhibited a mild increase in anthocyanin but not as strong as that in the pap1-D transgenic plant with overexpression of MYB75 (Figures 10 and 11C).
Taken together, overexpression of GL3 or EGL3 was able to rescue trichome initiation dramatically (Figures 11A and 11B) and restore anthocyanin accumulation in coi1-2 slightly but not dramatically (Figure 11C). These results imply that GL3 and EGL3 may activate distinct downstream cascades and different feedback loops to regulate trichome initiation and anthocyanin accumulation separately.
DISCUSSION
The JAZ proteins (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007) are speculated to repress JA-regulated plant responses through interaction and attenuation of their downstream transcription factors (Katsir et al., 2008a; Chung et al., 2009; Fonseca et al., 2009a). The bHLH transcription factors MYC2/3/4 and R2R3 transcription factors MYB21/24 act as direct targets of JAZ proteins (Chini et al., 2007; Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011; Song et al., 2011) to mediate JA-regulated root growth inhibition and stamen development, respectively (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007; Song et al., 2011). Here, we identified three bHLH transcription factors (TT8, GL3, and EGL3) and two MYB transcription factors (MYB75 and GL1), essential components of WD-repeat/bHLH/MYB complexes, as new targets of JAZ proteins. We also showed that JAZ proteins attenuate the transcriptional function of WD-repeat/bHLH/MYB complexes to regulate JA-induced anthocyanin accumulation and trichome initiation negatively in Arabidopsis.
A bHLH protein (EGL3, GL3, or TT8), a R2R3 MYB protein (GL1, MYB75, MYB90, MYB113, MYB114, or MYB23) and the WD-repeat protein TTG1 form WD-repeat/bHLH/MYB complexes to regulate anthocyanin biosynthesis and trichome initiation (Zimmermann et al., 2004; Gonzalez et al., 2008; Zhao et al., 2008). Previous studies showed that JA induces anthocyanin accumulation through upregulation of late anthocyanin biosynthetic genes DFR, LDOX, and UF3GT (Shan et al., 2009) and that JA enhances trichome formation through influence on GL3 expression (Maes et al., 2008; Yoshida et al., 2009). It was still unclear whether and how the WD-repeat/bHLH/MYB complexes are regulated in the JA signaling pathway to stimulate anthocyanin accumulation and trichome initiation. In this study, we demonstrated that the WD-repeat/bHLH/MYB complexes are regulated through interaction of JAZ proteins with the bHLH members (TT8, GL3, and EGL3) and the R2R3 MYB members (GL1 and MYB75). It remains to be elucidated whether JAZ proteins interact with the remaining R2R3 MYB members (MYB23, MYB90, MYB113, and MYB114) of the complexes.
Previous studies showed that R3-type MYB transcription factors (such as TRY and MYBL2) repress trichome formation and anthocyanin accumulation through their interaction with bHLH members of WD-repeat/MYB/bHLH complexes: the R3 MYB transcription factors compete with R2R3 MYB members (GL1 or MYB75) for the interaction domain located at the N terminus of the bHLH members (GL3, EGL3, and TT8) to repress the transcriptional function of the WD-repeat/MYB/bHLH complexes (Schellmann et al., 2002; Esch et al., 2003; Zhang et al., 2003; Matsui et al., 2008). By contrast, JAZ proteins interact with C terminus of both R2R3 MYB and bHLH members to attenuate the transcriptional function of WD-repeat/bHLH/MYB complexes (Figures 4, 6, and 7). JA mediates anthocyanin accumulation and trichome initiation probably through activation of transcriptional function of WD-repeat/bHLH/MYB complexes by removal of the JAZ repression on R2R3 MYB and bHLH members (Figures 6 and 7). Molecular, genetic, and physiological analysis showed that the JAZ1△3A transgenic plant and the coi1-2 mutant, which highly accumulate JAZ proteins, exhibited repression on JA-induced expression of the anthocyanin biosynthetic genes (DFR, LDOX, and UF3GT) and the trichome initiation factor GL2 (Figure 7F), target genes of the WD-repeat/bHLH/MYB complexes, and displayed a reduction in JA-induced anthocyanin accumulation and trichome initiation (Figure 8). Consistent with this, overexpression of MYB75 in the coi1 mutant overcame the JAZ repression to restore JA-induced anthocyanin accumulation (Figure 10), and overexpression of GL3 or EGL3 in coi1-2 surmounted the JAZ repression to rescue trichome initiation (Figure 11).
GL3 and EGL3 play important roles in both anthocyanin accumulation and trichome formation (Zhang et al., 2003; Gonzalez et al., 2008). However, we found that overexpression of GL3 or EGL3 was able to rescue trichome initiation in coi1-2 fully (Figures 11A and 11B) but restore anthocyanin accumulation in coi1-2 only slightly, not obviously (Figure 11C), which suggests that GL3 and EGL3 mediate JA-regulated anthocyanin accumulation and trichome formation through different downstream signal cascades and distinct feedback loops. Consistent with this speculation, we found that the double mutants lacking both GL3 and EGL3 showed a complete loss in trichome initiation but just a moderate reduction in JA-induced anthocyanin accumulation (Figure 8) (Yoshida et al., 2009). Furthermore, the R3-MYB transcription factor MYBL2 (Dubos et al., 2008; Matsui et al., 2008), which was upregulated by GL3 (Morohashi and Grotewold, 2009), was found to function as a negative regulator of GL3, EGL3, and TT8 to inhibit GL3/EGL3/TT8-mediated anthocyanin accumulation but not to repress GL3/EGL3-mediated trichome initiation due to lack of MYBL2 expression in trichomes (Dubos et al., 2008; Matsui et al., 2008). It is possible that, in the coi1-2 GL3OE and coi1-2 EGL3OE lines, the elevated GL3 or EGL3 is able to activate fully the trichome cascade essential for trichome initiation but is unable to activate efficiently the distinct anthocyanin cascade.
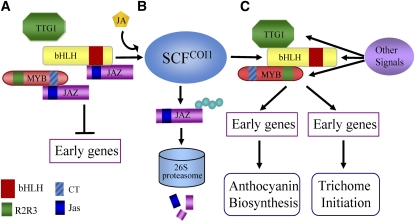
We propose a model to elucidate the molecular mechanism for JA function in regulating anthocyanin accumulation and trichome formation (Figure 12). JAZ proteins directly interact with C terminus of both bHLH (TT8, GL3, and EGL3) and R2R3 MYB components (MYB75 and GL1) of WD-repeat/bHLH/MYB complexes (Figures 1 to 5), attenuate their transcriptional function (Figures 6 and 7), and therefore repress JA-regulated anthocyanin accumulation and trichome initiation (Figure 8). Upon JA treatment or JA biosynthesis stimulated by diverse developmental and environmental signals, the JA signal induces degradation of JAZ proteins through the SCFCOI1-26S proteasome pathway (Chini et al., 2007; Thines et al., 2007; Chung et al., 2009); the WD-repeat/bHLH/MYB complexes are then released to activate their respective downstream transcriptional cascades, leading to anthocyanin biosynthesis and trichome formation. Many other signals, including environmental signals (cold, high light, and insect attack) and endogenous signals (cytokinins and gibberellin), were shown to regulate anthocyanin accumulation or trichome formation (Deikman and Hammer, 1995; Gan et al., 2007; Cominelli et al., 2008; Loreti et al., 2008; Rowan et al., 2009; Yoshida et al., 2009; Yu et al., 2010). These signals may influence anthocyanin accumulation and trichome initiation also through directly or indirectly regulating WD-repeat/bHLH/MYB complexes by virtue of bHLH and R2R3 MYB components as well as the WD-repeat protein TTG1.
Figure 12.
A Simplified Model for JA Action in Regulation of Anthocyanin Accumulation and Trichome Initiation in Arabidopsis.
(A) The Arabidopsis JAZ proteins (JAZ), by virtue of their C-terminal Jas domain, interact with the C-terminal domains of bHLH transcription factors (TT8, GL3, and EGL3) and MYB proteins (MYB75 and GL1), the essential components of WD-repeat/bHLH/MYB complexes, to attenuate transcriptional function of WD-repeat/bHLH/MYB complexes and therefore inactivate downstream genes (early genes), repressing JA-regulated anthocyanin biosynthesis and trichome initiation.
(B) Upon perception of JA signal, COI1 recruits JAZ proteins to the SCFCOI1 complex for ubiquitination and degradation through the 26S proteasome.
(C) Upon degradation of JAZ proteins, the bHLH and MYB components of WD-repeat/bHLH/MYB complexes are subsequently released to activate their respective downstream genes, which are essential for anthocyanin biosynthesis and trichome formation. Other signals (such as cytokinin, gibberellin, Suc, nutrition depletion, light, and cold) may also regulate anthocyanin biosynthesis and trichome initiation by directly or indirectly regulating WD-repeat/bHLH/MYB complexes through the bHLH and MYB components as well as the WD-repeat protein TTG1.
[See online article for color version of this figure.]
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana coi1-1 (Xie et al., 1998), coi1-2 (Xu et al., 2002), and JAZ1△3A (Thines et al., 2007) mutants were described as previously. Columbia-0 (Col-0) was used as the wild-type control. The mutants tt8 (Salk_082999 with T-DNA insertion in the second intron), gl3 (gl3-1) (Payne et al., 2000), egl3 (Salk_077439), ttg1 (ttg1-1) (Walker et al., 1999), pap1-D (Borevitz et al., 2000), and gl1 (Herman and Marks, 1989) were all obtained from the ABRC.
The JAZ1△3A transgene was maintained by genetic cross of the male-sterile JAZ1△3A transgenic plants with wild-type pollen grains. The harvested seeds from the cross were germinated to produce a population with 1:1 segregation of the wild type and JAZ1△3A transgene. The JAZ1△3A transgenic seedlings were identified by PCR amplification with the primer pair 35S promoter-F/JAZ1△3A-R (see Supplemental Table 1 online) when grown on Murashige and Skoog (MS) medium or screened by observation of JA-insensitive phenotypes (Thines et al., 2007) when grown on MS medium supplied with MeJA.
Arabidopsis seeds were disinfected and plated on MS medium (Sigma-Aldrich) supplemented with 3% sucrose, chilled for 3 d at 4°C, and transferred to a growth house under a 16-h-light (22-24°C)/8-h-dark (17-19°C) photoperiod. Nicotiana benthamiana was grown under a 16-h-light (28°C)/8-h-dark (22°C) photoperiod. All experiments were repeated at least three times.
Y2H Screening and Y2H Assays
The Matchmaker LexA two-hybrid system (Clontech) was used for Y2H screening of the Arabidopsis cDNA library (Xu et al., 2002). Arabidopsis JAZ1 cDNA was cloned into pLexA vector to fuse with DNA BD. Yeast transformants were exhaustively screened on 2% SD/Gal/Raf/X-gal (-Ura/-His/-Trp/-Leu) based on the manufacturer’s instructions (Clontech).
For Y2H assay, all the JAZ cDNAs and their domain derivatives were cloned into pLexA, and the full-length or domain deletion forms of EGL3/GL3/TT8/MYB75/GL1 were cloned into pB42AD. Primers used for the vector construction are presented in Supplemental Table 1 online. All the constructs were sequence verified. All the pLexA constructs were proved to have no self-activation by β-galactosidase assay in yeast. The indicated construct pairs were transformed into yeast (Saccharomyces cerevisiae) strain EGY48 and cultured at 30°C. Successful yeast transformants were cultured in liquid SD minimal medium (-Ura/-His/-Trp) at 230 rpm/30°C for ~24 h to the final concentration (OD60 0= 1.2 to ~1.5). Then, 1 mL of yeast strain was centrifuged and resuspended in 0.3 mL of distilled water. Five microliter of the suspension cells were plated on SD/Gal/Raf/X-gal (-Ura/-His/-Trp/-Leu) induction medium in 96-well plates. Y2H images were taken 3 d after incubation at 30°C. Experiments were repeated three times. The expressed proteins in different yeast strains were analyzed by immunoblot experiments. Proteins fused with the activation domain were detected using anti-HA antibody (Sigma-Aldrich). Proteins fused with the LexA DNA binding domain were detected using anti-LexA antibody.
BiFC Assays
Full-length coding sequences of Arabidopsis JAZ1, JAZ10, TT8, GL3, EGL3, MYB75, and GL1 were individually cloned into the pDONR207 vector (Invitrogen) through Gateway reaction (Invitrogen) and subsequently recombined into the binary YFP BiFC vectors (Song et al., 2011) so that they were fused with the N- or C-terminal fragment of YFP (nYFP or cYFP) to generate JAZ1/JAZ10/GL1/MYB75-nYFP and cYFP-TT8/GL3/EGL3/MYB75/GL1 plasmids. Primers used for plasmid construction are presented in Supplemental Table 1 online.
These constructs were transformed into Agrobacterium tumefaciens strain GV3101 through electroporation. The Agrobacterium strains containing different constructs were incubated, harvested, and resuspended in infiltration buffer (10 mM MES, 0.2 mM acetosyringone, and 10 mM MgCl2) to an ultimate concentration of OD600 = 0.5. Equal volumes of different combinations of Agrobacterium strains were mixed and coinfiltrated into the same N. benthamiana leaf with a needleless syringe. Plants were placed at 24°C for 50 h before detection of YFP fluorescence.
YFP fluorescence was detected with Zeiss microscope (LSM710) and analyzed by the ZEN 2009 software. For staining of the nuclei, 10 mg/mL 4′,6-diamidino-2-phenylindole (DAPI) was infiltrated into N. benthamiana leaves 2 h before observation. For the exogenous COR treatment in Figures 6G and 6H, infiltration buffer containing 5 μM COR (+COR) (or solvent buffer as control) was infiltrated into the N. benthamiana leaf area coexpressing the indicated constructs 10 h before YFP fluorescence observation. All experiments were repeated at least three times.
Transient Expression Assays
The transient expression assays (Matsui et al., 2008; Shang et al., 2010) were performed in N. benthamiana leaves. The DFR promoter was fused with luciferase reporter gene (ProDFR-LUC) through the KpnI and SmaI sites (Shang et al., 2010). JAZ1 was cloned into the modified pCAMBIA1300 vector (Cambia) with four CaMV35S promoters. TT8 and MYB75 were cloned into the pROK2 vector (Xu et al., 2002) through SmaI and SacI sites. Primers used for these constructs are shown in Supplemental Table 1 online. pBI121 vector (Clontech) was used for the transient expression of GUS.
The Agrobacterium strains containing different constructs were incubated, harvested, and resuspended in infiltration buffer (10 mM MES, 0.2 mM acetosyringone, and 10 mM MgCl2) to an ultimate concentration of OD600 = 0.5. Equal volumes of different combinations of Agrobacterium strains were mixed and coinfiltrated into N. benthamiana leaves by a needleless syringe. Plants were placed at 24°C for 50 h before CCD imaging. For the exogenous COR treatment in Figure 7E, the infiltration buffer containing 5 μM COR (+COR) (or solvent buffer as control) was infiltrated into the N. benthamiana leaf area coexpressing the indicated constructs 10 h before CCD imaging.
We used a low-light cooled CCD imaging apparatus (Andor iXon) to acquire the image. The leaves were sprayed with 100 μM luciferin and were placed in darkness for 5 min before luminescence detection. The computational method for counting luminescence intensity was described previously (Shang et al., 2010). Five independent determinations were assessed. Error bars represent se. All experiments were repeated with five independent biological replicates with similar results.
Generation of GL3 and EGL3 Transgenic Plants
To obtain Arabidopsis GL3 and EGL3 overexpression transgenic plants in wild-type and coi1-2 background, the full-length coding sequences of GL3 and EGL3 were amplified using the primers indicated in Supplemental Table 1 online and cloned into the modified pCAMBIA1300 vector (Cambia) containing a CaMV35S promoter through the SalI and SpeI sites. These constructs were introduced into the COI1/coi1-2 heterozygous plants using the Agrobacterium-mediated flower dip method (Clough and Bent, 1998).
Three overexpression transgenic lines for each gene (GL3 and EGL3) in wild-type and coi1-2 background were identified and characterized. Results from one transgenic line of GL3 (and EGL3) in wild-type and coi1-2 background was representatively displayed in the article.
In Vitro Pull-Down Assays
The coding region of JAZ1 was cloned into the pMAL-c2x (NEB) vector using the primers presented in Supplemental Table 1 online through EcoRI and SalI sites. This construct was transformed into E. coli to express MBP-JAZ1 protein. MBP and MBP-JAZ1 proteins were purified from E. coli using Ni affinity chromatography according to Yan et al. (2009).
Full-length CDSs of GL3 and TT8 were cloned into the modified pROK2-myc vector (Xu et al., 2002) through SmaI and SacI sites for fusion with six myc tags. These constructs were sequence verified and transformed into Agrobacterium. The Agrobacterium strains were cultured, harvested, and resuspended in infiltration buffer (10 mM MES, 0.2 mM acetosyringone, and 10 mM MgCl2) and then infiltrated into N. benthamiana leaves.
After 50 h, 5 g of the N. benthamiana leaves transiently expressing myc-GL3 or myc-TT8 were harvested for total protein extraction in a RB buffer (100 mM NaCl, 50 mM Tris-Cl, pH 7.8, 25 mM imidazole, 0.1% [v/v] Tween 20, 10% [v/v] glycerol, EDTA-free complete miniprotease inhibitor cocktail, and 20 mM 2-mercaptoethanol). The extracted total protein was concentrated to 300 μL.
Coomassie Brilliant Blue was used to confirm protein amount. About 50 μg of purified MBP and MBP-JAZ1 were incubated with 100 μL amylose resin beads for 2 h at 4°C. These amylose resin beads were then washed five times with 1 mL RB buffer and incubated with 100μL of concentrated total proteins containing myc-GL3 or myc-TT8 for 2 h. After washing five times with 1 mL RB buffer, the mixture was resuspended in SDS loading buffer, boiled for 5 min, separated on 15% SDS-PAGE, and immunoblotted using anti-c-myc antibody (Roche).
Anthocyanin Measurement
Arabidopsis seeds were plated on MS medium without or with 25 μM MeJA for 11 d and then were used for anthocyanin measurement with the method described previously (Shan et al., 2009). To show the quantity of anthocyanin, (A535-A650) per gram fresh weight was used. All experiments were repeated at least three times.
Real-Time PCR Analysis
For real-time PCR analysis, Arabidopsis seedlings were grown on MS medium for 3 weeks and then were treated with or without 100 μM MeJA for 0, 1, 2, 8, and 12 h. The treated seedlings were harvested for RNA extraction and subsequent reverse transcription. Real-time PCR analyses were performed with the RealMasterMix (SYBR Green I) (Takara) using the ABi7500 real-time PCR system. ACTIN8 was used as the internal control. The primers used for real-time PCR analysis are presented in Supplemental Table 1 online.
For real-time PCR analysis of JAZ gene expression, trichomes with epidermis were isolated from 4-week-old wild-type Arabidopsis leaves using sharp forceps under a microscope and further used for RNA extraction and subsequent reverse transcription. ACTIN8 was used as the internal control.
For real-time PCR analysis of EGL3 and GL3 in transgenic plants overexpressing of EGL3 and GL3, respectively, 3-week-old seedlings of transgenic plants were used for RNA extraction and subsequent reverse transcription. ACTIN8 was used as the internal control.
The N. benthamiana leaves for the BiFC assays and the transient assays were also used for RNA extraction and subsequent expression analysis after coinfiltration of construct combinations for 50 h. Nicotiana Actin was used as the internal control. All the primers specifically used for real-time PCR analysis are presented in Supplemental Table 1 online.
All the real-time PCR analyses were repeated for three biological replicates with similar results. Error bars represent se.
Trichome Measurement
The method for trichome measurement was modified according to Yoshida et al. (2009). Arabidopsis seeds on MS medium were chilled for 3 d at 4°C and transferred to a growth room under a 16-h-light (~22°C)/8-h-dark (~17°C) photoperiod for 6 d. The seedlings were subsequently transferred onto fresh MS medium in 15-cm-diameter plates. In the center of the 15-cm-diameter plate, a piece of filter paper was placed into an eppendorf tube cap. MeJA (150 nmol) was then dropped onto the filter paper for evaporation. The seedlings were cultured for two additional weeks. The fifth truth leaf was used for calculation of total trichome numbers by environmental scanning electron microscopy (FEI Quanta 200). At least 12 leaves for each genotype were analyzed. The experiments were repeated three times.
Accession Numbers
The Arabidopsis Genome Initiative numbers for the genes mentioned in this article are as follows: JAZ1 (AT1G19180), JAZ2 (AT1G74950), JAZ3 (AT3G17860), JAZ4 (AT1G48500), JAZ5 (AT1G17380), JAZ6 (AT1G72450), JAZ7 (AT2G34600), JAZ8 (AT1G30135), JAZ9 (AT1G70700), JAZ10 (AT5G13220), JAZ11 (AT3G43440), JAZ12 (AT5G20900), MYC2 (AT1G32640), TT8 (AT4G09820), GL3 (AT5G41315), EGL3 (AT1G63650), MYB75 (AT1G56650), MYB90 (AT1G66390), MYB113 (AT1G66370), MYB114 (AT1G66380), GL1 (AT3G27920), GL2 (AT1G79840), TTG1 (AT5G24520), DFR (AT5G42800), LDOX (AT4G22880), UF3GT (AT5G54060), ACTIN8 (AT1G49240), and Nicotiana Actin (EU938079).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Immunoblot Analysis of 12 Arabidopsis JAZ Proteins in Yeast.
Supplemental Figure 2. Negative Controls for the BiFC Experiments.
Supplemental Figure 3. Interaction of Arabidopsis TT8 with MYB75 in BiFC Assays.
Supplemental Figure 4. GUS Protein Does Not Affect the Transcriptional Function of TT8 and MYB75.
Supplemental Figure 5. Gene Expression Analysis of Transgenic Plants.
Supplemental Table 1. Primers Used for Vector Construction and Gene Expression Analysis.
Acknowledgments
We thank John Browse for providing JAZ1△3A transgenic plants, Yule Liu for the BiFC vectors, and the ABRC for mutants. We thank Claus Wasternack for helpful suggestions. This work was financially supported by the grants from the National Basic Research Program of China (973 Program 2011CB915404), the National Science Foundation of China (91017012), and the Ministry of Agriculture (2008ZX08009-004) to D.X.
References
- Allan A.C., Hellens R.P., Laing W.A. (2008). MYB transcription factors that colour our fruit. Trends Plant Sci. 13: 99–102 [DOI] [PubMed] [Google Scholar]
- Baudry A., Caboche M., Lepiniec L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46: 768–779 [DOI] [PubMed] [Google Scholar]
- Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39: 366–380 [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Lee M.M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439 [DOI] [PubMed] [Google Scholar]
- Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P. (2005). Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8: 272–279 [DOI] [PubMed] [Google Scholar]
- Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Sun L., Qi T., Zhang B., Peng W., Liu Y., Xie D. (2011). The bHLH transcription factor MYC3 interacts with the jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4: 279–288 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Chico J.M., Fernández-Calvo P., Solano R. (2009). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59: 77–87 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Koo A.J., Gao X., Jayanty S., Thines B., Jones A.D., Howe G.A. (2008). Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Niu Y., Browse J., Howe G.A. (2009). Top hits in contemporary JAZ: An update on jasmonate signaling. Phytochemistry 70: 1547–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cominelli E., Gusmaroli G., Allegra D., Galbiati M., Wade H.K., Jenkins G.I., Tonelli C. (2008). Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 165: 886–894 [DOI] [PubMed] [Google Scholar]
- Creelman R.A., Mullet J.E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 355–381 [DOI] [PubMed] [Google Scholar]
- Deikman J., Hammer P.E. (1995). Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana. Plant Physiol. 108: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H.K., Robbins T.P., Jorgensen R.A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25: 173–199 [DOI] [PubMed] [Google Scholar]
- Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., Routaboul J.M., Alboresi A., Weisshaar B., Lepiniec L. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Esch J.J., Chen M., Sanders M., Hillestad M., Ndkium S., Idelkope B., Neizer J., Marks M.D. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130: 5885–5894 [DOI] [PubMed] [Google Scholar]
- Farmer E.E. (2001). Surface-to-air signals. Nature 411: 854–856 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I., Wasternack C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53: 275–297 [DOI] [PubMed] [Google Scholar]
- Feys B., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chico J.M., Solano R. (2009a). The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009b). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Franceschi V.R., Grimes H.D. (1991). Induction of soybean vegetative storage proteins and anthocyanins by low-level atmospheric methyl jasmonate. Proc. Natl. Acad. Sci. USA 88: 6745–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y., Liu C., Yu H., Broun P. (2007). Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134: 2073–2081 [DOI] [PubMed] [Google Scholar]
- Gonzalez A. (2009). Pigment loss in response to the environment: a new role for the WD/bHLH/MYB anthocyanin regulatory complex. New Phytol. 182: 1–3 [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53: 814–827 [DOI] [PubMed] [Google Scholar]
- Gould K.S. (2004). Nature's swiss army knife: The diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 2004: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne J.B., Williams C.A. (2000). Advances in flavonoid research since 1992. Phytochemistry 55: 481–504 [DOI] [PubMed] [Google Scholar]
- Herman P.L., Marks M.D. (1989). Trichome development in Arabidopsis thaliana. II. Isolation and complementation of the GLABROUS1 Gene. Plant Cell 1: 1051–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A., Lightner J., Browse J., Ryan C.A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Katsir L., Chung H.S., Koo A.J.K., Howe G.A. (2008a). Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008b). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.H., Park S.H., Kim J.K. (2009). Methyl jasmonate triggers loss of grain yield under drought stress. Plant Signal. Behav. 4: 348–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Lee M.M., Wester K., Herrmann U., Zheng Z., Oppenheimer D., Schiefelbein J., Hulskamp M. (2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132: 1477–1485 [DOI] [PubMed] [Google Scholar]
- Kubasek W.L., Shirley B.W., McKillop A., Goodman H.M., Briggs W., Ausubel F.M. (1992). Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 4: 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J.C., Young N., Prigge M., Marks M.D. (1996). The control of trichome spacing and number in Arabidopsis. Development 122: 997–1005 [DOI] [PubMed] [Google Scholar]
- Lepiniec L., Debeaujon I., Routaboul J.M., Baudry A., Pourcel L., Nesi N., Caboche M. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo C., Lea U.S., Ruoff P. (2008). Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant Cell Environ. 31: 587–601 [DOI] [PubMed] [Google Scholar]
- Liu F., Ni W., Griffith M.E., Huang Z., Chang C., Peng W., Ma H., Xie D. (2004). The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell 16: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E., Povero G., Novi G., Solfanelli C., Alpi A., Perata P. (2008). Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 179: 1004–1016 [DOI] [PubMed] [Google Scholar]
- Maes L., Inze D., Goossens A. (2008). Functional specialization of the TRANSPARENT TESTA GLABRA1 network allows differential hormonal control of laminal and marginal trichome initiation in Arabidopsis rosette leaves. Plant Physiol. 148: 1453–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Umemura Y., Ohme-Takagi M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55: 954–967 [DOI] [PubMed] [Google Scholar]
- McConn M., Browse J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M., Creelman R.A., Bell E., Mullet J.E., Browse J. (1997). Jasmonate is essential for insect defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M., Mecey C., Niu Y., Chung H.S., Katsir L., Yao J., Zeng W., Thines B., Staswick P.E., Browse J., Howe G.A., He S.Y. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y., Nagel C., Taylor L.P. (1992). Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. USA 89: 7213–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., Grotewold E. (2009). A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., Zhao M., Yang M., Read B., Lloyd A., Lamb R., Grotewold E. (2007). Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 145: 736–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., Lepiniec L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibbe M., Hilpert B., Wasternack C., Miersch O., Apel K. (2002). Cell death and salicylate- and jasmonate-dependent stress responses in Arabidopsis are controlled by single cet genes. Planta 216: 120–128 [DOI] [PubMed] [Google Scholar]
- Niu Y., Figueroa P., Browse J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer D.G., Herman P.L., Sivakumaran S., Esch J., Marks M.D. (1991). A myb (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493 [DOI] [PubMed] [Google Scholar]
- Pauwels L., Morreel K., De Witte E., Lammertyn F., Van Montagu M., Boerjan W., Inzé D., Goossens A. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C.T., Zhang F., Lloyd A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D., Vachon G., Herzog M. (1998). Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis Plant Physiol. 117: 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M., Hülskamp M. (2009). One, two, three...models for trichome patterning in Arabidopsis? Curr. Opin. Plant Biol. 12: 587–592 [DOI] [PubMed] [Google Scholar]
- Pires N., Dolan L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27: 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay N.A., Glover B.J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Rao M.V., Lee H., Creelman R.A., Mullet J.E., Davis K.R. (2000). Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Han C., Peng W., Huang Y., Peng Z., Xiong X., Zhu Q., Gao B., Xie D. (2009). A leaky mutation in DWARF4 reveals an antagonistic role of brassinosteroid in the inhibition of root growth by jasmonate in Arabidopsis. Plant Physiol. 151: 1412–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Pan J., Peng W., Genschik P., Hobbie L., Hellmann H., Estelle M., Gao B., Peng J., Sun C., Xie D. (2005). Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J. 42: 514–524 [DOI] [PubMed] [Google Scholar]
- Reymond P., Weber H., Damond M., Farmer E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan D.D., Cao M., Lin-Wang K., Cooney J.M., Jensen D.J., Austin P.T., Hunt M.B., Norling C., Hellens R.P., Schaffer R.J., Allan A.C. (2009). Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 182: 102–115 [DOI] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jürgens G., Hülskamp M. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Howe G.A. (2005). Systemic signaling in the wound response. Curr. Opin. Plant Biol. 8: 369–377 [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.S., Joo J., Kim M.J., Kim Y.K., Nahm B.H., Song S.I., Cheong J.J., Lee J.S., Kim J.K., Choi Y.D. (2011). OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65: 907–921 [DOI] [PubMed] [Google Scholar]
- Serna L., Martin C. (2006). Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci. 11: 274–280 [DOI] [PubMed] [Google Scholar]
- Shan X., Wang J., Chua L., Jiang D., Peng W., Xie D. (2011). The role of Arabidopsis rubisco activase in jasmonate-induced leaf senescence. Plant Physiol. 155: 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X., Zhang Y., Peng W., Wang Z., Xie D. (2009). Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 60: 3849–3860 [DOI] [PubMed] [Google Scholar]
- Shang Y., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley B.W., Kubasek W.L., Storz G., Bruggemann E., Koornneef M., Ausubel F.M., Goodman H.M. (1995). Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Solfanelli C., Poggi A., Loreti E., Alpi A., Perata P. (2006). Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovchenko A., Schmitz-Eiberger M. (2003). Significance of skin flavonoids for UV-B-protection in apple fruits. J. Exp. Bot. 54: 1977–1984 [DOI] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springob K., Nakajima J., Yamazaki M., Saito K. (2003). Recent advances in the biosynthesis and accumulation of anthocyanins. Nat. Prod. Rep. 20: 288–303 [DOI] [PubMed] [Google Scholar]
- Staswick P.E., Su W., Howell S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89: 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Browse J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Ishihara H., Huep G., Barsch A., Mehrtens F., Niehaus K., Weisshaar B. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Sun J., et al. (2009). Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21: 1495–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamari G., Borochov A., Atzorn R., Weiss D. (1995). Methyl jasmonate induces pigmentation and flavonoid gene expression in petunia corollas: A possible role in wound response. Physiol. Plant. 94: 45–50 [Google Scholar]
- Teng S., Keurentjes J., Bentsink L., Koornneef M., Smeekens S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tohge T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traw M.B., Bergelson J. (2003). Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol. 133: 1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan P., Shockey J., Lévesque C.A., Cook R.J., Browse J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7209–7214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T., Pollak P., Tarlyn N., Taylor L.P. (1994). Pollination- or wound-induced kaempferol accumulation in petunia stigmas enhances seed production. Plant Cell 6: 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G.J., Wang E., Shepherd R.W. (2004). New approaches for studying and exploiting an old protuberance, the plant trichome. Ann. Bot. (Lond.) 93: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Dai L., Jiang Z., Peng W., Zhang L., Wang G., Xie D. (2005). GmCOI1, a soybean F-box protein gene, shows ability to mediate jasmonate-regulated plant defense and fertility in Arabidopsis. Mol. Plant Microbe Interact. 18: 1285–1295 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinthal D., Tzfira T. (2009). Imaging protein-protein interactions in plant cells by bimolecular fluorescence complementation assay. Trends Plant Sci. 14: 59–63 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Xiao S., Dai L., Liu F., Wang Z., Peng W., Xie D. (2004). COS1: An Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence. Plant Cell 16: 1132–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.X., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Sano R., Wada T., Takabayashi J., Okada K. (2009). Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 136: 1039–1048 [DOI] [PubMed] [Google Scholar]
- Yu N., Cai W.-J., Wang S., Shan C.-M., Wang L.-J., Chen X.-Y. (2010). Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22: 2322–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Turner J.G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Morohashi K., Hatlestad G., Grotewold E., Lloyd A. (2008). The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135: 1991–1999 [DOI] [PubMed] [Google Scholar]
- Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40: 22–34 [DOI] [PubMed] [Google Scholar]