The localization of fluorescently tagged Arabidopsis MAP65-1 and MAP65-2 is monitored in live epidermal hypocotyl cells. Cortical microtubules are shown to bundle via a microtubule-microtubule templating mechanism, and MAP65-1 and MAP65-2 are shown to contribute to the mechanism of axial cell growth in expanding hypocotyls.
Abstract
The Arabidopsis thaliana MAP65-1 and MAP65-2 genes are members of the larger eukaryotic MAP65/ASE1/PRC gene family of microtubule-associated proteins. We created fluorescent protein fusions driven by native promoters that colocalized MAP65-1 and MAP65-2 to a subset of interphase microtubule bundles in all epidermal hypocotyl cells. MAP65-1 and MAP65-2 labeling was highly dynamic within microtubule bundles, showing episodes of linear extension and retraction coincident with microtubule growth and shortening. Dynamic colocalization of MAP65-1/2 with polymerizing microtubules provides in vivo evidence that plant cortical microtubules bundle through a microtubule-microtubule templating mechanism. Analysis of etiolated hypocotyl length in map65-1 and map65-2 mutants revealed a critical role for MAP65-2 in modulating axial cell growth. Double map65-1 map65-2 mutants showed significant growth retardation with no obvious cell swelling, twisting, or morphological defects. Surprisingly, interphase microtubules formed coaligned arrays transverse to the plant growth axis in dark-grown and GA4-treated light-grown map65-1 map65-2 mutant plants. We conclude that MAP65-1 and MAP65-2 play a critical role in the microtubule-dependent mechanism for specifying axial cell growth in the expanding hypocotyl, independent of any mechanical role in microtubule array organization.
INTRODUCTION
Microtubules (MTs) play a critical role in the control of plant cell growth and morphogenesis (Baskin, 2001; Wasteneys and Fujita, 2006; Ehrhardt and Shaw, 2006; Lucas and Shaw, 2008; Sedbrook and Kaloriti, 2008). Interphase MTs form arrays at the cell cortex adjacent to the plasma membrane (Hardham and Gunning, 1978, 1979). Disruption of these interphase MT arrays with drugs produces severe growth control defects (Baskin et al., 1994; Corson et al., 2009), while mutations in tubulin and MT-associated proteins (MAPs) lead to a wide range of cellular phenotypes related to growth (Buschmann and Lloyd, 2008; Lucas and Shaw, 2008; Sedbrook and Kaloriti, 2008).
Severe disorganization of the interphase MT cytoskeleton typically causes the loss of anisotropic cell expansion, which is characterized by radial cell swelling (Whittington et al., 2001; Sugimoto et al., 2003; Bannigan et al., 2006). Milder defects in MT array organization manifest as organ-level defects, including the twisting of roots and petioles (Burk et al., 2001; Thitamadee et al., 2002; Sedbrook et al., 2004; Ishida and Hashimoto, 2007). Genetic lesions in some plant MAPs, such as CLASP and FragileFiber1 (FRA1), yield more subtle defects in cell growth and morphogenesis, without dramatic alterations to interphase MT array organization (Zhong et al., 2002; Ambrose et al., 2007; Kirik et al., 2007).
The molecular mechanisms by which interphase MTs affect cell growth are presently unclear. The pattern of cortical MT array organization has been correlated in many cases with the overlying pattern of cellulose microfibrils in the cell wall (Baskin, 2001; Paradez et al., 2006; Emons et al., 2007; Lloyd and Chan, 2008). Net-like MT arrays are often observed in cells that exhibit isotropic cell expansion and have less-ordered microfibrils. Parallel MT arrays, which are organized transversely to the plant growth axis, are associated with transverse cellulose microfibril organization and anisotropic cell expansion (e.g., axial cell growth). Imaging studies demonstrated that cellulose synthase (cesa) complexes use cortical MTs as tracks for patterning cellulose into the cell wall (Paredez et al., 2006; Chan et al., 2010; Chen et al., 2010). Further studies showed that MTs also guide cesa insertion into the plasma membrane (Crowell et al., 2009; Gutierrez et al., 2009). Additional roles for cortical MTs related to pectin secretion (Young et al., 2008) and patterning of extracellular matrix proteins have also been suggested (Roudier et al., 2005).
Electron microscopy (Hardham and Gunning, 1978, 1979) and time-lapse observations of live cells (Shaw et al., 2003; Dixit et al., 2006; Chan et al., 2007) indicate extensive bundling of the interphase cortical MTs. MT bundles form through the interaction of treadmilling MTs (Shaw et al., 2003) and through direct nucleation of newly formed MTs into a bundled state (Chan et al., 2003; Murata et al., 2005; Nakamura et al., 2010). Early biochemical assays for plant MT-associated proteins led to the discovery of MAP65 (Chang-Jie and Sonobe, 1993), the founding member of a large eukaryotic gene family that includes ANAPHASE SPINDLE ELONGATION1 (Ase1) from fission yeast and protein required for cytokinesis (PRC1) in mammals. Subsequent work demonstrated that MAP65 dimers bundle purified MTs in vitro (Chan et al., 1996, 1999; Smertenko et al., 2004; Gaillard et al., 2008), localize to MTs in vivo (Van Damme et al., 2004; Chang et al., 2005; Sasabe and Machida, 2006; Smertenko et al., 2008), and hyperbundle MTs into transverse arrays when overexpressed in plant cells (Van Damme et al., 2004; Mao et al., 2005a, 2006). MAP65-2 has additionally been reported to stabilize MTs in vitro (Li et al., 2009). Based on these observations, MAP65s have been proposed as key regulators of cortical MT array organization through control of MT bundling (Chan et al., 1999; Smertenko et al., 2000, 2004; Mao et al., 2006; Lloyd and Chan, 2008; Sasabe and Machida, 2006).
The Arabidopsis thaliana genome contains nine MAP65-related genes (Hussey et al., 2002; Sasabe and Machida, 2006). Mutations in the Arabidopsis MAP65-3 gene revealed a critical role for this MAP65 family member in mitosis and cell division (Müller et al., 2004; Caillaud et al., 2008), consistent with proposed roles for mammalian PRC1 and fungal Ase1 orthologs in maintaining spindle structure (Mollinari et al., 2002; Schuyler et al., 2003; Loïodice et al., 2005). However, no interphase phenotypes have been reported for the plant MAP65 gene family. MAP65-1 and MAP65-2 form a distinct clade in the Arabidopsis MAP65 gene family (Smertenko et al., 2008; Guo et al., 2009) and show a remarkable similarity in coding sequence (87% nucleotide and 89% amino acid) and gene structure (Figure 1A). MAP65-1 proteins have been localized to the cortical MT array in epidermal cells using immunofluorescence microscopy (Smertenko et al., 2004), suggesting a possible function in the interphase cortical array.
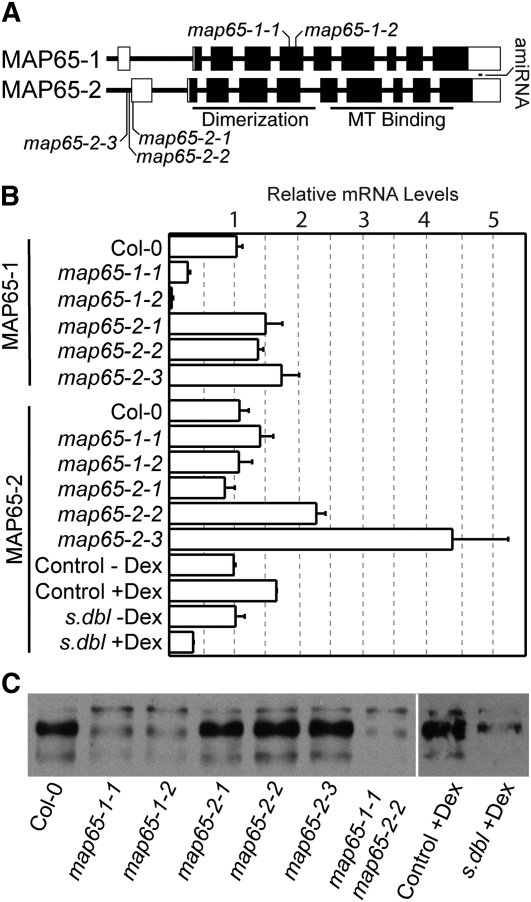
Figure 1.
MAP65-1 and MAP65-2 Gene Structure and the Relative mRNA Levels of map65 Mutant Alleles.
(A) The MAP65-1 and MAP65-2 gene structure with tDNA insertion sites marked in both genes and the position of the amiRNA65-2 sequence in the 3′ UTR of MAP65-2. Closed boxes are exons, and open boxes are untranslated regions.
(B) The relative amount of mRNA transcript for map65-1 and map65-2 tDNA alleles and the synthetic double mutant (s. dbl) assessed by qPCR and displayed as mean ± sd.
(C) Immunoblot of protein extracts made from wild-type and mutant plant lines (see Supplemental Figure 1 online for loading controls) probed with antibodies raised against MAP65-1 with cross-reactivity to MAP65-2 proteins.
In this study, we investigate the in planta role of MAP65-1 and MAP65-2 in the MT-dependent axial growth of the Arabidopsis hypocotyl (Refrégier et al., 2004; Derbyshire et al., 2007; Lucas and Shaw, 2008). We examine hypocotyl growth in a series of map65 mutants and determine that MAP65-2 is specifically required for proper axial extension. Live-cell imaging studies, using native promoter–driven fluorescent MAP65-1 and MAP65-2 fusions proteins, revealed preferential in vivo interaction with antiparallel MT bundles at the cell cortex. Dynamical studies provide evidence that these MAP65 proteins associate and dissociate with cortical MTs as the MTs undergo growth and shortening events within bundles, without altering the rate of polymerization or depolymerization. map65 mutants with severe hypocotyl growth defects maintained transversely organized cortical MT arrays, similar to wild-type seedlings. We conclude that the cortical MT array requires MAP65-1 and MAP65-2 for proper axial cell growth, independently of any mechanical function of MAP65-1 and MAP65-2 in MT array organization.
RESULTS
Isolation of map65-1 and map65-2 Mutants
We obtained Arabidopsis lines with tDNA insertions in the MAP65-1 and MAP65-2 genes from publically available seed collections. We confirmed multiple alleles for both genes by DNA sequencing. SALK_006083.45.75 (map65-1-1) and SALK_118225.51.80 (map65-1-2) contain tDNA insertions in the fifth exon of MAP65-1 94 and 132 nucleotides from the left exon border, respectively, and upstream of the MT binding site (Figure 1A). We obtained three tDNA insertion lines for MAP65-2 and identified their insertion sites as SALK_061943.49.20 (map65-2-1) 19 nucleotides into the reported 5′ untranslated region (UTR), and SAIL_1214_G02 (map65-2-2) and SAIL_378_A01 (map65-2-3), both positioned within 25 nucleotides upstream of the 5′ UTR.
The relative amounts of MAP65-1 and MAP65-2 mRNA present in the tDNA alleles were measured using quantitative RT-PCR (qPCR). Both map65-1 alleles produced significantly less full-length MAP65-1 mRNA than wild-type seedlings (Figure 1B). The amount of MAP65-2 mRNA was not significantly reduced in the map65-2-1 allele and appeared to be increased in the map65-2-2 and map65-2-3 alleles. Immunoblots performed using antibodies raised against MAP65-1 that show cross-reactivity to MAP65-2 (Smertenko et al., 2004, 2008) revealed markedly lower MAP65-1/2 protein levels in map65-1-1 and map65-1-2 and a slight reduction in map65-2-1 (Figure 1C; see Supplemental Figure 1 online). MAP65-1/2 protein levels do not appear markedly different for the map65-2-2 and map65-2-3 alleles. The double map65-1-1 map65-2-2 mutant shows a substantial reduction in total MAP65-1/2 protein level despite the apparent increase in MAP65-2 mRNA for the map65-2-2 allele.
We created an artificial microRNA (Schwab et al., 2006) to the 3′ UTR of MAP65-2 (amiRNA65-2) and placed it under a dexamethasone (dex)–inducible promoter (Aoyama and Chua, 1997). We transformed the amiRNA65-2 into the map65-1-2 mutant background to create a synthetic double mutant. Single amiRNA65-2 lines are not reported owing to the possibility of secondary targeting of MAP65-1 mRNA due to the high sequence similarity between the two genes. We observed that continuous dex treatment produces a >50% knockdown in MAP65-2 mRNA for 5-d-old seedlings compared with solvent-treated controls (Figure 1B). Dex treatment had no apparent effect on MAP65 protein levels in control plants, and the synthetic double mutant had reduced levels of MAP65-1/2 protein as indicated by immunoblots (Figure 1C).
MAP65-2 Promotes Hypocotyl Cell Elongation
We investigated the role of MAP65-1 and MAP65-2 in axial cell growth by comparing hypocotyl length in mutant and wild-type seedlings (Figure 2A). Seedlings were grown at 22°C either on agar plates or in liquid culture for 10 d in complete darkness to evaluate maximum hypocotyl extension (Derbyshire et al., 2007). Neither of the homozygous map65-1 tDNA alleles showed a significant difference in hypocotyl length when compared with wild-type plants in this terminal growth assay (Figure 2B; see Supplemental Figure 2 online). The map65-2-1 tDNA allele showed a trend toward shorter (95% of the wild type) hypocotyl length. The map65-2-2 and map65-2-3 mutants reached only 80 and 85% of wild-type length, respectively, both significant (t test P < 0.0001) when compared with wild-type plants (Figure 2B).
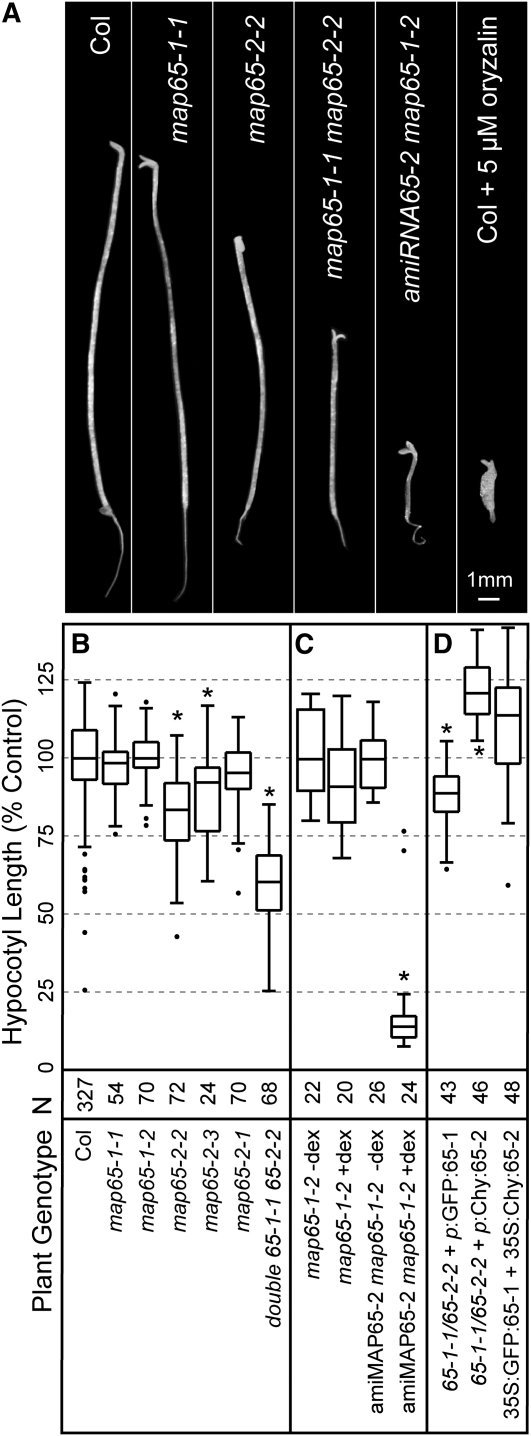
Figure 2.
Terminal Hypocotyl Length of map65-1 and map65-2 Mutant Alleles.
(A) Dark-grown seedlings from the wild type, map65-1, map65-2, map65-1 map65-2, and synthetic double mutant lines after 10 d in liquid culture. The amiRNA65-2 map65-1-2 plant was continuously treated with dex. A wild-type seedling and wild type continuously treated with 5 μM oryzalin are included for comparisons.
(B) Terminal hypocotyl length of etiolated wild-type and mutant seedlings. Hypocotyl length was normalized to the mean control value (percent of control), to account for differences between experimental growth conditions, and reported as means with 25th and 75th percentiles (box) and range (whiskers). n = number of plants measured.
(C) Hypocotyl length of the etiolated synthetic double mutant amiRNA65-2 map65-1-2 plants grown in liquid culture with and without dex induction. The map65-1-2 mutant was grown in liquid culture for 10 d as a control.
(D) Complementation of etiolated hypocotyl growth phenotypes by transgenic expression of GFP:MAP65-1 or GFP:MAP65-2 from native promoters in the double map65-1 map65-2 mutant. Expression of GFP:MAP65-1 and mCherry:MAP65-2 from constitutive cauliflower mosaic virus 35S promoters are used for comparison. n = plants measured in at least three different experiments. Asterisks denote t tests with P values < 0.0001.
To test for genetic interactions between MAP65-1 and MAP65-2, we crossed map65-2-2 and map65-1-1 alleles to create a homozygous double mutant line. Double map65-1-1 map65-2-2 mutant hypocotyls were significantly shorter than those measured in dark-grown wild-type plants (t test P < 0.0001), reaching only 60% of Columbia-0 (Col-0) length (Figure 2B). Hypocotyl length in the double mutant is also significantly shorter than hypocotyl length in the map65-2-2 allele (t test P < 0.0001), indicating a genetic interaction (Figure 2B). Light-grown hypocotyl length in the double map65-1-1 map65-2-2 mutant (see Supplemental Figure 2 online) was also significantly shorter than in wild-type plants (80% of the wild type, t test P < 0.0001).
Continuous dex induction of amiRNA65-2 in the map65-1-2 background further reduced hypocotyl length to <25% of wild-type length after 10 d of growth (Figure 2C). Synthetic double mutant plants grown without dex treatment were not different in length from wild-type plants (Figure 2C). Dex treatment had no measurable effect on hypocotyl length in wild-type Col-0 or in plants expressing green fluorescent protein (GFP) from the same dex-inducible vector (see Supplemental Figure 3 online).
To determine whether the hypocotyl phenotype was attributable to interphase growth or to defects in cell division, we counted the number of epidermal cells per file in map65-1-1 map65-2-2 double mutant hypocotyls. Cell number per file did not differ between wild-type and double mutant plants (wild type = 20.5 ± 5.0, n = 73 files; map65-1-1/map65-2-2 = 19.2 ± 4.3, n = 59 files). These data show that MAP65-2 is required for proper axial extension of the hypocotyl cells and that MAP65-1 plays a contributing role in the absence of MAP65-2. The general growth habit and size of map65 mutant alleles grown on agar plates for >16 d in continuous light did not differ substantively from wild-type plants except in the case of the synthetic double mutant grown on dex-supplemented media (see Supplemental Figure 4 online).
Fluorescent Fusion Proteins Complement Gene Function
We tested the causal relationship between the verified tDNA insertions and the hypocotyl growth phenotypes by creating transgenic plants expressing MAP65-1 or MAP65-2 as fluorescent protein fusions. We created N-terminal fluorescent protein fusions using genomic DNA sequences for both MAP65-1 and MAP65-2 driven by 2.6- and 2.1-kb fragments of genomic DNA cloned upstream of the respective genes. To evaluate fusion protein function, we transformed both fusion constructs into the map65-1-1 map65-2-2 double mutant and subjected the transgenic lines to hypocotyl elongation assays. The double tDNA mutant was used because single map65-1 alleles lack a phenotype and the single map65-2 alleles show a weak phenotype.
Expression of GFP:MAP65-1 in the double mutant (four independent lines) restored etiolated hypocotyl elongation from 60 to 85% of wild-type length (Figure 2D; see Supplemental Figure 2 online). This hypocotyl length is significantly shorter than that of wild-type plants (t test, P < 0.0001) and similar to that of the map65-2-2 single mutant allele. Expression of mCherry:MAP65-2 in the double mutant (four independent lines) produced hypocotyls that were slightly longer (t test, P < 0.0001) than those of wild-type plants (Figure 2D). We additionally coexpressed both GFP:MAP65-1 and mCherry:MAP65-2 constitutively from cauliflower mosaic virus 35S promoters in wild-type plants and found no additional gain in hypocotyl length beyond that found with expression of the native promoter mCherry:MAP65-2 in the double mutant background (Figure 2D). Together, these data provide evidence that the mutant growth phenotype is due to the tDNA insertions in the respective MAP65-1 and MAP65-2 genes and indicate that the GFP:MAP65-1 and mCherry:MAP65-2 fusion proteins are functional in plants.
Sensitizing Treatments Do Not Alter the map65 Growth Phenotype
Our terminal hypocotyl growth assays indicated that MAP65-2 is required for axial hypocotyl extension and that MAP65-1 may have an auxiliary role. To further probe the function of MAP65-1 and MAP65-2 in axial hypocotyl extension, we placed additional stress on plants and evaluated growth under these sensitized conditions. We grew wild-type and double tDNA map65-1-1 map65-2-2 mutant plants in a concentration series of the MT polymerization inhibitor oryzalin (Figure 3A) or the cellulose synthase inhibitor isoxoben (Figure 3B), both previously shown to exaggerate growth defects in plants with cytoskeletal defects (Baskin et al., 1994; Paredez et al., 2008). We observed that both drugs affect wild-type and mutant plants starting at the same drug concentrations and have proportionally similar effects on hypocotyl length across all drug concentrations. We found no evidence for supersensitivity to either drug.
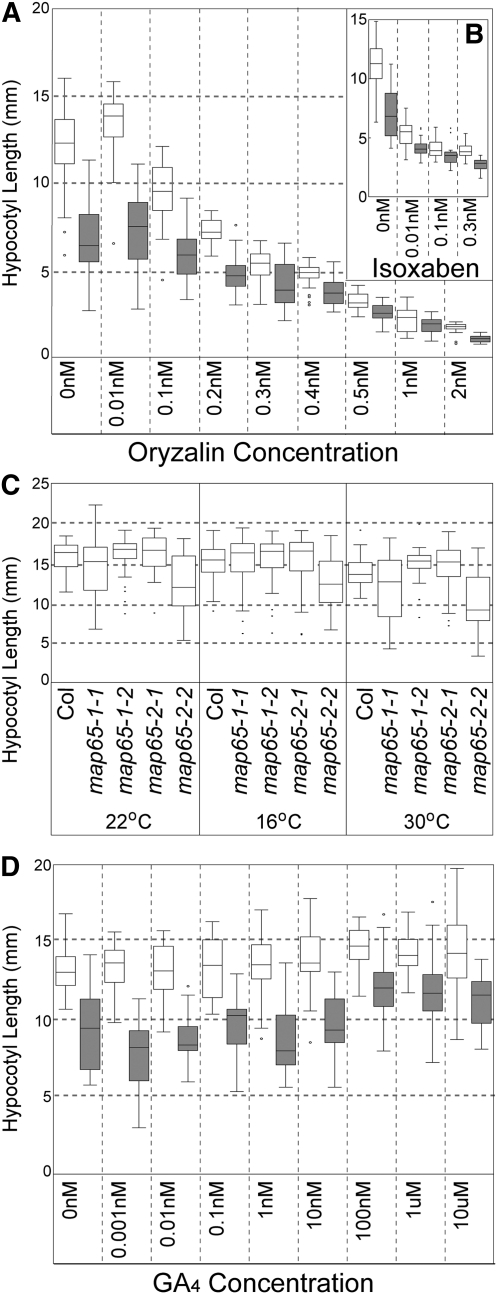
Figure 3.
Effect of Sensitizing Treatments on Hypocotyl Growth in map65 Mutants.
(A) Terminal hypocotyl length of etiolated wild-type and map65 double mutant plants grown in the MT polymerization inhibitor oryzalin over a range of concentrations for 10 d.
(B) Terminal hypocotyl length for etiolated wild type and map65 double mutants grown in the cellulose synthase inhibitor isoxoben over a range of concentrations for 10 d.
(C) Terminal hypocotyl length was measured for etiolated single map65-1 and map65-2 mutant alleles grown on agar plates for 10 d over a range of temperatures.
(D) Terminal hypocotyl lengths of etiolated wild-type and map65-1 map65-2 double mutant plants grown in liquid culture over a range of GA4 concentrations. Open boxes represent the wild type, and gray boxes represent the mutant in (A), (B), and (D).
Plant growth rate and cytoskeletal dynamics are both influenced by ambient temperature (Gray et al., 1998; Kawamura et al., 2006), and in vitro studies have suggested that MAP65-1 can stabilize MTs against depolymerization at low temperatures (Mao et al., 2005b; Meng et al., 2010). To determine if MAP65-1 or MAP65-2 are attuned to modulating growth at a specific temperature range, we grew wild-type, map65-1-1, and map65-2-2 mutant plants at normal (22°C), cold (16°C), and warm (30°C) temperatures for 10 d in the dark and compared hypocotyl lengths. We observed that growth at different temperatures does not change the proportional growth of the wild-type and mutant plants under these conditions (Figure 3C). Our terminal growth assays indicated that these map65-1 and map65-2 mutant alleles do not show a supersensitivity phenotype to temperature.
Gibberellic acid (GA4) increases the rate of hypocotyl growth in Arabidopsis seedlings (Taylor and Cosgrove, 1989; Collett et al., 2000) and has been shown to bias cortical MT orientation toward transverse coalignment (Lloyd et al., 1996). We assayed dark-grown wild-type and double tDNA map65-1-1 map65-2-2 mutant seedlings for hypocotyl growth effects across a range of GA4 concentrations (Figure 3D). We observed that 10 d of GA4 treatment increases the terminal length of both wild-type and mutant hypocotyls. Wild-type plants reached maximum lengths that were 10% longer than untreated plants, while mutant hypocotyls increased from 60% of the length of untreated wild-type hypocotyls to 80% (Figure 3D). These data indicated that the mutant growth phenotype is not exaggerated by the stress of GA4 addition, that the mutant has the capacity for further hypocotyl elongation, and that GA4 does not fully rescue the hypocotyl growth defect.
map65-1 and map65-2 Mutants Exhibit Cellular Growth Arrest
Mutations in MAPs and application of MT-specific small molecule inhibitors often lead to defects in cellular morphogenesis (Baskin, 2001; Paradez et al., 2006; Buschmann and Lloyd, 2008). To visualize the hypocotyl surface in live plants without chemical fixation or dehydration, we stained cell walls with propidium iodide and imaged the cells using confocal laser scanning microscopy. We created surface projections from each axial image series using software (see Methods). Seedlings were imaged at 4 d postgermination to show the details of wild-type and mutant cells on a comparable scale.
Hypocotyl cells in the single and double map65-1 map65-2 tDNA mutants formed distinct cell files in dark-grown seedlings and showed no obvious swelling or cellular deformation (Figures 4A to 4D). We observed no twisting or rotation of hypocotyls in the etiolated tDNA mutant alleles or the dex-induced synthetic double mutant (Figures 4A to 4E). The epidermal cells are shorter than wild-type cells as evidenced by the position of the cross walls. Due to the severe growth retardation, we compared cell morphology of the dex-induced synthetic double mutant (Figure 4E) to younger (2 d postgermination) wild-type plants having a similar epidermal cell size (Figure 4F). We observe that the 10-d-old synthetic double mutant has a subtle qualitative difference in cell shape but no obvious morphological defects, suggesting that cell growth is arrested rather than aberrant.
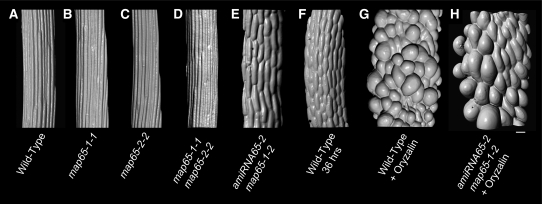
Figure 4.
Hypocotyl Cell Morphology and Cell File Organization.
Surface renderings from serial optical sections of propidium iodide-stained 4-d-old etiolated seedlings showing cells just below the hypocotyl-petiole junction. The wild type (A) and map65 tDNA mutants ([B] to [D]) show narrow epidermal cells in regular cell files terminated by transverse cross-walls. The amiRNA65-2 map65-1 synthetic double mutant (E) was grown in liquid culture with dex prior to imaging. A wild-type plant at 2 d of growth (F) is presented for comparison of cell size and shape to the amiRNA65-2 map65-1 synthetic double mutant (E). The etiolated wild type (G) and dex-induced synthetic double mutant (H) were grown for 4 d in the MT depolymerizing drug oryzalin (5 μM) to examine the cell expansion when cortical MTs are prevented from organizing. Bar = 30 μm.
We additionally compared our synthetic double mutant to etiolated wild-type plants that were continuously treated with 5 μm oryzalin (Figure 4G). The oryzalin-treated wild-type plants reach about the same hypocotyl length as our synthetic double mutant (Figure 2A) but contain highly distorted epidermal cells that are larger than those in the dex-treated synthetic double mutant (Figures 4E and 4G). Continuous oryzalin treatment of the dex-induced synthetic double mutant produced epidermal cells with the same obvious cell swelling observed for wild-type plants, with cells that were visibly much larger than cells in the map65 mutant not treated with oryzalin (Figures 4E and 4H). These data show that the map65 mutant has not lost the capacity for cell growth per se and that MTs are required for the retardation of growth observed in these map65 mutants. Together, these experiments indicate that the principal observed defect in map65 mutant plants is a MT-dependent growth arrest and not a defect in creating the correct cell shape.
MAP65-1 and MAP65-2 Are Expressed throughout Hypocotyl Development
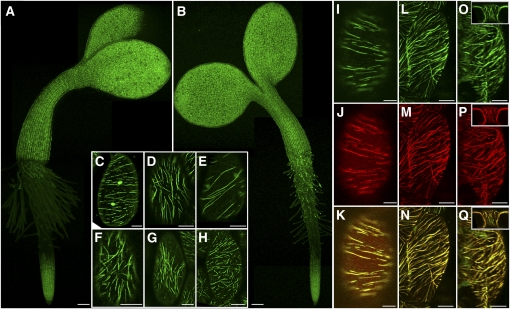
To determine the spatiotemporal expression of MAP65-1 and MAP65-2 during hypocotyl growth, we transformed the native promoter GFP:MAP65-1 (four lines) and GFP:MAP65-2 (four lines) constructs into map65-1-2 and map65-2-1 alleles, respectively, yielding genomic fusion protein expression from the native promoters in the appropriate mutant backgrounds (GFP:MAP65-1 and GFP:MAP65-2). Using these lines as translational gene expression reporters, we evaluated the pattern of GFP:MAP65-1 and GFP:MAP65-2 accumulation in living seedlings using confocal laser scanning microscopy (Figures 5A and 5B). Consistent with the growth phenotypes, both GFP:MAP65-1 and GFP:MAP65-2 were visible in all epidermal hypocotyl cells at all examined times from germination to 10 d postgermination. Localization was observed in both light- and dark-grown seedlings with no marked expression gradients or obvious cell-specific differences. We find that GFP:MAP65-1 and GFP:MAP65-2 show completely overlapping expression patterns in the hypocotyl epidermis. The absence of GFP:MAP65-1 and GFP:MAP65-2 expression in the upper portions of the root demonstrates tissue-specific control of MAP65:1 and MAP65-2 accumulation during seedling development (Figures 5A and 5B).
Figure 5.
Expression and Cellular Localization of MAP65-1 and MAP65-2 Fluorescent Fusion Proteins.
(A) and (B) Three-day-old light-grown seedlings reporting GFP:MAP65-2 (A) or GFP:MAP65-1 (B) protein expressed from native reporters in the respective tDNA mutant alleles. Expression is also observed in the cotyledons and base of the root. Note the absence of expression in the upper portion of the root, indicating developmental regulation of protein accumulation. All regions of expression were verified for label incorporation into linear filaments. Apparent root hair expression is autofluorescence. Bars = 100 μm.
(C) to (H) Cellular localization of fusion proteins in the hypocotyl epidermis of light-grown seedlings indicating labeling for transverse (C), longitudinal (D), oblique (E), and less defined organizations ([F] to [H]). GFP:MAP65-1 ([C], [D], and [F]) and GFP:MAP65-2 ([E], [G], and [H]).
(I) to (K) Double transgenic plants indicating that GFP:MAP65-1 (I) and mCherry:MAP65-2 (J) exactly overlap (K) in their intracellular localization.
(L) to (Q) Expressed GFP:MAP65-1 ([L] and [O]) and mCherry:MAP65-2 ([M] and [P]) show overlapping localization ([N] and [O]) throughout the cell cortex, including peridermal face ([L] and [M]), the anticlinal side ([O] to [Q]), and end faces (insets) of the cell.
Bars = 10 μm in (C) to (Q).
Intracellular Localization of Arabidopsis MAP65-1 and MAP65-2
We examined the intracellular localization of GFP:MAP65-1 and GFP:MAP65-2 in their respective mutant backgrounds using confocal laser scanning microscopy (Figure 5). We observed linear structures, presumed to be MT bundles, organized into transverse, oblique, longitudinal, and less organized array patterns in cells from light-grown plants (Figures 5C to 5H). Using genomic promoters, we found no evidence for hyperbundling of arrays (Mao et al., 2005a, 2006) and no obvious preference for transverse array orientation to the growth axis of the cell in these light-grown plants (Van Damme et al., 2004).
To determine whether MAP65-1 and MAP65-2 labeled different structures in the same cell, we created transgenic plants that coexpress GFP:MAP65-1 and mCherry:Map65-2 under native promoters in the double mutant background (Figures 5I to 5Q). We created emission bandwidths with the microscope spectral detector to eliminate crosstalk between GFP and mCherry signal and verified channel spatial alignment with double-labeled fluorescent particles prior to imaging. Observations on three independently derived transgenic lines revealed complete colocalization of MAP65-1 and MAP65-2 in all epidermal hypocotyl cells and in all MT array organizations (Figures 5I to 5K).
We observed MAP65-1 and MAP65-2 reporters on all cell faces using reconstructions of serial optical sections from light-grown epidermal cells. The anticlinal side and end faces were labeled with MAP65-1 and MAP65-2 (Figures 5L to 5Q). Localization was continuous from periclinal (surface) to anticlinal (side) structures in many cases where the density of labeling allowed for unambiguous tracing of the filaments (Figures 5L to 5N).
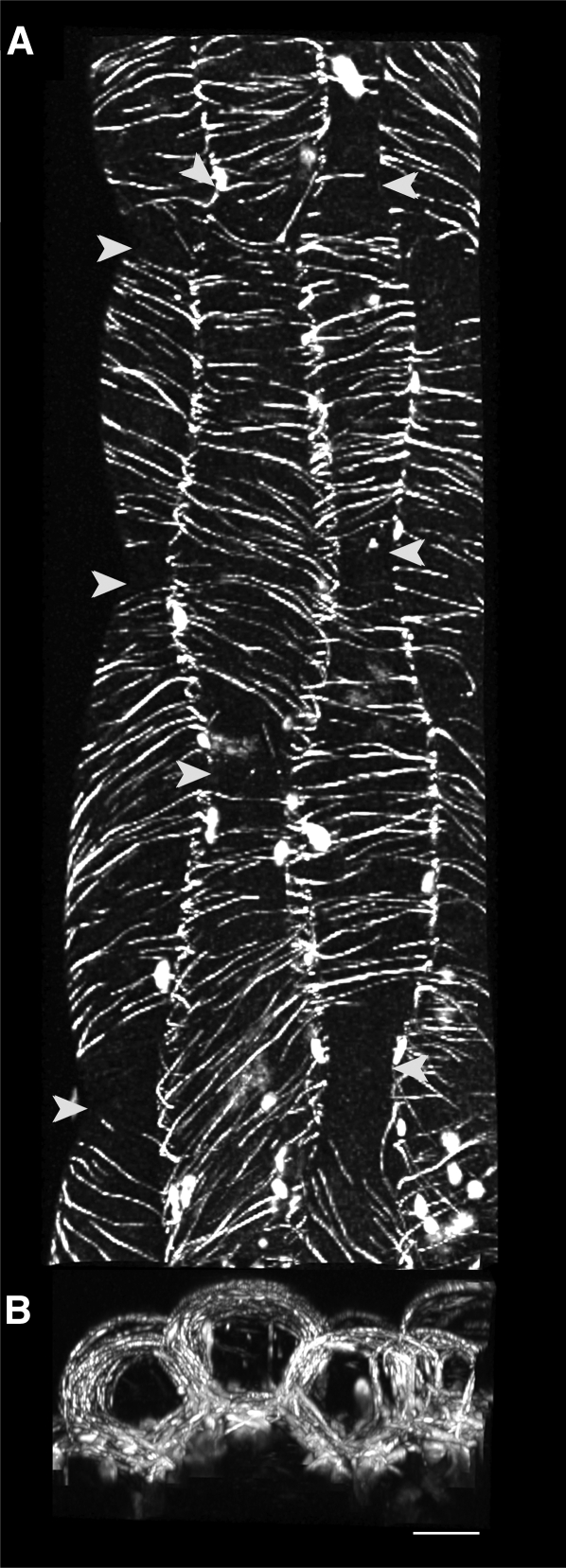
One critical exception to the observation that MAP65-1 and MAP65-2 are present on all cell faces is found in young light-grown seedlings. Hypocotyl cells at 2 to 3 d postgermination displayed transversely coaligned arrays of MAP65-1 and MAP65-2 on the surface and anticlinal side faces of the cell (Figure 6A). In these cases, we observe that MAP65-1 and MAP65-2 are absent at the apical and basal anticlinal end faces (Figure 6B). When viewed along the plant growth axis, reconstructions of these epidermal cells show no observable MAP65-1/2 labeling at the interface between sequential cells in the cell file.
Figure 6.
MAP65 Is Absent from the End Faces of Cells with Transverse MT Arrays.
(A) A maximum intensity projection of serially acquired images showing the outer peridermal arrays labeled with GFP:MAP65-1 in four cell files (right-most file is clipped in half) from 3-d-old light-grown seedlings. End walls are visible due to the constricted cell shape (arrowheads).
(B) A maximum projection from a 90° rotation of the image series capturing the entire volume of the epidermal cells along the length of the cell files shown in (A). GFP:MAP65-1 labeling is visible in the cortex adjacent to the side walls and peridermal cortices. Labeling is absent from end walls in cells with transverse arrays. Bar = 10 μm.
MAP65-1 and MAP65-2 Associate with MT Bundles
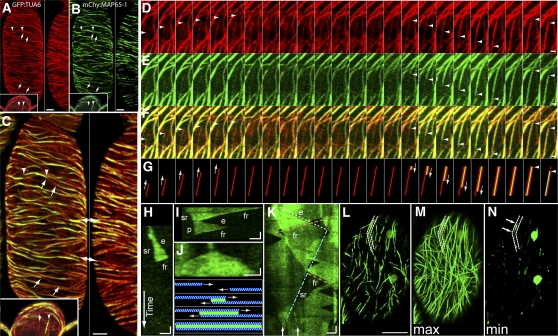
To covisualize MAP65 with MTs, we created double transgenic plant lines expressing either mCherry:MAP65-1 or mCherry:MAP65-2 and a constitutively expressed GFP-α-tubulin-6 transgene (GFP:TUA6). We found that mCherry:MAP65-1 and mCherry:MAP65-2, when expressed from native promoters and in their respective tDNA mutant backgrounds, localized exclusively to MTs (Figures 7A to 7C) throughout the cortical array, including the anticlinal side and end (insets in Figures 7A to 7C) faces of cells. We observed that not all MTs were MAP65 labeled (arrows in Figures 7A to 7C). Unbundled MTs, identified by their dynamic exposed free ends, exhibited barely detectable levels of mCherry:MAP65-1 or mCherry:MAP65-2 (Figures 7A to 7C). We also observed regions of GFP:TUA6 having higher relative brightness than unbundled MTs that were not labeled with MAP65-1 or MAP65-2 (Figures 7A to 7C). These unlabeled regions often appeared as gaps within a linear trace of MAP65-1 or MAP65-2 labeling.
Figure 7.
MAP65-1 and MAP65-2 Dynamically Label MT Bundles.
(A) to (C) Transgenic plants expressing GFP:TUA6 (red; [A]) and mCherry:MAP65-2 (green; [B]) imaged by confocal laser scanning microscopy ([C]; merged image at higher magnification). Reconstructions from serial optical sections reveal MAP65-1 localization to a subset of cortical MTs throughout the cortical MT array. Arrowheads in the left panels indicate regions of overlap in the peridermal (surface) array, and insets show the anticlinal end array. Arrows delimit regions where MTs are not labeled with mCherry:MAP65-2. An anticlinal side cortex is shown in the right panels, and double-headed arrows indicate regions shown in both panels. Bars = 5 μm.
(D) to (G) Time-lapse observations (8-s intervals) of antiparallel MT bundling showing MT polymerization (D) followed by mCherry:MAP65-2 accumulation (E) at the site of antiparallel MT-MT overlap ([F], merge; [G], cartoon diagram). An unbundled MT polymerizing from bottom to top (left set of arrowheads in [D] and [F]) is joined by a second MT moving from top to bottom (middle set of arrowheads in [D] and [F]) coincident with mCherry:MAP65-2 association (arrowheads in [E] and [F]). Rightmost arrowheads show the depolymerization of a MT within the bundle ([D] and [F]) and the disassociation of MAP65-1 (E). Bars = 1 μm in (D) to (F).
(H) to (N) Kymographs of GFP:MAP65-1 ([H] to [K]) from time-lapse images (4-s interval over 9 min). Kymograph (H) showing GFP:MAP65-1 label elongation (e), slow retraction (sr), and fast retraction (fr). Kymograph (I) showing an apparent pause (p). Kymograph showing a triangular shape indicating antiparallel MT interactions ([J]; with diagrammed MTs in blue and MAP65 in green). Kymograph (K) taken from time-lapse ([L] to [N], dashed outline), indicating overlapping intrabundle events with highlighted extension (white dashed line, e), fast retraction (orange dashed line, fr), and slow retraction (stippled line, sr). Horizontal bars = 1 μm and vertical bars = 30 s. The first image of a 135-frame (4-s interval) time-lapse sequence of GFP:MAP65-1 (L). A maximum intensity projection of the 135-frame time-lapse series (M) shows the extent of GFP:MAP65-1 labeling over the 9-min interval. A minimum intensity projection of the same data (N) indicates that most of the labeled regions are devoid of MAP65-1 during some part of the time-lapse experiment. Arrows in (N) correspond to arrows in (K), denoting regions of residual GFP:MAP65-1 labeling. Bar = 5 μm for (L) to (N).
Consistent with in vitro observations (Gaillard et al., 2008), we observed mCherry:MAP65-1 and mCherry:MAP65-2 localized to regions of antiparallel MT overlap in vivo (Figures 7E to 7G). The antiparallel structure of the MT bundle was inferred from time-lapse observation of two unbundled MTs polymerizing toward each other to form the bundle (Figures 7D and 7F). We observed the MAP65 labeling to coincide spatially and temporally with the polymerizing end of the bundling MT (Figures 7D to 7F; see Supplemental Movie 1 online). While we found no direct evidence for MAP65-1 or MAP65-2 labeling of parallel MT bundles, the complexity of the labeling patterns precluded our strictly ruling out this possibility (Van Damme et al., 2004) or the possibility that some antiparallel MT overlaps remain unlabeled. We conclude from our observations that MAP65-1 and MAP65-2 label a subset of cortical array MT bundles, a major portion of which are antiparallel MTs.
MAP65 orthologs in mammalian and fungal systems work with other MAPs to modify the dynamic properties of bundled MTs (Bratman and Chang, 2007; Janson et al., 2007; Bieling et al., 2010; Subramanian et al., 2010). To examine the dynamic properties of the MAP65-labeled regions, we made time-lapse observations of the GFP:MAP65-1 and GFP:MAP65-2 expressing lines. The MAP65 labeling exhibited episodes of extension and retraction in time-lapse movies (Figures 7H to 7N; see Supplemental Movie 2 online). Kymographs made from linear traces of MAP65 labeling in time-lapse experiments (Figures 7H and 7I) show phases of extension (e), fast retraction (fr), and slow retraction (sr) consistent with previously defined phases of MT plus end growth, plus end shortening, and the slower minus end shortening associated with MT treadmilling, respectively. We additionally observed apparent transitions between phases (Figure 7H, e to fr) and evidence for paused (p) GFP:MAP65-1 label (Figure 7I) that could arise due to a paused MT end, intrabundle nucleation, or a junction site where a treadmilling MT has initiated a bundle.
Consistent with the time-lapse colocalization data for mCherry:MAP65-1 and GFP:TUA6 (Figures 7E to 7G), we found that the velocity of MT growth, measured from unbundled MTs (5.5 μm/min ± 1.35, n = 37 MTs), was equivalent to the rate of MAP65 label extension (5.1 μm/min ± 1.1, n = 53 events). Rates for fast retraction (17.2 μm/min ± 7.7, n = 35 events) and slow retraction (1.8 μm/min ± 0.17, n = 32 events) were also found to be equivalent to rates of plus end depolymerization (18.9 μm/min ± 7.2, n = 23 MTs) and minus end depolymerization (1.8 μm/min ± 0.6, n = 16 MTs) in these light-grown cells. Therefore, MAP65 does not appear to alter the growth or shortening velocities of MTs within bundles, and we find that antiparallel-bundled MTs appear to create and remove MAP65 binding sites coincident with polymerization and depolymerization, respectively. Intrabundle catastrophe and rescue (Shaw and Lucas, 2011) were not measured using the MAP65 probes because we cannot unambiguously discriminate between these events and MTs that are entering or leaving sites of antiparallel overlap within a bundle.
We commonly observed kymographs that had triangular shapes, consistent with MTs polymerizing in opposite directions to create antiparallel MT overlaps (Figure 7J). We also observed MAP65-labeled structures that had multiple and overlapping extension and retraction events within the related kymographs (Figure 7K), consistent with the presence of multiple dynamic MTs within the bundle. To determine how much of the MAP65-labeled portion of the MT array is dynamic, we compared maximum and minimum intensity projections from our time-lapse image data (Figures 7L to 7N; see Supplemental Movie 2 online). The maximum intensity projections indicate the total extent of GFP:MAP65-1 labeling in the array over a 9-min period (Figure 7M). When compared with the first image in the time-lapse series (Figure 7L), the maximum intensity image shows longer linear structures that cover a large portion of the cell face. The minimum intensity projection (Figure 7N) shows that nearly all MAP65-labeled structures (Figure 7M) do not remain labeled with MAP65 over the entire 9-min period.
Kymographs made from regions of the minimum intensity projection image with residual GFP:MAP65-1 label (e.g., dashed outline in Figures 7L to 7N corresponds to kymograph in Figure 7K) contained highly dynamic MAP65-1 labeling with evidence for overlapping extension and retraction events. These persistent MAP65-1–labeled regions appear to be dynamic sites where multiple overlapping MT bundles intersect rather than individual stabilized MT bundles. We find that the MAP65-labeled MT bundles in these cells (n = 10 movies of >9 min from light-grown seedlings, >10 kymographs per movie) are predominantly composed of dynamic MTs that create and remove MAP65 binding sites through intrabundle polymerization and depolymerization.
Biochemical and structural studies show that MAP65 proteins form homodimers that associate into MT bundles (Li et al., 2007; Smertenko et al., 2008; Subramanian et al., 2010). Noting the extensive cellular colocalization and gene similarity shared between MAP65-1 and MAP65-2, we used fluorescence redistribution after photobleaching (FRAP) to assess the likelihood that MAP65-1 and MAP65-2 form heterodimers that bind to MT bundles. We reasoned that if a significant fraction of the proteins heterodimerize before insertion into the bundle, their recovery time after bleaching (i.e., the rate of replacement in the bundle) should be equivalent or overlapping. Using a confocal laser scanning microscope, we photobleached GFP:MAP65-1, GFP:MAP65-2, or mCherry:MAP65-2 in 3- to 5-μm regions of individual bundles in single- or double-labeled transgenic plants where the native protein level is diminished. MAP65 accumulation on unbundled MTs was too low in vivo for FRAP measurement, suggesting that MAP65 has a relatively rapid off rate when not bridging MTs. We determined a mean half-time of recovery (t1/2) of 7.1 ±2.5 s for GFP:MAP65-1 (n = 13) in bundles, consistent with prior measurements (Smertenko et al., 2008). We find a t1/2 of 1.1 ± 0.5 s for mCherry:MAP65-2 (n = 12) in bundles, showing that MAP65-2 has a more transient interaction at bundle sites than MAP65-1. Our data are consistent with MAP65-1 and MAP65-2 forming homodimers that enter and leave bundles on different time scales (Smertenko et al., 2004; Subramanian et al., 2010).
The map65-1 map65-2 Mutant Forms Organized MT Arrays
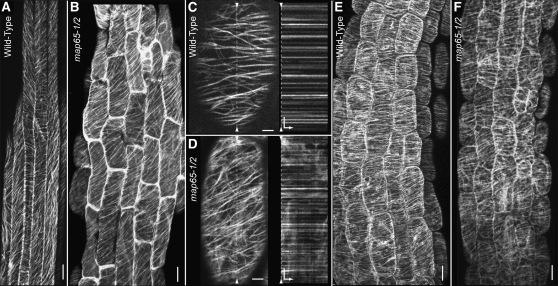
Axial growth of plant cells is strongly correlated with transversely coaligned organization of the cortical MT array (Baskin, 2001; Ehrhardt and Shaw, 2006). To assess cortical MT array organization in hypocotyl cells deficient for MAP65-1 and MAP65-2, we created plants expressing a GFP:TUA6 transgene in the amiRNA65-2 map65-1-1 synthetic double mutant. Since the 488-nm light used for GFP excitation signals transverse MT arrays to reorganize, we rapidly (<1 min) mounted and transferred dark-grown seedlings to the microscope for imaging. We observed that MT arrays in both the wild-type and dex-induced synthetic double mutant plants exhibit predominantly coaligned MT arrays oriented transversely or slightly oblique to the plant growth axis (Figures 8A and 8B). MT density, spacing, and relative orientation do not appear qualitatively different between wild-type and mutant arrays (n = 24 plants for each).
Figure 8.
Interphase MT Array Organization in the Synthetic Double amiRNA65-2 map65-1-2 Mutant.
(A) and (B) MT organization in 5-d-old hypocotyls from mock-treated wild type (A) and synthetic double mutants (B) visualized with GFP:TUA6. Dark-grown seedlings cultured in liquid and continuously treated with dex were rapidly mounted and imaged using confocal microscopy. MT arrays are coaligned and consistently organized into transverse or oblique patterns in both mock-treated (A) and mutant seedlings (B).
(C) and (D) MT arrays in light-grown cells from wild-type (C) and dex-treated amiRNA65-2 map65-1-2 (D) hypocotyls. Kymographs from the dashed line through (C) and (D) represent 3.5 min of time-lapse imaging at 4-s intervals and indicate no lateral MT movement in the wild type (C) or mutant (D) cortical arrays. Hypocotyls from 4-d-old, light-grown hypocotyls treated for 3 h with 10 μM GA4.
(E) and (F) MT arrays are both coaligned and oriented transverse to the plant growth axis in both wild-type (E) and dex-induced amiRNA65-2 map65-1-2 seedlings after GA4 treatment (F).
Bars = 10 μm for (A), (B), (E), and (F) and 5 μm for (C) and (D). Time scale is 30 s (arrow bar) in (C) and (D).
Observations of MT arrays in light-grown plants revealed no obvious differences in MT array organization when wild-type plants were compared with the dex-induced synthetic double mutant (Figures 8C and 8D). Consistent with prior observations of wild-type plants (Shaw et al., 2003), kymographs made from time-lapse images of GFP:TUA6 MTs (4-s interval, 3.5-min duration) in the wild type and synthetic double mutant indicate that the MTs show no obvious lateral movement on the cell cortex and do not slide apart in the absence of MAP65-1/2 (Figures 8C and 8D).
We used FRAP on 10-μm circular regions of cells expressing GFP:TUA6 (Shaw et al., 2003) to determine if the global MT dynamic properties are changed in the synthetic double map65 mutant. Recovery times of 46.2 s for wild-type plants and 49.5 s in dex-treated synthetic double mutants (n = 22 cells from more than three plants for each; see Supplemental Figure 5 online) reveal no substantive difference in MT dynamic properties in MAP65-1/2–deficient plants. Fluorescence recovery, normalized for the bleaching that occurs during time-lapse imaging, reached >95% for both wild-type and mutant MT arrays, suggesting that most of the MTs turn over in both mutant and wild-type plants in the 5-min duration of the experiment.
To functionally test the ability of the amiRNA65-2 map65-1-1 synthetic double mutants to form transverse and coaligned MT arrays, we treated 4-d-old light-grown seedlings with 10 μm GA4 and visualized the MT array by confocal microscopy 3 h after treatment (Shibaoka, 1994; Lloyd et al., 1996; Wenzel et al., 2000). For both the wild type and dex-induced synthetic double mutant, we found that GA4 treatment (n = 10 plants each) led to coaligned MT arrays, organized transverse to the plant growth axis (Figures 8E and 8F). We observed that the degree of MT coalignment was visually indistinguishable from control plants treated with GA4. These data indicate that reduction of MAP65-1 and MAP65-2 levels does not significantly impede the MTs from organizing into an array that is transversely coaligned to the plant growth axis.
DISCUSSION
MAP65-1 and MAP65-2 Modulate Hypocotyl Cell Growth
Our genetic analysis of map65-1 and map65-2 mutants revealed an MT-dependent function in the axial extension of Arabidopsis hypocotyl cells. The map65-2 mutants were shorter than wild-type plants (85 and 80%), and the phenotype was exaggerated when map65-1 was combined with a map65-2 mutation (~60%) or a microRNA targeted against MAP65-2 (~20%). These genetic data establish an interphase function for the plant MAP65-1 and MAP65-2 proteins in directing a specific class of plant cell growth. Surprisingly, deficiencies in MAP65-1/2 accumulation resulted in growth arrest and did not cause significant defects in cell or organ morphology other than size. We observed that cortical MTs organized into coaligned arrays that were transverse to the plant growth axis, even in map65 mutant hypocotyls that were severely retarded for axial growth. We conclude that these MAP65 proteins play a critical role in the MT-dependent mechanism leading to axial cell growth, independent of any mechanical role in organizing the MT array.
MAP65-1 and MAP65-2 and Cortical MT Array Organization
In vitro studies have demonstrated that purified MAP65-1 and MAP65-2 bundle MTs and may affect MT dynamics or stability (Smertenko et al., 2000, 2004, 2008; Van Damme et al., 2004; Chang et al., 2005; Mao et al., 2006; Sasabe and Machida, 2006; Li et al., 2009). To assess these potential functions in planta, we made observations on transgenic plants expressing MAP65-1 and MAP65-2 fusion proteins driven from native promoters in their respective mutant backgrounds. In these plant lines, the MAP65-1/2 proteins associated predominantly with antiparallel MT bundles and did not hyperbundle the MTs (Mao et al., 2006) or drive the array into a specific pattern (Van Damme et al., 2004). Our data show that etiolated map65-1 map65-2 mutant plants have severe axial growth defects, but the MT arrays remain organized and do not degenerate into disorganized tangles of unbundled polymers.
Our time-lapse imaging of interphase plant cells shows that MAP65-1 and MAP65-2 accumulate on MTs as they form bundles through either treadmilling-related interactions or by apparent intrabundle nucleation (Shaw et al., 2003; Dixit and Cyr, 2004; Nakamura et al., 2010). These data provide significant insight into the mechanism of cortical array bundling. We observe that the plus end of the bundling MT uses the interacting MT polymer as a template to direct the trajectory of MT polymerization as the MAP65-1/2 proteins are bound. This bundling mechanism differs significantly from the zippering mechanism observed in vitro, where two preexisting MTs laterally interact and are held together by the bridging interactions of the bundling proteins. The in planta bundling mechanism arises due to the extensive association of MTs with the cell cortex along their length (Hardham and Gunning, 1978; Lloyd et al., 1996). The cortical association immobilizes the MTs and prevents lateral movement that would displace the individual polymers from bundles (Figures 8C and 8D; Shaw et al., 2003). Due to this plasma membrane association, we propose that after a MT plus end has polymerized alongside an adjacent MT template to form a bundle, a physical MAP65 cross-bridge may not be absolutely required in vivo to maintain the coaligned position of MTs at the cell cortex.
Our observations of MAP65-1 and MAP65-2 association with dynamic cortical MT bundles indicate that MAP65-1/2 do not obligately stabilize bundled MTs in planta. MAP65-1/2 association into bundles occurred coincident with the intrabundle MT dynamics and did not affect MT growth or shortening velocities. While catastrophe and rescue were not measured directly in this study, recent work comparing the transition frequencies of unbundled and bundled MTs revealed little relevant difference between the populations (Shaw and Lucas, 2011). The minimum intensity projections of GFP:MAP65-1 time-lapse movies showed that MAP65-1–labeled structures were relatively transient in the cell cortex. This analysis suggests that most, if not all, of the MAP65-1/2–labeled MTs remain dynamic within bundles. Finally, our FRAP experiments show no substantive difference between the general level of MT dynamics in the wild type and map65 synthetic double mutants. These data are consistent with recent studies of intrabundle MT dynamics (Shaw and Lucas, 2011) and, taken together, suggest that the persistence of MT bundles at the plant cell cortex is due to the addition of new MTs to the bundle and not to stabilization of the constituent MTs.
We found that hypocotyl cells from dark-grown plants that were deficient in MAP65-1 and MAP65-2 formed coaligned MT arrays, oriented transverse to the plant growth axis. We additionally found that treating light-grown mutant plants with GA4 (Lloyd et al., 1996) resulted in a reorganization of the cortical MT arrays into a transverse coalignment. To test that MT array organization does not fail under stress in these map65 mutant alleles, we showed that hypocotyl growth was not supersensitive to oryzalin, isoxaben, or temperature extremes. Therefore, our overarching conclusion from these data is that loss of MAP65-1 and MAP65-2, and any mechanical role they may play in MT bundling, does not significantly impair the MT array from organizing into a coaligned pattern that recognizes and orients to the growth axis of the cell.
MAP65-1 and MAP65-2 Exhibit Partial Functional Redundancy
MAP65-1 and MAP65-2 show the highest degree of sequence similarity within the Arabidopsis MAP65 gene family. Therefore, we hypothesized that MAP65-1 and MAP65-2 would show significant functionally redundancy. We found that MAP65-1 and MAP65-2 are both expressed in the same epidermal cells and share completely overlapping localization on MT bundles. However, the map65-1 alleles exhibited no measurable growth phenotype across a range of growth temperatures and conditions while the map65-2 mutant alleles were consistently shorter than the wild type. Complementation studies of the double map65-1 map65-2 tDNA mutant with the fluorescent fusion proteins suggest that MAP65-1 does not fully recover MAP65-2 function in axial growth. Consistent with the proteins not being fully interchangeable, our FRAP experiments suggest that these proteins have different association times within bundles and do not form a significant population of heterodimeric molecules (i.e., MAP65-1/MAP65-2) prior to interactions with MT bundles in vivo. We conclude that MAP65-2 has a more significant role in affecting interphase cell growth and that MAP65-1 can contribute to the same cellular growth mechanism.
Potential Roles for MAP65-1/2 Proteins in Interphase Plant Cell Growth
We found that the cortical MT arrays organized transversely to the plant growth axis in our map65-1/2 mutants but were defective in their ability to support axial cell growth. We show that depolymerizing the MTs with oryzalin relieved the general growth arrest caused by the absence of MAP65-1/2, as evidenced by the larger cells with an aberrant morphology. These data show that MAP65-1/2 function in an MT-dependent mechanism to promote axial cell growth. The cortical MT cytoskeleton has two known functions in directing cell growth; namely, facilitating the localized insertion of cesa complexes into the plasma membrane (Crowell et al., 2009; Gutierrez et al., 2009) and guiding cesa complexes as they extrude cellulose at the plasma membrane (Paredez et al., 2006). Other mechanisms related to vesicle and protein targeting have also been suggested (Roudier et al., 2005; Young et al., 2008). The molecular mechanisms responsible for these MT array functions have not yet been elucidated. We propose that MAP65-1 and MAP65-2 may have a role in localizing the deposition of materials or growth-related factors. We observed that MAP65-1 and MAP65-2 highlight specific regions of the cell cortex where MT polymers will have a relatively long residency time. We reason that this property would make the cortical area an advantageous position for targeting of cell growth–related factors. We note that MAP65-1/2 were absent from the end faces of very young hypocotyl cells in light-grown seedlings (Figure 6). We speculate that these cells are still expanding axially but may not be growing at these end walls.
MAP65 orthologs in yeast (Ase1) and mammalian (Prc1) cells attract other MAPs to the MT bundle (Mollinari et al., 2002; Loïodice et al., 2005; Bieling et al., 2010; Subramanian et al., 2010). Ase1 interacts with CLASP1 to mediate MT dynamics (Bratman and Chang, 2007), and PRC1 loads a kinesin-4 protein, orthologous to the plant FRA1, onto MTs to create specific lengths of antiparallel MT overlap in vitro (Bieling et al., 2010). The observed map65-1/2 hypocotyl phenotype resembles the hypocotyl phenotypes reported for the Arabidopsis clasp1 (Ambrose et al., 2007; Kirik et al., 2007) and the fra1 mutants (Burk et al., 2001). We propose that MAP65-1 or MAP65-2 could act to localize the activity of these interphase plant MAPs to cortical bundles to facilitate targeting of new materials or protein complexes for axial cell growth.
METHODS
Plant Materials
Arabidopsis thaliana seeds were surface sterilized (<10 s) in an aqueous solution of 1.5% hydrogen peroxide and 80% ethanol. Seeds were sown on half-strength Murashige and Skoog (MS) media solidified with 0.8% agar (Sigma-Aldrich) with no added sugar source. All seeds were incubated for 2 d at 4°C in the dark before being placed in a 22°C Percival incubator with continuous light. Dark-grown plants were exposed to light for 2 to 6 h in the incubator and then wrapped in a double layer of aluminum foil or placed in a light tight box.
Transgenic Lines
Full-length genomic clones for MAP65-1 and MAP65-2 genes were PCR amplified from Arabidopsis Col-0 DNA, verified by complete sequencing, and inserted into pPZP211x binary plasmid vectors (Hajdukiewicz et al., 1994). mCherry or eGFP sequences were amplified without the 3′ termination codon and cloned upstream of both genes. Genomic sequence upstream of both MAP65-1 (2.6 kb) and MAP65-2 (2.1 kb) was cloned and inserted upstream of the fluorescent protein sequence to drive gene expression. Inducible artificial microRNA constructs were created by PCR amplification of distinct sequence regions from MAP65-2 according to Schwab et al. (2006) and insertion between flanking regions of the MIR319 microRNA. The artificial microRNAs were subsequently cloned into the pTA7001 binary vector (Aoyama and Chua, 1997) for dex-inducible transcription. All sequence-confirmed constructs were transformed into Agrobacterium tumefacians strain GV3001 for use in plant transformations.
Arabidopsis plants were transformed via the floral dip method (Clough and Bent, 1998). Prior to transformation, bacterial cultures were resuspended in a virulence induction medium containing 0.1 mM acetosyringone for 4 to 8 h. Bacteria were resuspended in water containing 5% Suc and 0.02% Silwet L-77 immediately prior to dipping. Transgenic plants were selected on half-strength MS plates supplemented with 10 μg/mL carbenicillin and either 25 μg/mL kanamycin or 5 μg/μL hygromycin.
Hypocotyl Growth Assays
Seeds were incubated on plates for 4 d at 4°C in darkness, exposed to 2 to 6 h of light, wrapped in foil, and placed vertically in an incubator. Plants were grown for 10 d at 22°C before measurement. Oryzalin (5 μM) supersensitivity and dex induction (5 μM) assays were performed in 5 mL of liquid half-strength MS culture. Ethanol (95%) was used for mock treatment of dex. Hypocotyl length was measured using ImageJ (NIH freeware) from images taken on a stereoscopic microscope. Graphs were created with Kaleidograph (Synergy Software).
Confocal Microscopy
Seedlings were mounted in liquid half-strength MS between a glass slide and a cover slip held together with silicone vacuum grease. Mounted seedlings were rested for >20 min before imaging. Fluorescence images were created using a Leica SP5 microscope with an acousto-optical beam splitter system for creating spectral emission windows. We used either a ×10 0.40 numerical aperture (N.A.), ×20 water immersion 0.70 N.A., ×40 oil immersion 1.25 N.A., or ×63 water immersion 1.2 N.A. lens using 488-nm excitation for GFP and 561-nm excitation for mCherry probes.
FRAP was performed using the Leica live-cell FRAP tool for one- or two-color channels on the Leica SP5 and setting initial detector values to 80% of maximum. For MAP65 fusion proteins, regions of individual bundles were highlighted for bleaching and recovery using flanking regions of the bundle for estimation of photobleaching. Single exponential functions were fit to photobleach-corrected average pixel intensity data, and the half time to recovery (t1/2) was calculated from a least squares best fit model (Walczak et al., 2010). For MT arrays, 10-μm-diameter circles were bleached and recovery was plotted from time-lapse frames taken at 9-s intervals for 5 min (Shaw et al., 2003). For fitting, all data were normalized for imaging-related photobleaching during postbleach time lapse (Walczak et al., 2010), and the average of all experimental traces from the wild type or synthetic double mutant was fit with a single exponential curve.
Surface rendering of hypocotyls was created from through-focal series of propidium iodide–stained hypocotyls. Hypocotyls were stained for 2 h in 20 μg/mL propidium iodide and 0.002% Silwet in half-strength liquid MS media. The 488-nm laser line was used to excite the stain, and the acousto-optical beam splitter was set to capture 550- to 650-nm light emission. Optical stacks of images were rendered using Imaris Bitplane software.
qPCR
RNA was extracted from 5-d-old light-grown seedlings using the Spectrum plant total RNA kit (Sigma-Aldrich). cDNA was produced from 3 ng of total RNA using Superscript III (Invitrogen) reverse transcriptase. Relative RNA amounts were determined by qPCR using the Brilliant II SYBR Green qPCR Master Mix and MxPro3000 qPCR system (Stratagene). A comparative Ct method was used to determine relative quantities (relative quantity = 2(−((Ct GOI unknown − Ct normalizer unknown) − (Ct GOI calibrator – Ct normalizer calibrator))), where GOI is the gene of interest. qPCR was performed in triplicate on three different biological samples. A clathrin adapter subunit was used for normalization (Czechowski et al., 2005). Transcript-specific primer sequences are as follows: MAP65-1 F1, 5′-GCCTTGAGATAGAGAGCCCATAAAG-3′; MAP65-1 R1, 5′-TGCCTTGCCTCTTGGGCGC-3′; MAP65-2 F1, 5′-CGGGCAAGCAAGTCCAAGAGAGACAG-3′; MAP65-2 R1, 5′-AAGCCATCACTGGGTCAGCACGAG-3′; At4g24550 F1, 5′-TGGTAATCCAGAAATCCGTTTAGC-3′; At4g24550 R1, 5′-ACCCCTGAACCTGAAGAACTCC-3′.
Immunoblotting
Protein extracts were created by grinding 20 seedlings in liquid nitrogen for 30 s using plastic pestles in 2.0 mL centrifuge tubes before adding 2% SDS solubilization buffer. Solubilization buffer contained a protease inhibitor cocktail (Sigma-Aldrich) and 0.01% DTT. Samples were boiled for 10 min followed by maximum speed centrifugation for 10 min. Protein samples and BenchMark Prestained Protein Ladder (Invitrogen) were loaded onto 10% polyacrylimade gels and subjected to electrophoresis. Protein loading was assessed by comparing lanes from Commassie Brilliant Blue–stained gels and adjusting load volumes for immunoblots. For blotting, protein was transferred electrophoretically to nitrocellulose membrane, and the membrane was blocked overnight in nonfat milk and incubated with anti-MAP65-1 antibody (that also recognizes MAP65-2) diluted 1:1000 (kind gift from Andrei Smertenko) for >2 h followed by incubation in secondary anti-rabbit sera >2 h. Immunoblots were developed using the Pierce ECL reagent (Thermo Scientific) and Classic Blue Autoradiography BX film (Midsci).
MT Velocities and MAP65-2 Extension and Retraction
Kymographs were generated from time-lapse images (3.98-s interval) of cells expressing both mCherry:MAP65-2 and GFP:TUA6 (ImageJ). From kymographs, the rate of growth and shortening was determined by measuring the slope of the excursions and correcting for pixels per micrometer. Measurements for mCherry:MAP65-2 and GFP:TUA6 were made in the same cells and were generated from 13 cells total taken from three different plants.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MAP65-1 (AT5G55230), MAP65-2 (AT4G26760), and Clathrin adapter protein (At4g24550).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Protein Gel Blots and Loading Controls for Determining Relative MAP65-1/2 Protein Levels in Mutant Alleles.
Supplemental Figure 2. Terminal Hypocotyl Length for Plant Lines Used.
Supplemental Figure 3. Dex-Dependent Expression and Control for Dex Effects on Hypocotyl Growth.
Supplemental Figure 4. Light-Grown map65-1, map65-2, and Combinatorial Mutants.
Supplemental Figure 5. Fluorescence Redistribution after Photobleaching of MTs in the Wild Type and map65 Synthetic Double Mutant.
Supplemental Movie 1. Coincident mCherry:MAP65-1 Labeling of an Antiparallel Microtubule Interaction.
Supplemental Movie 2. GFP:MAP65-1 Time-Lapse Images.
Acknowledgments
We thank Craig Whippo, Geraint Parry, and Roger Hangarter for suggestions with the manuscript. We acknowledge the National Science Foundation (MCB-0920555 to S.L.S.), The Indiana Metabolomics and Cytomics Initiative (METACyt), and the Indiana University College of Arts and Sciences for funding. We thank James Powers, Frederic Pontvianne, Xuhong Yu, and the IU-Light Microscopy and Imaging Center for helpful assistance. We thank Andrei Smertenko for the MAP65-1/2 antibody.
References
- Ambrose J.C., Shoji T., Kotzer A.M., Pighin J.A., Wasteneys G.O. (2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19: 2763–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Bannigan A., Wiedemeier A.M., Williamson R.E., Overall R.L., Baskin T.I. (2006). Cortical microtubule arrays lose uniform alignment between cells and are oryzalin resistant in the Arabidopsis mutant, radially swollen 6. Plant Cell Physiol. 47: 949–958 [DOI] [PubMed] [Google Scholar]
- Baskin T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215: 150–171 [DOI] [PubMed] [Google Scholar]
- Baskin T.I., Wilson J.E., Cork A., Williamson R.E. (1994). Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol. 35: 935–942 [PubMed] [Google Scholar]
- Bieling P., Telley I.A., Surrey T. (2010). A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell 142: 420–432 [DOI] [PubMed] [Google Scholar]
- Bratman S.V., Chang F. (2007). Stabilization of overlapping microtubules by fission yeast CLASP. Dev. Cell 13: 812–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk D.H., Liu B., Zhong R., Morrison W.H., Ye Z.H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13: 807–827 [PMC free article] [PubMed] [Google Scholar]
- Buschmann H., Lloyd C.W. (2008). Arabidopsis mutants and the network of microtubule-associated functions. Mol. Plant 1: 888–898 [DOI] [PubMed] [Google Scholar]
- Caillaud M.C., Lecomte P., Jammes F., Quentin M., Pagnotta S., Andrio E., de Almeida Engler J., Marfaing N., Gounon P., Abad P., Favery B. (2008). MAP65-3 microtubule-associated protein is essential for nematode-induced giant cell ontogenesis in Arabidopsis. Plant Cell 20: 423–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Calder G., Fox S., Lloyd C. (2007). Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat. Cell Biol. 9: 171–175 [DOI] [PubMed] [Google Scholar]
- Chan J., Calder G.M., Doonan J.H., Lloyd C.W. (2003). EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat. Cell Biol. 5: 967–971 [DOI] [PubMed] [Google Scholar]
- Chan J., Crowell E., Eder M., Calder G., Bunnewell S., Findlay K., Vernhettes S., Höfte H., Lloyd C. (2010). The rotation of cellulose synthase trajectories is microtubule dependent and influences the texture of epidermal cell walls in Arabidopsis hypocotyls. J. Cell Sci. 123: 3490–3495 [DOI] [PubMed] [Google Scholar]
- Chan J., Jensen C.G., Jensen L.C., Bush M., Lloyd C.W. (1999). The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc. Natl. Acad. Sci. USA 96: 14931–14936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Rutten T., Lloyd C. (1996). Isolation of microtubule-associated proteins from carrot cytoskeletons: A 120 kDa map decorates all four microtubule arrays and the nucleus. Plant J. 10: 251–259 [DOI] [PubMed] [Google Scholar]
- Chang H.Y., Smertenko A.P., Igarashi H., Dixon D.P., Hussey P.J. (2005). Dynamic interaction of NtMAP65-1a with microtubules in vivo. J. Cell Sci. 118: 3195–3201 [DOI] [PubMed] [Google Scholar]
- Chang-Jie J., Sonobe S. (1993). Identification and preliminary characterization of a 65 kDa higher-plant microtubule-associated protein. J. Cell Sci. 105: 891–901 [DOI] [PubMed] [Google Scholar]
- Chen S., Ehrhardt D.W., Somerville C.R. (2010). Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc. Natl. Acad. Sci. USA 107: 17188–17193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collett C.E., Harberd N.P., Leyser O. (2000). Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol. 124: 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corson F., Hamant O., Bohn S., Traas J., Boudaoud A., Couder Y. (2009). Turning a plant tissue into a living cell froth through isotropic growth. Proc. Natl. Acad. Sci. USA 106: 8453–8458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell E.F., Bischoff V., Desprez T., Rolland A., Stierhof Y.D., Schumacher K., Gonneau M., Höfte H., Vernhettes S. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire P., Findlay K., McCann M.C., Roberts K. (2007). Cell elongation in Arabidopsis hypocotyls involves dynamic changes in cell wall thickness. J. Exp. Bot. 58: 2079–2089 [DOI] [PubMed] [Google Scholar]
- Dixit R., Chang E., Cyr R. (2006). Establishment of polarity during organization of the acentrosomal plant cortical microtubule array. Mol. Biol. Cell 17: 1298–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R., Cyr R. (2004). Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell 16: 3274–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt D.W., Shaw S.L. (2006). Microtubule dynamics and organization in the plant cortical array. Annu. Rev. Plant Biol. 57: 859–875 [DOI] [PubMed] [Google Scholar]
- Emons A.M., Höfte H., Mulder B.M. (2007). Microtubules and cellulose microfibrils: How intimate is their relationship? Trends Plant Sci. 12: 279–281 [DOI] [PubMed] [Google Scholar]
- Gaillard J., Neumann E., Van Damme D., Stoppin-Mellet V., Ebel C., Barbier E., Geelen D., Vantard M. (2008). Two microtubule-associated proteins of Arabidopsis MAP65s promote antiparallel microtubule bundling. Mol. Biol. Cell 19: 4534–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Ostin A., Sandberg G., Romano C.P., Estelle M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ho C.M., Kong Z., Lee Y.R., Qian Q., Liu B. (2009). Evaluating the microtubule cytoskeleton and its interacting proteins in monocots by mining the rice genome. Ann. Bot. (Lond.) 103: 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M., Ehrhardt D.W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hardham A.R., Gunning B.E. (1978). Structure of cortical microtubule arrays in plant cells. J. Cell Biol. 77: 14–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardham A.R., Gunning B.E. (1979). Interpolation of microtubules into cortical arrays during cell elongation and differentiation in roots of Azolla pinnata. J. Cell Sci. 37: 411–442 [DOI] [PubMed] [Google Scholar]
- Hussey P.J., Hawkins T.J., Igarashi H., Kaloriti D., Smertenko A. (2002). The plant cytoskeleton: Recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol. Biol. 50: 915–924 [DOI] [PubMed] [Google Scholar]
- Ishida T., Hashimoto T. (2007). An Arabidopsis thaliana tubulin mutant with conditional root-skewing phenotype. J. Plant Res. 120: 635–640 [DOI] [PubMed] [Google Scholar]
- Janson M.E., Loughlin R., Loïodice I., Fu C., Brunner D., Nédélec F.J., Tran P.T. (2007). Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell 128: 357–368 [DOI] [PubMed] [Google Scholar]
- Kawamura E., Himmelspach R., Rashbrooke M.C., Whittington A.T., Gale K.R., Collings D.A., Wasteneys G.O. (2006). MICROTUBULE ORGANIZATION 1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the Arabidopsis root. Plant Physiol. 140: 102–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Herrmann U., Parupalli C., Sedbrook J.C., Ehrhardt D.W., Hülskamp M. (2007). CLASP localizes in two discrete patterns on cortical microtubules and is required for cell morphogenesis and cell division in Arabidopsis. J. Cell Sci. 120: 4416–4425 [DOI] [PubMed] [Google Scholar]
- Li H., Mao T., Zhang Z., Yuan M. (2007). The AtMAP65-1 cross-bridge between microtubules is formed by one dimer. Plant Cell Physiol. 48: 866–874 [DOI] [PubMed] [Google Scholar]
- Li H., Zeng X., Liu Z.Q., Meng Q.T., Yuan M., Mao T.L. (2009). Arabidopsis microtubule-associated protein AtMAP65-2 acts as a microtubule stabilizer. Plant Mol. Biol. 69: 313–324 [DOI] [PubMed] [Google Scholar]
- Lloyd C., Chan J. (2008). The parallel lives of microtubules and cellulose microfibrils. Curr. Opin. Plant Biol. 11: 641–646 [DOI] [PubMed] [Google Scholar]
- Lloyd C., Shaw P.J., Warn R.M., Yuan M. (1996). Gibberellic-acid-induced reorientation of cortical microtubules in living plant cells. J. Microsc. 181: 140–144 [Google Scholar]
- Loïodice I., Staub J., Setty T.G., Nguyen N.P., Paoletti A., Tran P.T. (2005). Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol. Biol. Cell 16: 1756–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas J., Shaw S.L. (2008). Cortical microtubule arrays in the Arabidopsis seedling. Curr. Opin. Plant Biol. 11: 94–98 [DOI] [PubMed] [Google Scholar]
- Mao G., Buschmann H., Doonan J.H., Lloyd C.W. (2006). The role of MAP65-1 in microtubule bundling during Zinnia tracheary element formation. J. Cell Sci. 119: 753–758 [DOI] [PubMed] [Google Scholar]
- Mao G., Chan J., Calder G., Doonan J.H., Lloyd C.W. (2005a). Modulated targeting of GFP-AtMAP65-1 to central spindle microtubules during division. Plant J. 43: 469–478 [DOI] [PubMed] [Google Scholar]
- Mao T., Jin L., Li H., Liu B., Yuan M. (2005b). Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiol. 138: 654–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q., Du J., Li J., Lü X., Zeng X., Yuan M., Mao T. (2010). Tobacco microtubule-associated protein, MAP65-1c, bundles and stabilizes microtubules. Plant Mol. Biol. 74: 537–547 [DOI] [PubMed] [Google Scholar]
- Mollinari C., Kleman J.P., Jiang W., Schoehn G., Hunter T., Margolis R.L. (2002). PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157: 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S., Smertenko A., Wagner V., Heinrich M., Hussey P.J., Hauser M.T. (2004). The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr. Biol. 14: 412–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T., Sonobe S., Baskin T.I., Hyodo S., Hasezawa S., Nagata T., Horio T., Hasebe M. (2005). Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 7: 961–968 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Ehrhardt D.W., Hashimoto T. (2010). Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 12: 1064–1070 [DOI] [PubMed] [Google Scholar]
- Paradez A., Wright A., Ehrhardt D.W. (2006). Microtubule cortical array organization and plant cell morphogenesis. Curr. Opin. Plant Biol. 9: 571–578 [DOI] [PubMed] [Google Scholar]
- Paredez A.R., Persson S., Ehrhardt D.W., Somerville C.R. (2008). Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiol. 147: 1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Refrégier G., Pelletier S., Jaillard D., Höfte H. (2004). Interaction between wall deposition and cell elongation in dark-grown hypocotyl cells in Arabidopsis. Plant Physiol. 135: 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F., Fernandez A.G., Fujita M., Himmelspach R., Borner G.H., Schindelman G., Song S., Baskin T.I., Dupree P., Wasteneys G.O., Benfey P.N. (2005). COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17: 1749–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe M., Machida Y. (2006). MAP65: A bridge linking a MAP kinase to microtubule turnover. Curr. Opin. Plant Biol. 9: 563–570 [DOI] [PubMed] [Google Scholar]
- Schuyler S.C., Liu J.Y., Pellman D. (2003). The molecular function of Ase1p: Evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 160: 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook J.C., Ehrhardt D.W., Fisher S.E., Scheible W.R., Somerville C.R. (2004). The Arabidopsis sku6/spiral1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. Plant Cell 16: 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook J.C., Kaloriti D. (2008). Microtubules, MAPs and plant directional cell expansion. Trends Plant Sci. 13: 303–310 [DOI] [PubMed] [Google Scholar]
- Shaw S.L., Kamyar R., Ehrhardt D.W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Shaw S.L., Lucas J. (2011). Intrabundle microtubule dynamics in the Arabidopsis cortical array. Cytoskeleton (Hoboken) 68: 56–67 [DOI] [PubMed] [Google Scholar]
- Shibaoka H. (1994). Plant hormone-induced changes in the orientation of cortical microtubules: Alterations in the cross-linking between microtubules and the plasma membrane. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45: 527–544 [Google Scholar]
- Smertenko A., Saleh N., Igarashi H., Mori H., Hauser-Hahn I., Jiang C.J., Sonobe S., Lloyd C.W., Hussey P.J. (2000). A new class of microtubule-associated proteins in plants. Nat. Cell Biol. 2: 750–753 [DOI] [PubMed] [Google Scholar]
- Smertenko A.P., Chang H.Y., Wagner V., Kaloriti D., Fenyk S., Sonobe S., Lloyd C., Hauser M.T., Hussey P.J. (2004). The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell 16: 2035–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A.P., Kaloriti D., Chang H.Y., Fiserova J., Opatrny Z., Hussey P.J. (2008). The C-terminal variable region specifies the dynamic properties of Arabidopsis microtubule-associated protein MAP65 isotypes. Plant Cell 20: 3346–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R., Wilson-Kubalek E.M., Arthur C.P., Bick M.J., Campbell E.A., Darst S.A., Milligan R.A., Kapoor T.M. (2010). Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell 142: 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Himmelspach R., Williamson R.E., Wasteneys G.O. (2003). Mutation or drug-dependent microtubule disruption causes radial swelling without altering parallel cellulose microfibril deposition in Arabidopsis root cells. Plant Cell 15: 1414–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A., Cosgrove D.J. (1989). Gibberellic acid stimulation of cucumber hypocotyl elongation: Effects on growth, turgor, osmotic pressure, and cell wall properties. Plant Physiol. 90: 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee S., Tuchihara K., Hashimoto T. (2002). Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193–196 [DOI] [PubMed] [Google Scholar]
- Van Damme D., Van Poucke K., Boutant E., Ritzenthaler C., Inzé D., Geelen D. (2004). In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol. 136: 3956–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.E., Rizk R.S., Shaw S.L. (2010). The use of fluorescence redistribution after photobleaching for analysis of cellular microtubule dynamics. Methods Cell Biol. 97: 35–52 [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O., Fujita M. (2006). Establishing and maintaining axial growth: Wall mechanical properties and the cytoskeleton. J. Plant Res. 119: 5–10 [DOI] [PubMed] [Google Scholar]
- Wenzel C.L., Williamson R.E., Wasteneys G.O. (2000). Gibberellin-induced changes in growth anisotropy precede gibberellin-dependent changes in cortical microtubule orientation in developing epidermal cells of barley leaves. Kinematic and cytological studies on a gibberellin-responsive dwarf mutant, M489. Plant Physiol. 124: 813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington A.T., Vugrek O., Wei K.J., Hasenbein N.G., Sugimoto K., Rashbrooke M.C., Wasteneys G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613 [DOI] [PubMed] [Google Scholar]
- Young R.E., McFarlane H.E., Hahn M.G., Western T.L., Haughn G.W., Samuels A.L. (2008). Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20: 1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Burk D.H., Morrison W.H., III, Ye Z.H. (2002). A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14: 3101–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]