A direct molecular link between ethylene signaling component EIN3/EIL1 and the iron deficiency response regulator FIT is shown. EIN3 and EIL1 were found to be required for Fe deficiency responses and FIT abundance. In the absence of EIN3/EIL1, FIT was more prone to proteasomal degradation. The ein3 eil1 transcriptome was affected to a greater extent upon iron deficiency compared with the wild type.
Abstract
Understanding the regulation of key genes involved in plant iron acquisition is of crucial importance for breeding of micronutrient-enriched crops. The basic helix-loop-helix protein FER-LIKE FE DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT), a central regulator of Fe acquisition in roots, is regulated by environmental cues and internal requirements for iron at the transcriptional and posttranscriptional levels. The plant stress hormone ethylene promotes iron acquisition, but the molecular basis for this remained unknown. Here, we demonstrate a direct molecular link between ethylene signaling and FIT. We identified ETHYLENE INSENSITIVE3 (EIN3) and ETHYLENE INSENSITIVE3-LIKE1 (EIL1) in a screen for direct FIT interaction partners and validated their physical interaction in planta. We demonstrate that the ein3 eil1 transcriptome was affected to a greater extent upon iron deficiency than normal iron compared with the wild type. Ethylene signaling by way of EIN3/EIL1 was required for full-level FIT accumulation. FIT levels were reduced upon application of aminoethoxyvinylglycine and in the ein3 eil1 background. MG132 could restore FIT levels. We propose that upon ethylene signaling, FIT is less susceptible to proteasomal degradation, presumably due to a physical interaction between FIT and EIN3/EIL1. Increased FIT abundance then leads to the high level of expression of genes required for Fe acquisition. This way, ethylene is one of the signals that triggers Fe deficiency responses at the transcriptional and posttranscriptional levels.
INTRODUCTION
Iron (Fe) is one of the most important micronutrients for many organisms because of its reduction-oxidation role as cofactor in multiple essential proteins and enzymes (Marschner, 1995; de Benoist et al., 2008). Limited Fe bioavailability, due to low solubility under alkaline or calcareous conditions in the soil, is a challenge for plants and can only be overcome by efficient mobilization of this micronutrient. Since Fe reactivity can also confer toxicity (Briat et al., 2010), Fe uptake requires controlled balancing in tight response to the availability and internal requirement. Understanding the regulation of key genes involved in plant Fe uptake is of crucial importance for plant breeding of more nutritious and micronutrient-enriched crops as well as of crops optimized for cultivation on calcareous and alkaline soils.
One of the prime research targets in this respect is the control leading to Fe acquisition in the root. Dicotyledonous plants mobilize Fe via the so-called Strategy I, based on soil acidification and Fe reduction for acquisition of Fe (Marschner and Römheld, 1994), including the model plant Arabidopsis thaliana (Kim and Guerinot, 2007; Walker and Connolly, 2008). In Arabidopsis, three genetic components have been demonstrated to play essential roles in Strategy I. Fe3+ is reduced in outer root cells by the plasma membrane–bound FERRIC REDUCTION OXIDASE2 (FRO2) (Robinson et al., 1999). After reduction, Fe is taken up through the plasmalemma of the root epidermis via the divalent metal transport protein IRON-REGULATED TRANSPORTER1 (IRT1) (Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002). Finally, FRO2 and IRT1 are regulated at the transcriptional level and induced upon Fe deficiency by the central basic helix-loop-helix (bHLH) transcription factor FER-LIKE FE DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005; Bauer et al., 2007).
Control of FRO2 and IRT1 activity is crucial for the plant to regulate Fe uptake into the root. Understanding the regulatory mechanisms that act upon FIT may ultimately allow us to gain insight into the signals by which plants sense their environment and internal requirement for Fe uptake. We previously reported that FIT and its functional homolog (FER) from tomato (Solanum lycopersicum) are regulated by the iron deficiency status of the plant through transcriptional and posttranscriptional mechanisms (Jakoby et al., 2004; Brumbarova and Bauer, 2005). Further indications for regulation of FIT activity came from observations that bHLH038 and bHLH039 can physically interact with FIT in plants and upregulate Fe acquisition responses upon overexpression (Yuan et al., 2008). BHLH038 and BHLH039 together with two other subgroup Ib BHLH genes, BLHL100 and BHLH101, are highly induced by Fe deficiency (Heim et al., 2003; Wang et al., 2007).
A variety of signaling molecules and plant hormones modulate the Fe deficiency response in a positive manner, such as nitric oxide (Graziano et al., 2002; Graziano and Lamattina, 2007; Chen et al., 2010; García et al., 2010) and auxin (Schikora and Schmidt, 2001; Chen et al., 2010), or in a negative manner, like cytokinin (Séguéla et al., 2008). Experiments with the plant hormone ethylene in different dicot plants indicated a physiological connection between ethylene and iron deficiency signaling. Ethylene is produced upon Fe deficiency (Romera et al., 1999; Li and Li, 2004; Zuchi et al., 2009). Applying ethylene precursors such as 1-aminocyclopropane-1-carboxylic acid or ethephon to plants could mimic morphological growth responses of Fe-deficient plants (Romera and Alcantara, 1994; Schmidt et al., 2000). This treatment increased molecular-physiological Fe deficiency responses, like the activation of IRT1 and FRO2 gene expression (Lucena et al., 2006; Waters et al., 2007; García et al., 2010). Conversely, plant treatment with ethylene inhibitors abolished some of the Fe deficiency responses (Romera and Alcantara, 1994) and diminished the mRNA levels of FRO2 and IRT1 (Lucena et al., 2006; García et al., 2010). These experiments suggested that ethylene is able to upregulate Fe acquisition.
ETHYLENE INSENSITIVE3 (EIN3) and ETHYLENE INSENSITIVE3-LIKE1 (EIL1) are two members out of a small family of plant-specific transcription factors that are activated through the ethylene signaling pathway (Chao et al., 1997). These two proteins that are highly related in their amino acid sequence then regulate a series of ethylene responses from the seedling stage to reproduction (Solano et al., 1998; An et al., 2010). EIN3/EIL1 regulation is attributed essentially to posttranscriptional regulation. A major mechanism to regulate EIN3/EIL1 activity acts via controlled proteolysis by the 26S proteasome, which is mediated through recognition of EIN3/EIL1 by Skp, Cullin, F-box–containing complexes with EIN3 BINDING F-BOX PROTEINS1 and 2 (SCFEBF1/EBF2) complexes (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004). Upon ethylene signaling, EBF1 and EBF2 function is prevented so that EIN3/EIL1 are stabilized for inducing downstream ethylene responses (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004). In addition to protein degradation, which seems to be the major pathway regulated by ethylene signaling, differential phosphorylation through a mitogen-activated protein kinase cascade has also been reported, although it remains unclear whether or not phosphorylation depends on the same signaling pathway as proteolysis (Yoo et al., 2008; An et al., 2010). EIN3 and/or EIL1 were shown to bind to promoters of downstream target genes involved in a multitude of responses ranging from biotic stress defense (Chen et al., 2009; Boutrot et al., 2010) and chlorophyll biosynthesis (Zhong et al., 2009) to ethylene signaling (Solano et al., 1998; Konishi and Yanagisawa, 2008).
Although the physiological link between ethylene and Fe deficiency responses was an important observation, the molecular basis of this phenomenon remained elusive. Here, we show a molecular link between Fe deficiency responses and hormone signaling, and by this, we also present evidence for a mechanism that regulates FIT abundance. We demonstrate that EIN3/EIL1 physically interact with FIT, are required for full FIT accumulation, and contribute to full FIT downstream target gene expression. We propose that ethylene is thereby one of the signals that triggers Fe deficiency responses at the transcriptional and posttranscriptional levels.
RESULTS
EIN3 and EIL1 Transcription Factors of the Ethylene Signaling Pathway Were Identified as FIT Interaction Partners
To unravel the network of signaling components involved in iron deficiency responses and in particular to identify components that may alter FIT activity, we screened for protein interaction partners using a yeast two-hybrid assay. The screen was realized under growth conditions that would allow yeast colonies to grow if an activation of the GAL4 promoter occurred due to interaction of the C-terminal FIT peptide fused to the GAL4 DNA binding domain (FIT-C-BD, excluding the N terminus and bHLH domain of FIT) with an expressed peptide from a root –Fe cDNA library fused to the GAL4 activation domain.
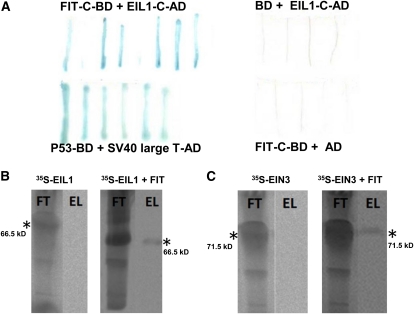
One candidate cDNA was identified three times in the screen out of a total of 14 putative candidate cDNAs identified in the yeast two-hybrid screen (see Methods for further details). This cDNA fragment was found to represent the last 250-bp coding sequence of the EIL1 transcription factor, which plays a role in the ethylene response pathway (Chao et al., 1997). After retransformation of yeast cells with the EIL1 cDNA clone (EIL1-C-AD), positive interaction signals with FIT-C-BD were still obtained (Figure 1A). The interaction between FIT and EIL1 was further verified by in vitro protein pull-down assays. In the final eluates containing the retained proteins, we were able to detect radioactive bands, confirming interactions between full-length FIT and full-length EIL1 (Figure 1B). In a further step, we could also identify in vitro interaction of full-length FIT with the closest related homolog of EIL1, namely, EIN3 (Figure 1C). Taken together, we could thus demonstrate that FIT can physically interact with EIL1 in yeast and in vitro and with EIN3 in vitro.
Figure 1.
Yeast and in Vitro Protein Interaction of FIT with EIL1 and EIN3.
(A) In yeast; lacZ assays after retransformation. FIT-C fused to Gal4 DNA binding domain (FIT-C-BD) with EIL1-C peptide fused to activation domain (EIL1-C-AD) (top left, positive interaction resulting in blue staining); unfused BD with EIL1-C-AD (top right, negative control, white staining); P53-BD with SV40 large T antigen-AD (bottom left, positive control, blue staining); and FIT-C-BD with unfused AD (bottom right, negative control, white staining).
(B) In vitro protein pull down of 35S-labeled EIL1 by S-tagged FIT, detected by immunoblot and phosphor imaging (right side), and in the absence of S-tagged FIT (left side, negative control).
(C) As in (B) with 35S-labeled EIN3. FT, flow-through fraction after binding and loading onto S-agarose column; EL, eluate fraction containing radioactive protein interaction partner. Asterisk indicates the respective positions and sizes of the radiolabeled proteins.
[See online article for color version of this figure.]
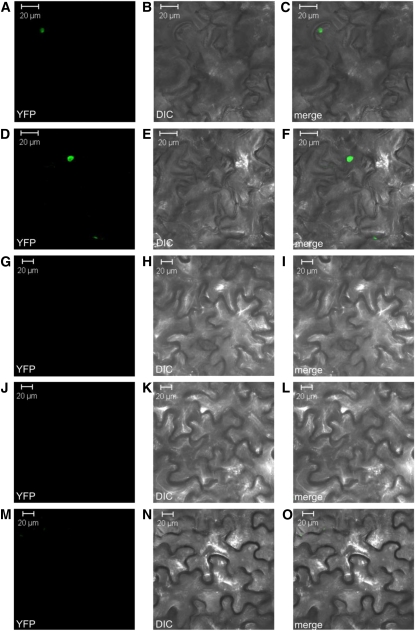
To address the interaction between FIT and EIN3/EIL1 in planta, we performed bimolecular fluorescence complementation (BiFC). The full-length coding sequences of FIT, EIN3, and EIL1 were tested for interaction in reciprocal combinations with the N-terminal (YN) or C-terminal (YC) part of the yellow fluorescent protein (YFP), respectively, in transient Agrobacterium tumefaciens–mediated Nicotiana benthamiana leaf infiltration assays. Protein interactions were revealed due to fluorescence of reconstituted YFP fluorescent protein using confocal microscopy. Fluorescent signals were observed in the nuclei of the epidermal cells with all combinations of FIT and EIN3 as well as of FIT and EIL1. The most intense fluorescence signals were obtained when combining YN-EIN3 or YN-EIL1 with YC-FIT, respectively (Figures 2A to 2F). Empty vector controls and combinations of empty vector controls with the respective partial YFP fusion protein did not result in any YFP signals (Figures 2G to 2O).
Figure 2.
In Planta Protein Interaction of FIT and EIN3/EIL1.
BiFC of YFP in transiently transformed tobacco leaf epidermal cells. Left column ([A], [D], [G], [J], and [M]), YFP fluorescent signal detection by confocal microscopy; middle column ([B], [E], [H], [K], and [N]), differential interference contrast (DIC) microscopy; right column ([C], [F], [I], [L], and [O]), merge of fluorescent signal and DIC. YC-FIT plus YN-EIN3 ([A] to [C]); YC-FIT plus YN-EIL1 ([D] to [F]); negative control YC plus YN-EIN3 ([G] to [I]); YC-FIT plus negative control YN ([J] to [L]); negative control YC plus negative control YN ([M] to [O]). For details on the number of experiments and fluorescence signals, refer to Methods.
Taken together, our results confirmed a physical protein interaction between the bHLH transcription factor FIT and the ethylene signaling transcription factors EIN3 and EIL1 in the nucleus of plant cells.
The protein interaction studies suggest that an Fe deficiency is not required for the interaction of FIT with EIN3/EIL1. EIN3 and EIL1 can act in light-grown seedlings (Smalle et al., 1997). To confirm a potential action in the light, we investigated whether EIL1-GFP (green fluorescent protein) could be detected in 6-d-old light-grown seedlings using EIL1-GFP plants. EIL1-GFP signals were detectable in the nuclei of plant cells in the roots of seedlings grown in the presence (+) and absence (−) of Fe (see Supplemental Figure 1 online). Hence, EIL1-GFP was found to accumulate in the EIL1-GFP plants in the light at + and −Fe. We deduce that in root cells of light-grown seedlings EIL1 and its associate EIN3 are thus likely able to interact with FIT under + and –Fe.
FIT Protein Levels and FIT Downstream Responses Were Affected by Ethylene Inhibition
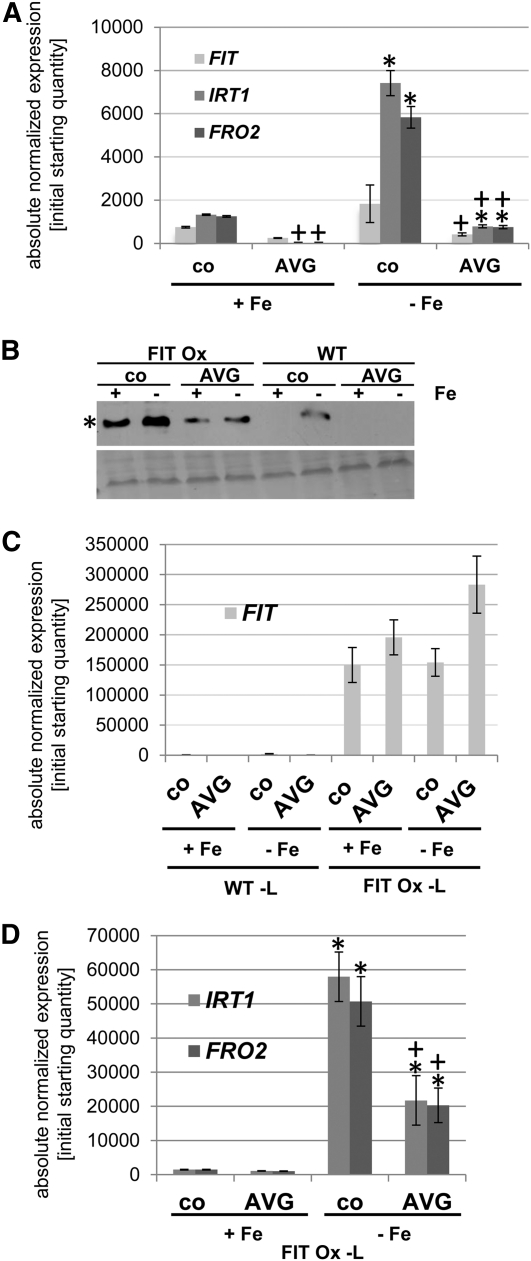
The presence of these protein interactions suggests that ethylene is involved in the regulation of molecular Fe deficiency responses. It was previously reported that the application of ethylene synthesis inhibitor aminoethoxyvinylglycine (AVG) suppresses Fe acquisition responses (Romera and Alcantara, 1994; Lucena et al., 2006; García et al., 2010). Therefore, we first tested whether such a repressing effect on Fe acquisition responses was also detectable in our 6-d growth system. Seedlings were germinated in the presence or absence of AVG and Fe, respectively. Monitoring of gene expression showed that IRT1 and FRO2 were induced by –Fe in the absence and presence of AVG. Expression levels were lower in the presence of AVG than in the control (Figure 3A). Expression of FIT was also reduced upon AVG treatment compared with the control at –Fe. Hence, we conclude that in our seedling growth system, ethylene was necessary for full upregulation of Fe deficiency responses.
Figure 3.
Reduced Fe Deficiency Gene Expression and FIT Abundance in Response to AVG.
Gene and protein expression analysis in response to Fe supply (+ and –Fe) and 10 μM AVG (co means control without AVG), showing that Fe deficiency gene expression and FIT protein abundance were reduced by AVG. In (A), (C), and (D), asterisk indicates significant change versus +Fe (P < 0.05); + indicates significant change versus no AVG control (P < 0.05).
(A) qRT-PCR analysis of FIT, IRT1, and FRO2 in wild-type seedlings; 6-d seedling growth assay, n = 2. sd were calculated for two biological replicates.
(B) FIT protein detection by immunoblot in FIT Ox and wild-type (WT) seedlings with anti-FIT-C antibody (top image; asterisk indicates 35 kD position of FIT protein). Ponceau S-staining of proteins on the same membrane (bottom image).
(C) qRT-PCR analysis of FIT in wild-type and FIT Ox leaves.
(D) qRT-PCR analysis of IRT1 and FRO2 in FIT Ox leaves.
We used the wild type and a FIT overexpression line (2xProCaMV35S:FIT, named FIT Ox; Jakoby et al., 2004) to monitor the effect of AVG on FIT protein level. In wild-type roots, FIT protein was only detectable at –Fe in the control, but not at + Fe, while in FIT Ox plants, FIT protein was present in + and −Fe control conditions (Figure 3B). The FIT Ox line was thus useful for further analysis since the constitutive FIT mRNA expression allowed analysis of protein expression uncoupled from FIT gene transcription. Upon AVG treatment, FIT protein was no longer detectable in the wild type. In FIT Ox roots, FIT protein levels were reduced to 30 and 40% of control levels by AVG under + and –Fe conditions, respectively (Figure 3B). In these plants, FIT mRNA levels were at least as high upon AVG treatment in leaves as in the controls (Figure 3C). Therefore, FIT protein downregulation in AVG-treated plants was independent of transcriptional regulation of FIT. AVG treatment resulted in a lower gene expression level of IRT1 and FRO2 in the FIT Ox leaves (Figure 3D).
As a control, we also tested the effect of ethylene inhibition by 20 μM aminooxoacetic acid (AOA) and 200 μM silver thiosulfate (STS) on FIT protein accumulation (see also Romera and Alcantara, 1994). AOA is an inhibitor of ethylene synthesis, and STS is an inhibitor of ethylene perception. We observed that AOA and STS treatment resulted in reduced FIT protein levels, to 80 and 50% of those in control FIT Ox plants, respectively (AVG treatment caused a reduction to 40% of the control level in this experiment; see Supplemental Figure 2 online).
In conclusion, AVG did not only inhibit Fe deficiency response gene expression as reported in previous studies (Romera and Alcantara, 1994; Lucena et al., 2006; García et al., 2010) but also FIT protein accumulation. These findings suggest that ethylene regulates Fe acquisition responses also via FIT protein stability.
ein3 eil1 Mutations Regulated the Expression of Genes Involved in Fe Acquisition
EIN3 and EIL1 are central redundant transcription factors in the ethylene signaling pathway and are able to physically interact with FIT in root cells. We asked whether elimination of functional EIN3 and EIL1 had any measurable effect on the molecular iron deficiency responses that are regulated by FIT.
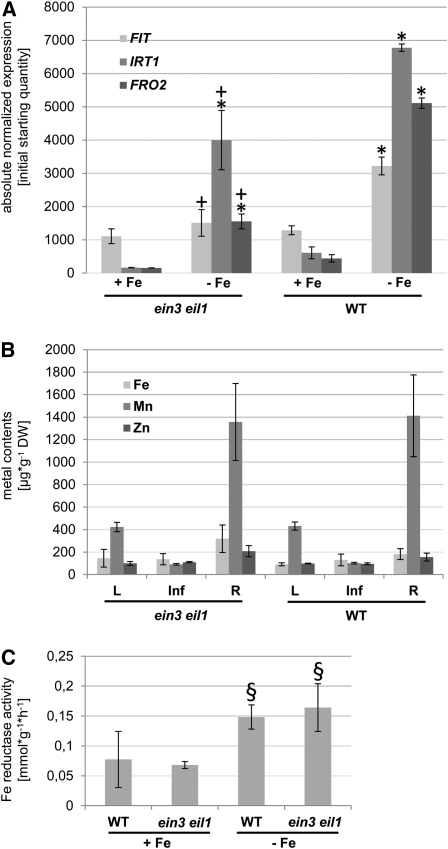
ein3 eil1 double mutant plants together with wild-type control plants were grown in the 6-d growth assay on either + Fe or –Fe. Quantitative RT-PCR (qRT-PCR) analysis was performed to investigate the expression of Fe acquisition genes. FIT was induced threefold upon –Fe in the wild type (Figure 4A), while IRT1 and FRO2 were induced to a higher extent, namely 15- and 10-fold, respectively, demonstrating that induction of expression in response to Fe deficiency took place (Figure 4A). We found that IRT1 and FRO2 transcript levels were reduced to 40% in the ein3 eil1 double mutant compared with the wild type under both Fe supply conditions (Figure 4A; –Fe, P < 0.05; +Fe, P = 0.07 and 0.08, respectively). Induction of IRT1 and FRO2 by –Fe compared with +Fe was still observable in ein3 eil1. Compared with the wild type, FIT decreased to half the levels in the double ethylene signaling mutant upon –Fe, whereas no decrease was obvious under +Fe (Figure 4A). FIT was therefore not differently expressed in ein3 eil1 and wild-type plants grown upon +Fe, but a clear induction at –Fe failed in ein3 eil1 (Figure 4A). These observations indicate a role of ethylene and EIN3 and EIL1 as positive regulators of the Fe deficiency response in Arabidopsis seedlings.
Figure 4.
Altered Physiological Responses of ein3 eil1 Mutants to Fe Deficiency.
Fe deficiency gene expression was reduced in ein3 eil1 compared with the wild type, while metal content and Fe reduction were not significantly different. In (A), asterisk indicates significant change versus +Fe (P < 0.05); + indicates significant change versus wild-type control (P < 0.05). In (C), § indicates difference versus +Fe (with P = 0.07 to 0.08).
(A) qRT-PCR analysis of FIT, IRT1, and FRO2 in ein3 eil1 and wild-type (WT) seedlings; 6-d seedling growth assay, n = 2. sd were calculated for two biological replicates.
(B) Metal contents of leaves (L), inflorescences (Inf), and roots (R) of 4-week-old plants (n = 5). sd were calculated for five biological replicates. DW, dry weight.
(C) Fe reductase activity of wild-type and ein3 eil1 seedling roots; 6-d seedling growth assay (n = 3).
Due to their lower expression of Fe acquisition genes, we suspected that perhaps ein3 eil1 mutants take up less Fe and/or other metals than wild-type plants. To test this idea, we determined the metal contents in the leaves, inflorescences, and roots of ein3 eil1 and wild-type plants. We found that in all three organs, Fe, Mn, and Zn contents were similar between ein3 eil1 mutants and the wild type (Figure 4B). Therefore, we conclude that the lower expression of Fe deficiency genes in ein3 eil1 mutants did not result in lower Fe contents as was described for Fe deficiency mutants, such as fro2 (Yi and Guerinot, 1996), irt1 (Vert et al., 2002), or fit (Colangelo and Guerinot, 2004). This finding was confirmed by the observation that despite the lower gene expression of FRO2, Fe reductase activity was similar in ein3 eil1 and the wild type (Figure 4C). ein3 eil1 mutants are thus not typical Fe deficiency mutants, which often display enhanced root Fe reductase activity at +Fe or decreased activity at −Fe.
Transcriptome Changes in ein3 eil1 Mutants Were More Pronounced at –Fe Than at +Fe
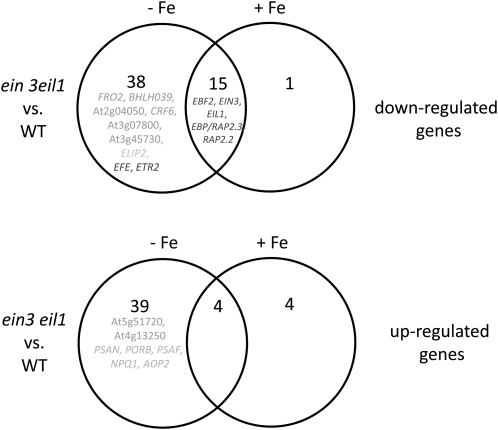
We further investigated the phenotype of ein3 eil1 seedling mutants versus the wild type in response to + and –Fe using microarray-based transcriptome analysis. Of 22,089 genes, 101 were found to be significantly differentially expressed between the wild type and mutant across the three biological replicates (see Supplemental Table 1 online; Figure 5). Interestingly, 19 genes were differentially regulated independently from Fe supply, of which 15 genes were downregulated in the mutant and four genes upregulated compared with the wild type. Among these 15 downregulated genes, five were related to the ethylene signaling pathway (EBF2, EIN3, EIL1, EBP/RAP2.3, and RAP2.2; see Supplemental Data Set 1 online; Figure 5, dark gray; in Supplemental Figure 3A online, EBF2 was used to validate microarray results by RT-PCR,). Five genes were found to be exclusively up- or downregulated in ein3 eil1 upon +Fe but not upon –Fe. Interestingly, the majority of differentially regulated genes showed this behavior only upon –Fe but not upon +Fe. Thirty-eight genes were downregulated in ein3 eil1 versus the wild type upon –Fe, while 39 genes were upregulated. Among the 38 downregulated genes, we could identify two characterized Fe deficiency response genes, FRO2 and BHLH039, and four additional genes that we have previously identified to be upregulated by −Fe in a robust manner in leaves, namely, At2g04050, CRF6, At3g07800, and At3g45730 (M. Schuler, A. Keller, C. Backes, K. Philippar, H.-P. Lenhof, and P. Bauer, unpublished data) (see Supplemental Table 1 online; Figure 5, medium intensity gray; in Supplemental Figure 3B online, FRO2 was used to validate microarray results by RT-PCR). Two ethylene signaling–related genes, EFE and ETR2, were also among the genes. Among the 39 genes upregulated in ein3 eil1, we could identify two genes that we described as repressed by –Fe in leaves, namely, At5g51720 and At4g13250 (M. Schuler, A. Keller, C. Backes, K. Philippar, H.-P. Lenhof, and P. Bauer, unpublished data). Thus, several of the genes differentially expressed in leaves in response to Fe supply are under control of EIN3/EIL1 when plants are grown in –Fe. We noted that among the genes differentially expressed in ein3 eil1 and the wild-type plants grown in –Fe, several have functions in light adaptation of photosynthesis, such as ELIP2, PSAN, PORB, PSAF, NPQ1, and AOP2 (see Supplemental Table 1 online; Figure 5, light gray). These findings suggest that EIN3/EIL1 might function in adapting the chloroplastic light responses to –Fe. The high number of deregulated genes in –Fe suggests that EIN3/EIL1 specifically act to adjust plant responses to Fe availability.
Figure 5.
Transcriptome Changes between ein3 eil1 Mutants and the Wild Type at + and –Fe.
The Venn diagrams represent the number of genes that were found to be significantly differentially expressed between ein3 eil1 and wild-type (WT) seedlings grown in the 6-d growth assay, upon – and +Fe (total number of differentially expressed genes was 101 out of a total of 22,089 genes, compare with gene list in Supplemental Data Set 1 online). The upper Venn diagram represents the number of genes that were found to be downregulated, and the lower Venn diagram depicts the number of genes that were found to be upregulated in the comparison of ein3 eil1 versus the wild type. Genes in dark gray have assigned functions in the ethylene signaling pathway. Genes in medium gray are markers for a differential regulation between − and +Fe in the wild type. Genes in light gray have an assigned function in the chloroplast and light responses. Differential expression of EBF2 and FRO2 was verified in RT-PCR experiments using the same RNA samples used for microarray hybridization (see Supplemental Figure 3 online).
The Expression of Genes Involved in Fe Did Not Reveal a Synergistic Interaction between FIT and EIN3/EIL1
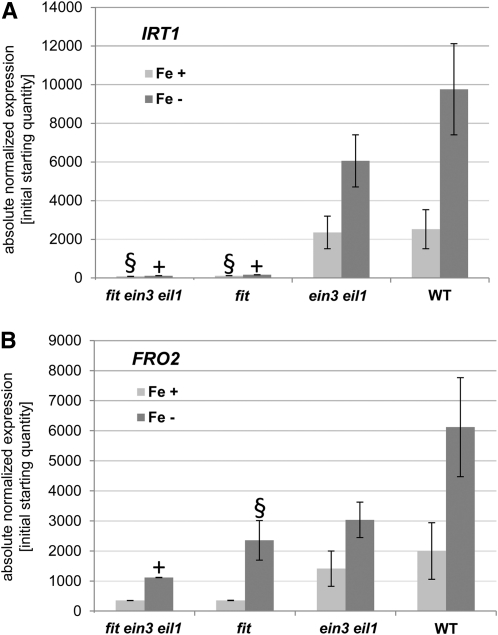
EIN3/EIL1 are required for the full upregulation of Fe acquisition response genes; however, in the fit mutant as well as in ein3 eil1 double mutants, an induction of IRT1 and FRO2 is still possible, although at a lower expression level than in the wild type (Jakoby et al., 2004). We asked whether FIT and EIN3/EIL1 may act in a synergistic manner, as would be expected if, for example, the two proteins acted jointly in a transcription factor complex to induce FIT downstream target genes. To test this, we produced a fit ein3 eil1 triple mutant, which was then analyzed for its expression of IRT1 and FRO2. In the fit ein3 eil1 triple mutant, IRT1 and FRO2 transcript amounts were 1 and 10% of the wild type in –Fe, respectively (Figures 6A and 6B). In the fit mutant, the transcript levels were 2% (IRT1) and 30% (FRO2) of wild-type levels in –Fe, respectively (Figures 6A and 6B). A significant difference of IRT1 and FRO2 expression between fit and fit ein3 eil1 with P < 0.05 was not found. IRT1 and FRO2 expression was lowered up to 40% of the wild-type levels in ein3 eil1 (Figures 4A, 6A, and 6B). Therefore, the combined effects of fit and ein3 eil1 mutations on the expression level of IRT1 and FRO2 were not found to be stronger than the sum of their individual effects. FIT and EIN3/EIL1 therefore did not act in a synergistic manner.
Figure 6.
Molecular Responses of Triple Mutant fit ein3 eil1 Plants.
qRT-PCR analysis of IRT1 (A) and FRO2 (B) in fit ein3 eil1, fit, ein3 eil1, and wild-type (WT) seedlings, showing that expression of IRT1 and FRO2 is not significantly different in fit ein3 eil1 and fit. This suggests that FIT and EIN3/EIL1 do not act in a synergistic manner to induce these genes. Six-day growth assay, n = 2; sd were calculated for two biological replicates; + indicates significant change versus wild-type control (P < 0.05); § indicates difference versus wild-type control (with P = 0.07 to 0.08).
Induction of Fe Acquisition Genes Did Not Require EIN3/EIL1 in FIT Ox Plants
FIT overexpression driven by a double cauliflower mosaic virus (CaMV) 35S promoter resulted in ectopic FIT expression and activity in leaves (Jakoby et al., 2004). Whereas gene expression from the strong constitutive promoter was independent of Fe supply, the ectopic induction of FRO2 and IRT1 in leaves mediated by the ectopic expression of FIT took place only under –Fe (Jakoby et al., 2004). These observations suggested that FIT activity was modulated by posttranscriptional regulation. One possible explanation for these observations is that EIN3/EIL1 proteins are needed to interact with FIT in leaves upon Fe deficiency to obtain FIT activity. To test this idea, we crossed the FIT Ox line (Jakoby et al., 2004) into the background of ein3 eil1 double mutants and performed gene expression analysis in leaves. As a control, FIT transcript levels were monitored and found to be present in FIT Ox and FIT Ox ein3 eil1 leaves but not in the absence of FIT Ox (see Supplemental Figure 4A online). We found that in FIT Ox leaves with wild-type EIN3/EIL1 and with an ein3 eil1 background, IRT1 and FRO2 expression levels were only detectable at –Fe but not at +Fe. While IRT1 expression was not significantly lowered in FIT Ox ein3eil1 compared with FIT Ox alone (with wild-type EIN3/EIL1), FRO2 expression was slightly lowered (P = 0.08).
Therefore, induction of IRT1 and FRO2 in FIT Ox leaves at –Fe did not require EIN3 and EIL1. These results further confirm that FIT can act independently from EIN3 and EIL1 and that FIT together with EIN3/EIL1 does not constitute a functional transcription factor complex relevant for induction of downstream IRT1 and FRO2 gene expression.
Induction of Subgroup Ib BHLH Genes upon –Fe Did Not Require EIN3/EIL1
One possible explanation for the reduced expression of Fe acquisition genes in ein3 eil1 mutants could be that in these mutants, BHLH038- and BHLH039-type genes are expressed at a lower level so that less of these bHLH proteins are available to interact with FIT (see Yuan et al., 2008) to induce Fe uptake. To test this idea, we investigated the expression levels of all four Fe deficiency-inducible subgroup Ib BHLH transcription factor genes. We found that expression levels of BHLH038, BHLH039, BHLH100, and BHLH101 were similar in ein3 eil1 and the wild type at +Fe and −Fe and that the genes were only strongly expressed at −Fe (see Supplemental Figures 5A to 5D online), in agreement with previous studies (Wang et al., 2007). In fit mutants, BHLH gene expression was induced at + and –Fe due to the strong Fe deficiency phenotype of the mutant (see Supplemental Figures 5A to 5D online; Wang et al., 2007). BHLH gene expression was not found to be significantly reduced in fit ein3 eil1 mutants compared with the fit mutant alone (see Supplemental Figures 5A to 5D online).
In FIT Ox leaves, the four BΗLH genes were also only upregulated at –Fe but not at +Fe (see Supplemental Figures 5E to 5G online). In FIT Ox –Fe leaves in an ein3 eil1 background, BHLH038, BHLH100, and BHLH101 were upregulated to a level similar as in FIT Ox leaves with a wild-type EIN3/EIL1 background (see Supplemental Figures 5E, 5G, and 5H online). Only BHLH039 expression was found to be reduced by half in FIT Ox leaves with an ein3 eil1 background (see Supplemental Figure 5F online). Thus, in FIT Ox plants, ein3 eil1 mutations resulted in similar expression levels of BHLH038, BHLH100, and BHLH101 genes compared with the wild type. BHLH039 expression was reduced in FIT Ox ein3 eil1 and reduced in ein3 eil1 in the microarray experiment. Since the strength of BHLH gene expression depends on the degree of Fe deficiency (Wang et al., 2007), one explanation may be that ein3 eil1 mutations decreased Fe deficiency signaling, which may have resulted in lower BHLH039 expression in some experiments. We conclude that EIN3/EIL1 cannot be the primary transcription factors for upregulating the Fe deficiency–inducible BHLH genes.
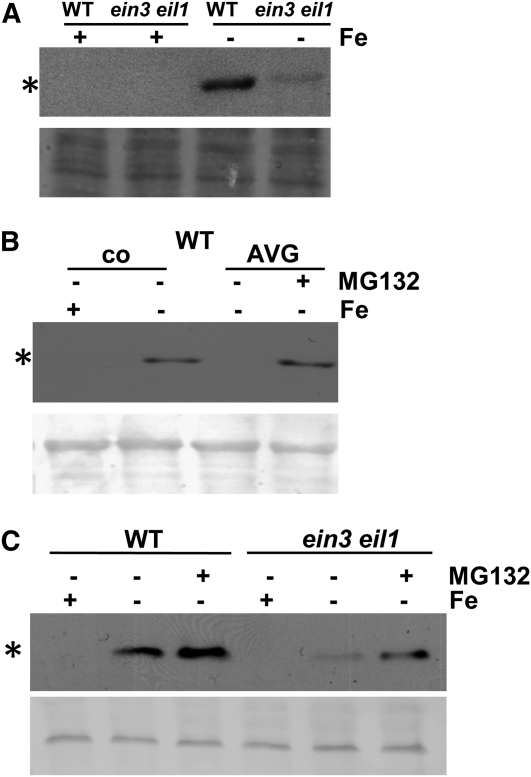
FIT Protein Abundance Was Reduced in ein3 eil1 Plants
To better understand the meaning of the combined action of upregulation of Fe deficiency responses by FIT and EIN3/EIL1 on one side and protein interaction between FIT and EIN3/EIL1 on the other side, we tested whether FIT protein levels may be affected by EIN3/EIL1. In wild-type roots, FIT was expressed at the protein level upon –Fe but not at +Fe (Figure 7A; see also Figure 3B). Interestingly, we found that FIT protein levels were reduced to 8% in the ein3 eil1 mutant compared with the wild type at −Fe. However, FIT gene expression levels had been reduced by half in ein3eil1 at –Fe (Figure 4A). The reduction was only observed in wild-type plants. In FIT Ox ein3 eil1 plants, the FIT protein level was not reduced compared with FIT Ox (in a wild-type EIN3/EIL1 background), while in the same experiment, FIT protein was reduced to 30% in ein3 eil1 versus the wild type at –Fe (see Supplemental Figure 6 online).
Figure 7.
FIT Protein Regulation in ein3 eil1 and upon MG132 Treatment.
(A) FIT protein abundance in seedling roots of the wild type (WT) and ein3 eil1, exposed to + or –Fe as indicated, showing that FIT protein abundance is reduced in the ein3 eil1 mutant.
(B) The effect of MG132 on FIT abundance in AVG-treated roots, showing that MG132 treatment restored FIT abundance upon AVG treatment. Wild-type seedlings, exposed to + and −Fe, treated with 10 μM AVG or untreated (co, control) and treated for 4 h with or without 100 μM MG132 (+ or −MG132).
(C) The effect of MG132 on FIT abundance in ein3 eil1, showing that MG132 restored FIT protein levels in ein3 eil1 mutants. Seedlings exposed to + and −Fe, treated for 4 h with or without 100 μM MG132 (+ or –MG132); immunoblot analysis with anti-FIT-C antibody (top image; asterisk indicates 35-kD position of FIT protein) and Ponceau S-staining of proteins on the same membrane (bottom image).
These experiments indicate that EIN3/EIL1 affect FIT accumulation in the wild type. We deduce that the interaction between FIT and EIN3/EIL1 may serve to increase the level of FIT protein.
Treatment with MG132 Restored FIT Protein Abundance
The downregulation of FIT levels in response to AVG and in the ein3 eil1 background suggests that perhaps FIT was destabilized and degraded by the proteasome in the absence of ethylene signaling. To test this idea, we incubated wild-type plants after their growth on AVG with the proteasome inhibitor MG132. In this experiment, FIT protein was reduced to 30% upon AVG treatment at –Fe compared with the control (Figure 7B). Upon treatment with 100 μM MG132, FIT protein levels were restored in plants exposed to AVG (Figure 7B).
We also tested the effect of MG132 on ein3 eil1 seedlings. In this experiment, the FIT protein level was 10% in the mutant versus wild type at –Fe. Upon treatment with 100 μM MG132, we observed a restoration of FIT protein abundance to 60% of the wild type at −Fe (Figure 7C).
Hence, these results indicate that in the absence of EIN3/EIL1, FIT protein was more prone to proteasome-dependent degradation than in their presence.
DISCUSSION
In this work, we demonstrated a direct molecular link between ethylene signaling and Fe deficiency response regulators at the protein level. We showed that EIN3/EIL1, central transcription factors of the ethylene pathway, physically interact with FIT, a master regulator of Fe deficiency responses.
Our results indicate the following flow of events: Ethylene production upon Fe deficiency positively affects FIT protein expression levels. Through physical interaction with EIN3/EIL1, proteasomal degradation of FIT is reduced. This in turn results in a higher level of Fe acquisition gene expression. These results agree well with previous findings on enhanced Fe deficiency gene expression in response to ethylene (Lucena et al., 2006). We propose that by controlling FIT expression, ethylene is one of the signals that triggers Fe deficiency responses at the transcriptional and posttranscriptional levels. We suggest that this EIN3/EIL1-mediated influence may serve to coadapt Fe acquisition and environmental responses, which otherwise may lead, for example, to photooxidative damage.
FIT Interacts Physically with EIN3/EIL1, Independently of Fe
EIN3/EIL1 can undergo a direct interaction with FIT in root cells. The strongest evidence for this is provided by the fact that ein3 eil1 mutants had measurable phenotypes in particular upon –Fe. The mutant phenotypes indicate that the two proteins were active in our growth conditions in the wild type. In addition, EIL1-GFP signals were detectable in roots upon + and –Fe. bHLH transcription factors bind DNA as homo- or heterodimers. An important domain for their interaction is the helix-loop-helix motif. EIN3/EIL1 belong to a different transcription factor family unrelated to bHLH, Myb, or Myc proteins, which have been generally described as common interaction partners for bHLH proteins (Carretero-Paulet et al., 2010). Since EIN3/EIL1 do not contain any helix-loop-helix domain, it is unlikely that interaction involved this domain in FIT. This assumption is confirmed by the fact that we found interaction not only between full-length FIT and full-length EIN3/EIL1, but also between partial fragments devoid of the bHLH domain. Initially, we used for the screen a partial C-terminal FIT peptide not containing the bHLH domain. This FIT fragment was sufficient for binding the EIL1 fragment in yeast. Hence, we deduce that the interaction between FIT and EIN3/EIL1 occurred between domains that were not involved in the DNA binding process itself.
In the BiFC experiments, tobacco plants were grown under regular conditions in soil. Since BiFC experiments showed a positive interaction between FIT and EIN3/EIL1 in the nucleus, we can speculate that most likely this interaction was not influenced by Fe supply in this overexpression situation. Both the yeast and the in vitro pull-down protein interaction studies worked under standard conditions, suggesting that specific modifications or additional factors acting in plant cells upon Fe deficiency were not needed for the physical interaction of the proteins. Altogether, these results indicate that the FIT-EIN3/EIL1 physical interaction does not have a requirement for Fe deficiency.
EIN3/EIL1 May Increase the Sensitivity for Fe Deficiency
Mutant analysis showed that EIN3/EIL1 can positively affect the expression levels of FIT, IRT1, FRO2, and BHLH039. All of these genes are inducible by Fe deficiency. It was previously shown that different pathways lead to induction of these genes under Fe deficiency. FIT, IRT1, and FRO2 expression and induction involve the FIT protein, whereas induction of the Fe deficiency–inducible BHLH gene is independent of FIT and much increased in the absence of FIT (Wang et al., 2007). It is therefore surprising to note that both FIT and BHLH039 were found to be upregulated in the presence of EIN3/EIL1 compared with their absence. These observations suggest that EIN3/EIL1 may act upstream of BHLH039 and FIT. From the analysis of fit ein3 eil1 triple mutants, we know that the elevated BHLH gene expression in the fit mutant background was not caused by EIN3/EIL1. Therefore, yet other still unknown transcription factors may be responsible for high-level expression of the BHLH genes.
ein3 eil1 mutants are not classical Fe deficiency mutants, which may show either increased constitutive Fe deficiency responses due to reduced internal Fe utilization (Rogers and Guerinot, 2002; Klatte et al., 2009) or low Fe acquisition due to a defect in the regulatory mechanisms and genes performing Fe uptake (Ling et al., 2002; Colangelo and Guerinot, 2004; Li et al., 2004). One possible explanation for this finding is that EIN3/EIL1 may cause a higher sensitivity of plants to respond to Fe deficiency so that Fe deficiency responses are amplified. This explanation is in agreement with reports of Lucena et al. (2006) that ethylene enhanced Fe acquisition gene expression if low concentrations of Fe were provided in the growth medium.
Our data suggest that FIT and EIN3/EIL1 do not act jointly on the promoters of IRT1 and FRO2 and therefore do not, for example, form part of a transcription factor complex (see model depicted in Figure 8). We explain the higher expression levels of IRT1 and FRO2 in the presence of EIN3/EIL1 by the increased stability and presumably also activity of FIT as detailed in the next paragraph.
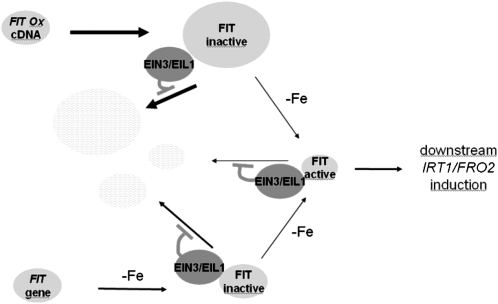
Figure 8.
Model Explaining the Interaction between FIT and EIN3/EIL1.
FIT is produced in root cells upon –Fe in wild-type plants or, alternatively, independently of Fe supply in transgenic FIT Ox plants (FIT gene, FIT Ox construct, and FIT protein are represented as light-gray ellipses). The activity of FIT is regulated. Upon –Fe, FIT is activated and induces downstream Fe deficiency responses, like IRT1 and FRO2 expression. It has been shown that FIT protein can dimerize with bHLH038 and bHLH039 (Yuan et al., 2008; not depicted in the model). From the relative IRT1 and FRO2 expression levels and the levels of FIT protein, we propose that only a small pool of FIT protein is active, while a large pool remains inactive (represented by the thickness of arrows and ellipses). We propose from our results that inactive and active FIT proteins are not stable inside cells and can be degraded, indicated by arrows pointing to ellipses with light gray–white shading. Fe deficiency leads to ethylene production (Romera et al., 1999; Li and Li, 2004; Zuchi et al., 2009). EIN3/EIL1 activated in the ethylene signaling pathway physically interact with FIT, which inhibits proteasomal degradation of FIT (EIN3/EIL1 are represented as dark-gray ellipses; FIT binding to EIN3/EIL1 does not involve the bHLH domain of FIT). According to our model, EIN3/EIL1 do not primarily participate in conjunction with FIT to induce IRT1 and FRO2. We favor the hypothesis that EIN3/EIL1 function to amplify Fe acquisition through stabilization of FIT.
EIN3/EIL1 Affect FIT Abundance
We present evidence that FIT abundance can be modulated in plants and that this modulation can be achieved by hormonal cues. We showed in two independent experimental assays that ethylene affected the levels of FIT. We demonstrated that the presence of the ethylene inhibitor AVG resulted in lower FIT amounts in plant seedlings than in the control. Pharmacological treatment using ethylene inhibitors is considered suboptimal, since the available substances have side effects. However, in combination with mutant analysis of the signaling pathway (ein3 eil1), the pharmacological results gained significant support. We presented evidence that in the absence of EIN3/EIL1, namely, in ein3 eil1 mutants, FIT was present at lower levels than in the wild-type control.
FIT levels were not found to be proportional to the amounts of measured FIT transcripts. In the wild-type situation, FIT was detectable at –Fe but not at +Fe. On the other hand, the difference in FIT expression between + and –Fe was only two- to threefold in the wild type. In the ein3 eil1 background, FIT levels were decreased in a stronger manner (reduction to 8, 10, and 30% of wild-type levels in different experiments) than FIT mRNA levels (reduction to 30 to 50%).
Two conclusions can be drawn from our results: The level of FIT is a target of regulation in plant cells, and ethylene signaling positively affects FIT levels. We thus propose in our model (Figure 8) that FIT is not stable in plant cells. One explanation is that ethylene signaling via EIN3/EIL1 results in the production of an unknown factor that is needed for maintaining a high level of FIT. Another explanation is that EIN3/EIL1 themselves are the factors needed for maintaining the high level of FIT. Support for this latter explanation comes from the observed protein interaction between FIT and EIN3/EIL1. In the model (Figure 8), we propose that the physical protein interaction may thus serve to modulate the stability of FIT. Through the increase in FIT stability, EIN3/EIL1 can then indirectly, in a nonsynergistic manner with FIT, contribute to full expression of FIT downstream target genes. The question remained as to how EIN3/EIL1 interaction may affect FIT protein stability. An answer was suggested from experiments with the proteasomal inhibitor MG132. Application of this inhibitor could alleviate the downregulation of FIT abundance upon AVG treatment and in the ein3 eil1 background. It is therefore a likely possibility that FIT is targeted by the proteasome and that the interaction with EIN3/EIL1 upon ethylene signaling may counteract this effect, hence resulting in a stronger FIT protein abundance and action. Since it was shown that bHLH038 and bHLH039 can interact with FIT (Yuan et al., 2008), it will be interesting to investigate the posttranscriptional regulation of these bHLH factors as well and perhaps their capacity to interact with EIN3/EIL1.
Explanation for the Physiological Effects of EIN3/EIL1 Action upon –Fe
The next questions that arise are: What are the functions of EIN3/EIL1 action upon –Fe? What is the purpose of FIT interaction with EIN3/EIL1? With regard to the first question, we obtained insight from the transcriptome analysis. Indeed, the majority of differential gene expression changes occurred at –Fe but not at +Fe in ein3 eil1 mutants versus the wild type. Therefore, Fe deficiency caused a physiological precondition for enhanced action of EIN3/EIL1. Among the target genes of EIN3/EIL1 at –Fe were several leaf-expressed genes that are involved in the remodeling of the photosynthetic apparatus and in responses to reactive oxygen species (ROS). Such a reconstruction is needed during high irradiation that results in photooxidative damage (Triantaphylidès and Havaux, 2009). A deterioration of electron flow is also caused by insufficient functioning of the numerous Fe-dependent proteins and by an augmentation of the pool of ROS. The thylakoid photosynthetic protein apparatus is widely rebuilt upon Fe deficiency, and ROS degrading proteins are produced in roots and leaves (Andaluz et al., 2006; Brumbarova et al., 2008; Laganowsky et al., 2009). This indicates that by reorganizing photosystems and activating ROS degrading enzymes, plants may counteract the toxic influence of light upon Fe deficiency. From our microarray data, we suggest that this adaptation to −Fe may at least partially require EIN3/EIL1. In this respect, it is interesting to note that it was previously shown that EIN3/EIL1 function in the prevention of photooxidation and the greening of seedlings. In these studies, EIN3/EIL1 were shown to directly activate among their target genes chlorophyll biosynthesis genes PORA and PORB, which encode protochlorophyllide oxidoreductases (Zhong et al., 2009). In ein3 eil1 mutants, the PORA and PORB transcript levels were found to be reduced in these studies (Zhong et al., 2009). By contrast, in our study, the target gene PORB was found to be upregulated in ein3 eil1 upon –Fe, suggesting that EIN3/EIL1 functioned to suppress PORB at −Fe. On the other hand, a second target gene directly downstream of EIN3/EIL1, namely, EBF2, was downregulated in ein3 eil1 mutants at + and –Fe in our study, as might be expected from previous studies (Potuschak et al., 2003; Konishi and Yanagisawa, 2008). Therefore, the regulation of downstream target genes by EIN3/EIL1 is complex and affected in different manners by various external factors. However, a common theme may be that EIN3/EIL1 serve to reduce photooxidation damage. Among the various adjustments to reduce photooxidation upon Fe deficiency could be the simple effect of increasing the acquisition and uptake of Fe by roots. Hence, the function of EIN3/EIL1 upon –Fe and the reason for their interaction with FIT could be to increase Fe acquisition in order to diminish photooxidative damage.
METHODS
Yeast Two-Hybrid Screen and Analysis
The yeast two-hybrid screen was performed according to the protocol of the Matchmaker library construction and screening kit as described by the manufacturer (BD Biosciences).
The coding sequence of the C-terminal part of FIT (following the helix-loop-helix domain) was amplified by PCR from Columbia-0 (Col-0) Fe-deficient root cDNA and cloned into the pGBKT7 vector. This construct (pGBKT7:FIT-C, resulting in fusion with the GAL4 DNA binding domain, FIT-C-BD) was sequenced for verification and transformed into the yeast strain Y187. Transformants were selected on SD medium lacking Trp. FIT-C was used for the screen since full-length FIT carries an activation domain itself and could self-activate the reporter gene in the experimental system.
The cDNA library was constructed using RNA from Col-0 roots grown for 14 d on 50 μM Fe and then for 3 d on 0 μM Fe. First-strand cDNA synthesis was followed by second-strand synthesis. To obtain molecules of at least 200 bp in length, the double-stranded cDNA was purified with Chroma Spin columns (BD Biosciences). This purified cDNA mix was cotransformed with the vector pGADT7-Rec (resulting in a fusion of the encoded peptides with the GAL4 activation domain –AD) into cells of the yeast strain AH109. Selection for recombinants was performed on SD medium lacking Leu.
After mating of the transformed strains (Y187 containing pGBKT7:FIT-C and AH109 containing the cDNA library in pGADT7), yeast cells were plated and incubated for 11 d on SD medium lacking Trp, Leu, and His and containing 4 mM 3-aminotriazol. Under these conditions, cells were selected in which an interaction between FIT-C-BD and the library cDNA/peptide-AD had occurred. From day 4 to 11, colonies were selected, regrown, and used for lacZ filter lift assays to test the lacZ reporter gene activity, according to the manufacturer’s instructions (BD Biosciences). Almost 700 colonies grown on the selective minimal medium were obtained and evaluated for β-galactosidase activity by performing a lacZ filter lift assay. A total of 495 positive colonies were picked, and insertions were amplified by PCR and sequenced. Sequence analysis revealed 40 different putative candidates.
To verify the interaction, plasmids of 40 selected colonies representing the 40 different candidates were isolated and cotransformed with pGBKT7:FIT-C in fresh yeast cells that were then plated on selection medium as described above. lacZ filter lift assays were performed. Colonies were considered for further analysis if positive lacZ test signals were obtained in the retransformation assays and cDNA plasmid contained plant-specific sequences. By the end of the series of control experiments, 14 out of the 40 putative candidates had been verified.
S-Protein Pull-Down Assay
S-protein pull-down assays with EIN3/EIL1 and FIT were performed as described (Hsiao and Chang, 1999). FIT protein fused to an S-tag was expressed in Escherichia coli and affinity purified using S-agarose columns. 35S-labeled EIN3 or EIL1 full-length proteins were produced by in vitro transcription followed by in vitro translation and labeling with 35S-Met using the TNT T7 PCR Quick Master Kit as described by the manufacturer (Promega). Verification of protein synthesis was performed by SDS-PAGE and phosphor imaging.
For the pull-down assay, S-protein agarose (Merck) was loaded with the S-tagged FIT protein in binding buffer and incubated for 4 h. Negative controls were performed without prior loading of S-tagged FIT. After washing, the in vitro–synthesized and labeled protein was loaded onto the S-protein agarose in binding buffer followed by overnight incubation with overhead rotation. The mix was then loaded on Micro BioSpin columns (Bio-Rad). Flow through, washes, and elution with 2× Laemmli buffer were collected and separated by SDS-PAGE. For analysis, the proteins were transferred to nitrocellulose membranes, and after drying, radioactive signals were detected by phosphor imaging.
BiFC
Full-length coding sequences of FIT, EIN3, and EIL1 were amplified and cloned into pBiFP vectors 1 (N-ter C-YFP fusion), 2 (N-ter N-YFP fusion), 3 (C-ter C-YFP fusion), and 4 (C-ter N-YFP fusion) using the Gateway-compatible vector cloning system (F. Parcy). The following final vectors were obtained: pBiFP1:FIT, pBiFP1:EIN3, pBIFP1:EIL1, pBiFP2:FIT, pBiFP2:EIN3, pBiFP2:EIL1, pBiFP3:FIT, pBiFP3:EIN3, pBiFP3:EIL1, pBiFP4:FIT, pBiFP4:EIN3, and pBiFP4:EIL1. Agrobacterium tumefaciens strain GV2260 (pGV2260) cells were transformed with all plasmids, respectively. Combinations of transformed cultures containing plasmids for C-YFP and N-YFP, respectively, were used for leaf infiltration of Nicotiana benthamiana. After 36 to 48 h, YFP fluorescent signals were detected using a LSM510 confocal laser scanning microscope (Zeiss). The fluorescence signals were observed at an excitation wavelength of 488 nm and emission wavelength of 500 to 530 nm. The BiFC experiments were repeated in independent repetitions on a total of six to eight infiltrated leaves. In positive BiFC interactions, the observed leaf area (representing one-quarter of the infiltrated leaf area) revealed 10 to 15 cells with fluorescent nuclei. BiFC controls with empty vectors did not give fluorescence signals.
Plant Material
The Arabidopsis thaliana accession used was Col-0. Seeds of the ein3-1 eil1-3 mutant (hereafter named ein3 eil1) were multiplied and verified in the triple response assay (Chao et al., 1997; Binder et al., 2007). To obtain transgenic EIL1-GFP plants (2xProCaMV35S:EIL1-GFP), EIL1 cDNA was amplified without stop codon and with added PacI, NcoI, and AscI restriction sites. This cDNA fragment was transferred into the binary vector pMDC83 (Curtis and Grossniklaus, 2003). Transgenic plants were obtained by Agrobacterium-mediated floral dip transformation (Clough and Bent, 1998), and T3 plants were used for analysis. The fit-3 loss-of-function mutant (hereafter named the fit mutant) was verified due to strong leaf chlorosis (Jakoby et al., 2004), and the FIT overexpression line (2xProCaMV35S:FIT; hereafter named FIT Ox) was verified by PCR (Jakoby et al., 2004). Multiple fit ein3 eil1 and FIT Ox ein3 eil1 mutants were obtained by crossing, selfing, selection, and multiplication. Seeds were used after verification of homozygous lines (verification of genotypes as indicated above).
Plant Growth Conditions
Arabidopsis seeds were surface sterilized with 6% NaOCl and 0.1% Triton-X for 10 min and washed five times with distilled water. Seeds were stratified for 2 d in 0.1% plant agar in the dark at 4°C.
For the 6-d growth assay, seeds were placed on Hoagland agar medium containing 50 μM FeNaEDTA (+Fe) or 0 μM FeNaEDTA (−Fe), respectively, germinated, and grown for 6 d (for Hoagland medium, see Jakoby et al., 2004). On day 6, seedlings were harvested for microscopy, or RNA or protein analysis.
For the 2-week growth assay, seeds were germinated and seedlings grown for 14 d on Hoagland agar medium containing 50 μM FeNaEDTA and then transferred for 3 d to fresh medium containing either 0 μM FeNaEDTA (−Fe) or 50 μM FeNaEDTA (+Fe), respectively. Then, leaves and roots were harvested separately for RNA or protein analysis. For metal determination, plants were grown to the age of 4 weeks, and roots were washed with 0.1 mM calcium nitrate solution.
If indicated in the text, 10 μM 1-aminocyclopropane-1-carboxylic acid (Sigma-Aldrich), 10 μM aminoethoxyvinylglycine (AVG; Sigma-Aldrich), 200 μM STS, or 20 μM AOA (Sigma-Aldrich) was added to the growth medium. For MG132 treatment, 6-d-old seedlings were treated for 4 h in liquid Hoagland medium containing 100 μM MG132 (Calbiochem) and harvested for analysis.
qRT-PCR
A detailed protocol of the qRT-PCR procedure is described (Klatte and Bauer, 2009). Briefly, plant material was ground in liquid nitrogen to a fine powder. Total RNA extraction was performed using the Spectrum plant total RNA kit (Sigma-Aldrich) according to the manufacturer’s protocol. One microgram of total RNA was used per cDNA synthesis reaction. RNA was first treated with DNase I (Fermentas) to eliminate DNA contamination. mRNAs were then converted into cDNAs using M-MLV reverse transcriptase (Fermentas) with an oligo(dT)18 primer. cDNA was diluted 1:100, and 10 μL cDNA was used in a 20-μL qPCR reaction. Quantitative real-time PCR was performed using the MyIQ real-time detection system and analyzed using IQ5 software (Bio-Rad). Gene-specific primers for RT-PCR are described (Wang et al., 2007). For quantification, self-made quantity standards were prepared with predefined template amounts. Ten microliters of 2× PreMix ExTaq (Takara) plus SYBR Green was used for a 20-μL qPCR reaction. The PCR program consisted of one cycle (95°C, 4 min), 40 cycles (95°C, 10 s; 58°C, 18 s; 72°C, 18 s), and one cycle (72°C, 5 min), followed by a melting curve program (55 to 90°C in increasing steps of 1°C). All reactions were performed in duplicate and analyzed individually. Normalization was achieved using UBP and EF1a amplification as constitutive controls.
Statistical Analysis
P values were obtained via t test using the GraphPad software at http://www.graphpad.com/welcome.htm. Microarray statistics are described in the next paragraph.
Transcriptome Studies and Statistical Analysis of Microarray Data
Microarray analysis was performed at the Unité de Recherche en Génomique Végétale (Evry, France) using the CATMA arrays, which contain 31,776 gene-specific tags corresponding to 22,089 genes from Arabidopsis (Crowe et al., 2003; Hilson et al., 2004). Three independent biological replicates were produced. For each biological repetition, RNA samples were prepared from 60 seedling plants grown in the 6-d growth assay at + and –Fe, respectively. Total RNA was extracted using the RNeasy plant mini kit (Qiagen) according to the supplier’s instructions. For each comparison, one technical replicate with fluorochrome reversal was performed for each biological replicate (i.e., four hybridizations per comparison). The labeling of cRNAs with Cy3-dUTP or Cy5-dUTP (Perkin-Elmer-NEN Life Science Products), the hybridization to the slides, and the scanning were performed as described by Lurin et al. (2004).
Experiments were designed with the statistics group of the Unité de Recherche en Génomique Végétale. For each array, the raw data comprised the logarithm of median feature pixel intensity at wavelengths of 635 nm (red) and 532 nm (green), and no background was subtracted. An array-by-array normalization was performed to remove systematic biases. First, spots considered badly formed features were excluded. Then, a global intensity-dependent normalization using the loess procedure (Yang et al., 2002) was performed to correct the dye bias. Finally, for each block, the log-ratio median calculated over the values for the entire block was subtracted from each individual log-ratio value to correct print tip effects. Each comparison was performed in dye-swap on three biological replicates. Therefore, a technical replicate was available for each biological replicate. Differential analysis was based on the log ratios averaged on the dye-swap: The technical replicates were averaged to get one log-ratio per biological replicate, and these values were used to perform a paired t test. This method allowed consideration of the fact that the biological variability is greater than the technical variability.
A trimmed variance was calculated from spots that did not display extreme variance (see details in Gagnot et al., 2008). The raw P values were adjusted by the Bonferroni method, which controls the family-wise error rate to keep a strong control of the false positives in a multiple-comparison context. Probes with a Bonferroni P value of < 0.05 were considered to be differentially expressed.
Microarray data were deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession number GSE 26510) and at CATdb (http://urgv.evry.inra.fr/CATdb/; project AU10-14_Fer) according to the Minimum Information About a Microarray Experiment standards.
For validation, the same RNA preparations were tested for differential gene expression via RT-PCR of selected genes identified from the microarray analysis (see Supplemental Figure 3 online).
Immunological Detection
Total protein extracts were prepared from roots of 6-d-old plant seedlings following a described procedure (Scharf et al., 1998). Protein concentrations were determined using the Bradford assay (Sigma-Aldrich). Ten micrograms of protein was loaded per lane on a 10% SDS-PAGE gel and subsequently blotted to a nitrocellulose membrane. Immunoblot analysis was conducted according to standard procedures. A mouse FIT antiserum was produced and purified to obtain antibodies directed against the specific 15-kD C-terminal part of FIT following the helix-loop-helix domain, according to standard procedures. This purified antiserum tested positive on E. coli–expressed FIT peptide and on plant extracts expressing FIT (Figures 3B and 7; see Supplemental Figures 2 and 6 online). The anti-FIT-C antiserum was specific for FIT, since no band of the predicted FIT size was detected in fit mutant extracts (see Supplemental Figure 7 online). The primary antibodies were detected with anti-mouse IgG conjugated with horseradish peroxidase (Sigma-Aldrich). Detection signals were developed using an enhanced chemiluminescence detection kit (ECL; GE Healthcare) according to the manufacturer’s protocol. Relative quantification of protein bands detected in immunoblot experiments was calculated using ImageJ software (Abramoff et al., 2004) and normalized to the Ponceau S–stained bands.
Metal Determination
Ten milligrams of dried plant material of 4-week-old plants grown on 50 μM Fe-containing Hoagland agar medium was used for direct solid sampling graphite furnace absorption spectrometry (GF AAS 6; Analytik Jena) for determination of Fe, Zn, and Mn. Reference standards were used for quantification. Each biological sample was measured five times for each metal, and a mean value was calculated.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the accession numbers listed in Supplemental Data Set 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Localization of EIL1-GFP.
Supplemental Figure 2. Effect of AOA and STS Ethylene Inhibitors on FIT Levels.
Supplemental Figure 3. Validation of Microarray Data Using RT-PCR on Selected Genes Identified in the Microarray.
Supplemental Figure 4. Molecular Responses of FIT Ox ein3eil1 Plants.
Supplemental Figure 5. Gene Expression of Fe Deficiency-Inducible BHLH Genes in fit ein3 eil1 and FIT Ox ein3 eil1 Plants.
Supplemental Figure 6. FIT Protein Regulation in FIT Ox ein3 eil1 Plants.
Supplemental Figure 7. Specificity Control of the Anti-FIT-C Antiserum.
Supplemental Table 1. Selected Gene Chip Hybridization Data.
Supplemental Data Set 1. Selected Gene Chip Hybridization Data.
Acknowledgments
We thank Iris Fuchs and U. Müller (Saarland University) for production of anti-FIT antiserum. We thank F. Parcy (CNRS Grenoble) for BiFC vectors and J. Romera (University of Cordoba) for multiplication of ein3 eil1 seeds. We thank R. Ivanov (Saarland University) for fruitful discussions. Funding of the Deutsche Forschungsgemeinschaft to P.B. is greatly acknowledged. T.P. was supported by Grant ANR-07-BLAN-0219 (EIN3-REG).
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. (2004). Image processing with ImageJ. Biophotonics Int. 11: 36–42 [Google Scholar]
- An F., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andaluz S., López-Millán A.F., De las Rivas J., Aro E.M., Abadía J., Abadía A. (2006). Proteomic profiles of thylakoid membranes and changes in response to iron deficiency. Photosynth. Res. 89: 141–155 [DOI] [PubMed] [Google Scholar]
- Bauer P., Ling H.Q., Guerinot M.L. (2007). FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol. Biochem. 45: 260–261 [DOI] [PubMed] [Google Scholar]
- Binder B.M., Walker J.M., Gagne J.M., Emborg T.J., Hemmann G., Bleecker A.B., Vierstra R.D. (2007). The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F., Segonzac C., Chang K.N., Qiao H., Ecker J.R., Zipfel C., Rathjen J.P. (2010). Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc. Natl. Acad. Sci. USA 107: 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat J.F., Ravet K., Arnaud N., Duc C., Boucherez J., Touraine B., Cellier F., Gaymard F. (2010). New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Ann. Bot. (Lond.) 105: 811–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T., Bauer P. (2005). Iron-mediated control of the basic helix-loop-helix protein FER, a regulator of iron uptake in tomato. Plant Physiol. 137: 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T., Matros A., Mock H.P., Bauer P. (2008). A proteomic study showing differential regulation of stress, redox regulation and peroxidase proteins by iron supply and the transcription factor FER. Plant J. 54: 321–334 [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L., Galstyan A., Roig-Villanova I., Martínez-García J.F., Bilbao-Castro J.R., Robertson D.L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153: 1398–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J.R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen H., Xue L., Chintamanani S., Germain H., Lin H., Cui H., Cai R., Zuo J., Tang X., Li X., Guo H., Zhou J.M. (2009). ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.W., Yang J.L., Qin C., Jin C.W., Mo J.H., Ye T., Zheng S.J. (2010). Nitric oxide acts downstream of auxin to trigger root ferric-chelate reductase activity in response to iron deficiency in Arabidopsis. Plant Physiol. 154: 810–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colangelo E.P., Guerinot M.L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe M.L., et al. (2003). CATMA: A complete Arabidopsis GST database. Nucleic Acids Res. 31: 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Benoist B., McLean E., Egli I., Cogswell M. (2008). Worldwide Prevalence of Anaemia 1993–2005. (Geneva, Switzerland: World Health Organization; ). [DOI] [PubMed] [Google Scholar]
- Gagne J.M., Smalle J., Gingerich D.J., Walker J.M., Yoo S.D., Yanagisawa S., Vierstra R.D. (2004). Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. USA 101: 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnot S., Tamby J.P., Martin-Magniette M.L., Bitton F., Taconnat L., Balzergue S., Aubourg S., Renou J.P., Lecharny A., Brunaud V. (2008). CATdb: A public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 36(Database issue): D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M.J., Lucena C., Romera F.J., Alcántara E., Pérez-Vicente R. (2010). Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J. Exp. Bot. 61: 3885–3899 [DOI] [PubMed] [Google Scholar]
- Graziano M., Beligni M.V., Lamattina L. (2002). Nitric oxide improves internal iron availability in plants. Plant Physiol. 130: 1852–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano M., Lamattina L. (2007). Nitric oxide accumulation is required for molecular and physiological responses to iron deficiency in tomato roots. Plant J. 52: 949–960 [DOI] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Henriques R., Jásik J., Klein M., Martinoia E., Feller U., Schell J., Pais M.S., Koncz C. (2002). Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol. Biol. 50: 587–597 [DOI] [PubMed] [Google Scholar]
- Hilson P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14: 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao P.W., Chang C. (1999). Isolation and characterization of ARA160 as the first androgen receptor N-terminal-associated coactivator in human prostate cells. J. Biol. Chem. 274: 22373–22379 [DOI] [PubMed] [Google Scholar]
- Jakoby M., Wang H.Y., Reidt W., Weisshaar B., Bauer P. (2004). FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 577: 528–534 [DOI] [PubMed] [Google Scholar]
- Kim S.A., Guerinot M.L. (2007). Mining iron: Iron uptake and transport in plants. FEBS Lett. 581: 2273–2280 [DOI] [PubMed] [Google Scholar]
- Klatte M., Bauer P. (2009). Accurate real-time reverse transcription quantitative PCR. Methods Mol. Biol. 479: 61–77 [DOI] [PubMed] [Google Scholar]
- Klatte M., Schuler M., Wirtz M., Fink-Straube C., Hell R., Bauer P. (2009). The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol. 150: 257–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S. (2008). Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 55: 821–831 [DOI] [PubMed] [Google Scholar]
- Laganowsky A., Gómez S.M., Whitelegge J.P., Nishio J.N. (2009). Hydroponics on a chip: Analysis of the Fe deficient Arabidopsis thylakoid membrane proteome. J. Proteomics 72: 397–415 [DOI] [PubMed] [Google Scholar]
- Li L., Cheng X., Ling H.Q. (2004). Isolation and characterization of Fe(III)-chelate reductase gene LeFRO1 in tomato. Plant Mol. Biol. 54: 125–136 [DOI] [PubMed] [Google Scholar]
- Li X., Li C. (2004). Is ethylene involved in regulation of root ferric reductase activity of dicotyledonous species under iron deficiency. Plant Soil 261: 147–153 [Google Scholar]
- Ling H.Q., Bauer P., Bereczky Z., Keller B., Ganal M. (2002). The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc. Natl. Acad. Sci. USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena C., Waters B.M., Romera F.J., García M.J., Morales M., Alcántara E., Pérez-Vicente R. (2006). Ethylene could influence ferric reductase, iron transporter, and H+-ATPase gene expression by affecting FER (or FER-like) gene activity. J. Exp. Bot. 57: 4145–4154 [DOI] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. (1995). Mineral Nutrition of Plants. (Boston: Academic Press; ). [Google Scholar]
- Marschner H., Römheld V. (1994). Strategies of plants for acquisition of iron. Plant Soil 165: 261–274 [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Robinson N.J., Procter C.M., Connolly E.L., Guerinot M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rogers E.E., Guerinot M.L. (2002). FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell 14: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romera F.J., Alcantara E. (1994). Iron-deficiency stress responses in cucumber (Cucumis sativus L.) roots (A possible role for ethylene?). Plant Physiol. 105: 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romera F.J., Alcantara E., de la Guardia M.D. (1999). Ethylene production by Fe-deficient roots and its involvement in the regulation of Fe-deficiency stress responses by Strategy I plants. Ann. Bot. (Lond.) 83: 51–55 [Google Scholar]
- Scharf K.D., Heider H., Höhfeld I., Lyck R., Schmidt E., Nover L. (1998). The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell. Biol. 18: 2240–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikora A., Schmidt W. (2001). Acclimative changes in root epidermal cell fate in response to Fe and P deficiency: A specific role for auxin? Protoplasma 218: 67–75 [DOI] [PubMed] [Google Scholar]
- Schmidt W., Tittel J., Schikora A. (2000). Role of hormones in the induction of iron deficiency responses in Arabidopsis roots. Plant Physiol. 122: 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séguéla M., Briat J.F., Vert G., Curie C. (2008). Cytokinins negatively regulate the root iron uptake machinery in Arabidopsis through a growth-dependent pathway. Plant J. 55: 289–300 [DOI] [PubMed] [Google Scholar]
- Smalle J., Haegman M., Kurepa J., Van Montagu M., Straeten D.V. (1997). Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc. Natl. Acad. Sci. USA 94: 2756–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantaphylidès C., Havaux M. (2009). Singlet oxygen in plants: Production, detoxification and signaling. Trends Plant Sci. 14: 219–228 [DOI] [PubMed] [Google Scholar]
- Varotto C., Maiwald D., Pesaresi P., Jahns P., Salamini F., Leister D. (2002). The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J. 31: 589–599 [DOI] [PubMed] [Google Scholar]
- Vert G., Grotz N., Dédaldéchamp F., Gaymard F., Guerinot M.L., Briat J.F., Curie C. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E.L., Connolly E.L. (2008). Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 11: 530–535 [DOI] [PubMed] [Google Scholar]
- Wang H.Y., Klatte M., Jakoby M., Bäumlein H., Weisshaar B., Bauer P. (2007). Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226: 897–908 [DOI] [PubMed] [Google Scholar]
- Waters B.M., Lucena C., Romera F.J., Jester G.G., Wynn A.N., Rojas C.L., Alcántara E., Pérez-Vicente R. (2007). Ethylene involvement in the regulation of the H(+)-ATPase CsHA1 gene and of the new isolated ferric reductase CsFRO1 and iron transporter CsIRT1 genes in cucumber plants. Plant Physiol. Biochem. 45: 293–301 [DOI] [PubMed] [Google Scholar]
- Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P. (2002). Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y., Guerinot M.L. (1996). Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 10: 835–844 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Wu H., Wang N., Li J., Zhao W., Du J., Wang D., Ling H.Q. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Yuan Y.X., Zhang J., Wang D.W., Ling H.Q. (2005). AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 15: 613–621 [DOI] [PubMed] [Google Scholar]
- Zhong S., Zhao M., Shi T., Shi H., An F., Zhao Q., Guo H. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 106: 21431–21436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchi S., Cesco S., Varanini Z., Pinton R., Astolfi S. (2009). Sulphur deprivation limits Fe-deficiency responses in tomato plants. Planta 230: 85–94 [DOI] [PubMed] [Google Scholar]