Nonadditive expression of miR163 in allopolyploids is caused largely by cis-acting promoters and by trans-acting factors present in Arabidopsis arenosa and allopolyploids but absent in Arabidopsis thaliana. miR163 negatively regulates secondary metabolite pathways in defense response. This is one example of genome-wide cis- and trans-regulation that shapes interspecific hybrids and allopolyploids.
Abstract
MicroRNAs (miRNAs) play essential roles in plant and animal development, but the cause and effect of miRNA expression divergence between closely related species and in interspecific hybrids or allopolyploids are unknown. Here, we show differential regulation of a miR163-mediated pathway in allotetraploids and their progenitors, Arabidopsis thaliana and Arabidopsis arenosa. miR163 is a recently evolved miRNA in A. thaliana and highly expressed in A. thaliana, but its expression was undetectable in A. arenosa and repressed in resynthesized allotetraploids. Repression of A. arenosa MIR163 (Aa MIR163) is caused by a weak cis-acting promoter and putative trans-acting repressor(s) present in A. arenosa and allotetraploids. Moreover, ectopic Aa MIR163 precursors were processed more efficiently in A. thaliana than in resynthesized allotetraploids, suggesting a role of posttranscriptional regulation in mature miR163 abundance. Target genes of miR163 encode a family of small molecule methyltransferases involved in secondary metabolite biosynthetic pathways that are inducible by a fungal elicitor, alamethicin. Loss of miR163 or overexpression of miR163 in mir163 mutant plants alters target transcript and secondary metabolite profiles. We suggest that cis- and trans-regulation of miRNA and other genes provides a molecular basis for natural variation of biochemical and metabolic pathways that are important to growth vigor and stress responses in Arabidopsis-related species and allopolyploids.
INTRODUCTION
Gene expression differences can result from cis- and/or trans-regulatory changes, leading to epistatic effects (Carlborg and Haley, 2004). cis-regulatory changes can affect transcription initiation, rate, and/or transcript stability, whereas trans-regulatory changes can alter the activity or expression of factors that interact with cis-regulatory sequences (Carroll, 2005; Wray, 2007). cis- and trans-acting effects have been documented for several dozens of genes in Drosophila melanogaster interspecific hybrids (Wittkopp et al., 2004) and at a genome-wide scale in yeast (Saccharomyces cerevisiae) interspecific hybrids (Tirosh et al., 2009). However, it is difficult to test the consequences of these expression differences in morphological evolution because interspecific hybrids are generally sterile and cannot produce offspring. By doubling the chromosomes in interspecific hybrids, allotetraploids can be readily produced by cross-pollinating autotetraploid Arabidopsis thaliana with tetraploid Arabidopsis arenosa (Comai et al., 2000; Wang et al., 2004). The resulting allotetraploids provide a tractable genetic system for testing genetic and epigenetic effects on gene expression, growth, and development (Chen, 2007; Leitch and Leitch, 2008). As examples, allotetraploids and hybrids induce altered expression of circadian clock genes that regulate downstream genes and pathways in chlorophyll biosynthesis and starch metabolism, leading to growth vigor (Ni et al., 2009). Parental genome dosage and altered expression of several imprinted genes in the allotetraploids is predicted to cause seed lethality (Josefsson et al., 2006). Moreover, a subset of protein-coding and transcriptional factor genes is nonadditively expressed (different from midparent value), which correlates negatively with nonadditive accumulation of microRNAs (miRNAs) in the allotetraploids (Ha et al., 2009).
miRNAs play essential roles in plant and animal development by regulating target gene expression through translational repression or mRNA degradation (Ambros, 2004; Chen, 2009; Voinnet, 2009). Despite the importance of miRNAs in cellular growth and development, the role of conserved miRNAs in expression differentiation and phenotypic variation between related species is unknown (Ha et al., 2008). Major miRNA renovations occurred at the emergence of vertebrates and mammals (Niwa and Slack, 2007). Among many miRNA genes identified in humans and chimpanzees (Berezikov et al., 2006), some are conserved in vertebrates or primates, whereas others are human specific. Among conserved miRNAs between species, their expression patterns are subjected to spatial and temporal regulation (Niwa and Slack, 2007). Some miRNAs, such as miR-126 and miR-206, are expressed in vertebrate-specific organs (Wienholds et al., 2005), and several others, such as miR-454a and miR-145, displayed spatial expression differences between medaka and zebra fish (Ason et al., 2006). The data suggest that species-specific miRNAs as well as spatiotemporal regulation of conserved miRNAs play important roles in shaping morphological and developmental variation among related species during evolution (Niwa and Slack, 2007; Ha et al., 2008).
Genome-wide gene expression analysis has indicated that differential accumulation of miRNAs correlates negatively with the expression of corresponding target genes in Arabidopsis allopolyploids and between the related species (Ha et al., 2009). Among the differentially expressed miRNAs, miR163 is severely repressed in leaves and flowers of A. arenosa and allotetraploids (Ha et al., 2009). MIR163 is one of several recently evolved miRNA loci in A. thaliana (Allen et al., 2004). Target genes of miR163 encode members of the plant SABATH methyltransferase family (D’Auria et al., 2003; Zhao et al., 2008). Jasmonic acid carboxyl methyltransferase and benzoic acid/salicylic acid methyltransferase 1 are members of this family and act in plant defense responses (Seo et al., 2001; Chen et al., 2003). A chemical screen for potential substrates has revealed one of the miR163 targets, farnesoic acid methyltransferase (FAMT), which converts farnesoic acid (FA) to methyl farnesoate (MeFA) (Yang et al., 2006). MeFA is a precursor of insect juvenile hormone III, and the presence of MeFA in plants perturbs insect growth and development (Toong et al., 1988; Shinoda and Itoyama, 2003). In this study, we investigated how MIR163 is differentially expressed between A. thaliana and A. arenosa and in allotetraploids and how miR163 regulates abundance of target transcripts and gene products.
RESULTS
Differential Expression of miR163 in Related Species
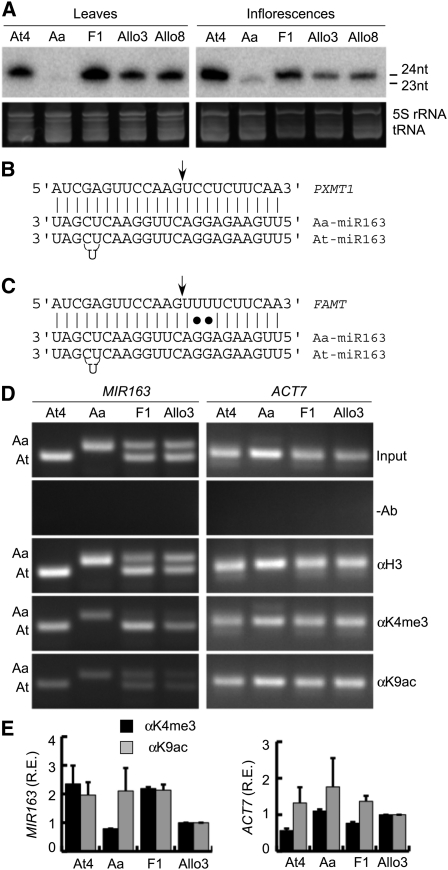
To test the consequence of miRNA expression diversity in Arabidopsis species and allopolyploids, we investigated miR163 and its target gene expression in A. thaliana and A. arenosa, two species that diverged ~6 million years ago (Koch et al., 2000). A. thaliana contains several nonconserved miRNA loci, such as MIR161 and 163. miR163 evolved recently from inverted repeats, and its precursors share high levels of sequence identity and structural similarity with their target genes (Allen et al., 2004). However, the regulation of miR163 and its target genes is largely unknown. miR163 levels were very high in A. thaliana but undetectable in A. arenosa leaves (Figure 1A). In A. arenosa flowers, a 23-nucleotide RNA accumulated at ~3% of A. thaliana miR163 levels. In the allotetraploids that are derived from A. thaliana and A. arenosa, miR163 levels in F1 were similar to those in A. thaliana leaves but lower than those in A. thaliana flowers. After eight generations of selfing, miR163 expression was reduced in two allotetraploids (Allo3 aka Allo733 and Allo8 aka Allo738), consistent with rapid epigenetic regulation of homoeologous loci in allotetraploids (Wang et al., 2004).
Figure 1.
Differential Accumulation of miR163 in Arabidopsis Species and Transcriptional Regulation of MIR163 by ChIP Analysis.
(A) Small RNA gel blots showing miR163 accumulation in mature leaves and inflorescences in tetraploid A. thaliana (At4), A. arenosa (Aa), and F1 and F8 (Allo3 and Allo8) generations of resynthesized allotetraploids. nt, nucleotide.
(B) Core sequence alignment of 24-nucleotide At and 23-nucleotide Aa miR163 with the target PXMT1.
(C) Core sequence alignment of 24-nucleotide At and 23-nucleotide Aa miR163 with the target FAMT.
(D) ChIP assays using antibodies against C terminus of histone H3 (H3), acetyl H3K9 (K9ac), and trimethyl H3K4 (K4me3). A negative control without antibody (-Ab) and a positive control with input DNA without IP (input) are included. Representative PCR results from ChIP and input (control) DNA of MIR163 promoter regions (266 bp in At and 286 bp in Aa) and the 5′ coding region of ACT7 (254 bp) are shown.
(E) Relative enrichment (R.E.) of MIR163 and ACT7 upstream regions for K4me3 (black) and K9ac (gray) was quantified by qPCR and normalized with the corresponding input DNA and histone H3 levels. Values are mean ± sd (from three biological replications).
A. thaliana MIR163 (At MIR163) and A. arenosa MIR163 (Aa MIR163) loci were cloned and sequenced (see Supplemental Figures 1 and 2 online). Compared with 24-nucleotide At-miR163 (Kurihara and Watanabe, 2004), mature Aa-miR163 had one nucleotide (U or T) deletion in the 3′ end and was 23 nucleotides long (Figures 1B and 1C). Six miR163 target genes present in the A. thaliana genome are grouped into two clades (see Supplemental Figure 3A online). One group consists of At3g44860 and At3g44870 duplicates that encode FAMT (Yang et al., 2006), and another group contains At1g66700 and At1g66690 encoding S-adenosylmethionine–dependent methyltransferases (PXMT1) (D’Auria et al., 2003; Zhao et al., 2008). The stem-loop sequence of At MIR163 (At1g66725) shared similarities with three of upstream target loci (At1g66690, At1g66700, and At1g66720) (Allen et al., 2004). Aa MIR163 had a higher degree of segmental similarities with its target gene (At1g66700) than At MIR163 (see Supplemental Figures 3B and 3C online). This suggests that after divergence between A. thaliana and A. arenosa, sequence mutations and deletions/insertions might have occurred in the At MIR163 stem-loop, leading to an overall high percentage of sequence complementarity between Aa MIR163 and its target sequences.
cis- and trans-Acting Effects on Transcriptional Regulation of MIR163 Loci
Differential accumulation of a mature miRNA may be regulated by transcription of miRNA precursors and/or biogenesis of mature miRNAs. miRNA genes are transcribed by RNA polymerase II (Lee et al., 2004), generating primary miRNA (pri-miRNA) that is processed by nuclear RNaseIII-like enzymes, such as Dicer and Drosha in animals and DICER-LIKE proteins (e.g., DCL1) in plants (Lee et al., 2003; Chen, 2009; Voinnet, 2009). Similar to the mature miR163 accumulation in leaves (Figure 1A), pri-miR163 was expressed at higher levels in A. thaliana than in A. arenosa (see Supplemental Figure 4 online). In addition, pri-miR163 expression was upregulated in F1 and reduced in F8 allotetraploids compared with A. thaliana. This suggests that differential miR163 accumulation between A. thaliana and A. arenosa is transcriptionally regulated. To test if MIR163 trancription is controlled by cis-regulatory changes in At MIR163 and Aa MIR163 loci, we isolated promoter fragments between the stop codon of the upstream gene (At1g66700) and the transcription start site of the MIR163 locus (At1g66725). A. thaliana and A. arenosa MIR163 promoter regions were isolated and designated as pAt-MIR163 (1439 bp) and pAa-MIR163 (1977 bp), respectively. A high level of sequence identity (~71%) was found in the proximal promoter regions (~450 bp upstream) between pAt-MIR163 (1439 bp) and corresponding pAa-MIR163 (1977 bp) promoters (see Supplemental Figure 1 online). Both promoters contained the canonical TATA sequence at the −35 position. Sequence divergence between the two promoters increased in the distal regions, including multiple insertions/deletions and nucleotide substitutions in pAa-MIR163 relative to pAt-MIR163. These regions contain multiple predicted negative and positive elements, including phytohormone, elicitor, and light-responsive elements (see Supplemental Table 1 online), suggesting a potential role for promoter divergence in cis-regulation of MIR163 expression.
Chromatin immunoprecipitation (ChIP) assays were used to test if histone modifications are associated with differential expression of endogenous MIR163 loci in A. thaliana, A. arenosa, and allotetraploids. The levels of permissive histone modifications at pAt- or pAa-MIR163 were evaluated using antibodies against the C terminus of histone H3 (H3), trimethylated H3 Lys 4, and acetylated H3 Lys 9 in the ChIP assay coupled with allele (At or Aa) specific PCR (Figure 1D; see Supplemental Table 2 online). The relative enrichment of histone modifications in the MIR163 promoter was quantified by quantitative PCR (qPCR) (Figure 1E). ACTIN7 (ACT7) promoter was included as a control. Nucleosomes with H3K4me3 are most often associated with actively transcribed genes (Santos-Rosa et al., 2002; Schübeler et al., 2004; Ruthenburg et al., 2007). Consistent with the miR163 abundance levels, the levels of H3K4me3 were higher in A. thaliana (2.4 ± 0.9) and F1 allotetraploids (2.2 ± 0.1) and lower in A. arenosa (0.8 ± 0.01) than in F8 allotetraploids. In F1 allotetraploids, a high level of H3K4me3 was found in the At MIR163 promoter allele (Figure 1D). This is consistent with a dominant accumulation of At-miR163 in the allotetraploids (Figure 1A). Similar enrichment levels of H3K9ac were detected in the MIR163 promoters in A. thaliana (2.0 ± 0.03), A. arenosa (2.1 ± 1.1), and F1 allotetraploids (2.1 ± 0.3). This high level of H3K9ac in the Aa MIR163 promoter is in contrast with the low Aa-miR163 abundance (Figure 1A), suggesting that the presence of a permissive (H3K9ac) nucleosomal state in the promoter is not sufficient for active transcription (Sterner and Berger, 2000; Ng et al., 2006). In Allo3, consistent with its low miR163 abundance, the levels of both H3K4me3 and H3K9ac in the MIR163 promoters were low, suggesting that it may take several generation in allopolyploids to establish certain chromatin states, such as H3K9ac (Wang et al., 2004). These data suggest that transcriptional regulation plays a role in mature miR163 abudance in A. arenosa, resynthesized F8 allotetraploids, and a natural allotetraploid Arabidopsis suecica (Figure 1A) (Ha et al., 2009), but other factors, including trans-acting factors, are responsible for mediating At or Aa MIR163 promoter activity.
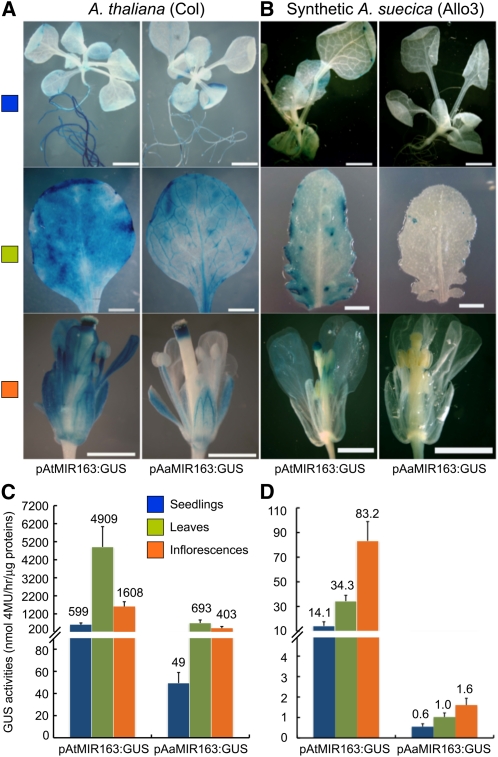
To directly measure promoter activity, we made fusion constructs of promoters (pAt-MIR163:GUS or pAa-MIR163:GUS) (see Supplemental Table 3 online) and produced 12 independent transgenic A. thaliana plants. β-Glucuronidase (GUS) staining and fluorometric assays (Jefferson et al., 1987) indicated that both constructs were expressed in seedlings, leaves, and flowers in A. thaliana (Figure 2A). The expression levels were 4- to 12-fold higher in pAt-MIR163:GUS transgenics than in pAa-MIR163:GUS transgenics in A. thaliana (Figure 2C; see Supplemental Table 4 online), indicating that Aa-miR163 has a relatively weak promoter. The expression difference between pAt-MIR163:GUS and pAa-MIR163:GUS is smaller in flowers (~4-fold) than in seedlings (~12-fold), suggesting spatial regulation of MIR163 expression.
Figure 2.
cis and trans-Acting Effects Revealed by At and Aa MIR163 Promoter-Driven GUS Expression in A. thaliana and Resynthesized Allotetraploids (Allo3).
(A) Representative GUS-stained seedlings, rosette leaves, and inflorescences in transgenic A. thaliana that expressed pAt-MIR163 and pAa-MIR163 driven GUS transgenes. Bars = 2 mm in seedlings and mature leaves and 1 mm in inflorescences.
(B) Same as (A) in transgenic allotetraploids that expressed pAt-MIR163 and pAa-MIR163 driven GUS transgenes.
(C) Fluorometric values of pAt-MIR163 and pAa-MIR163 driven GUS activities (nmol 4-MU per hour per microgram protein) in multiple independent transgenic A. thaliana lines (n = 12).
(D) Same as (C) in multiple independent transgenic allotetraploids (n = 11). Values are mean ± se.
The promoter activity alone (4- to 12-fold difference) cannot fully explain the >30-fold repression of endogenous miR163 in A. arenosa (Figure 1A) (Ha et al., 2009). Some trans-acting factors may affect miR163 expression in A. arenosa and allotetraploids. To test this, we generated 11 independent allotetraploid (F8, Allo3) lines using pAt-MIR163:GUS and pAa-MIR163:GUS (Figure 2B). The absolute values of GUS activities driven by both pAt-MIR163 and pAa-MIR163 were ~100-fold lower in the allotetraploid transgenics than in the A. thaliana transgenics (Figures 2C and 2D), suggesting a repression mechanism of allopolyploidy for both homoeologous MIR163 genes. Moreover, the GUS activities derived from pAt-MIR163 were 24- to 52-fold higher than that from pAa-MIR163 (Figure 2D), suggesting that repression of Aa MIR163 is also caused by one or more putative trans-acting repressors that are present in A. arenosa and allotetraploids but absent in A. thaliana.
Differential Processing of At- and Aa-miR163
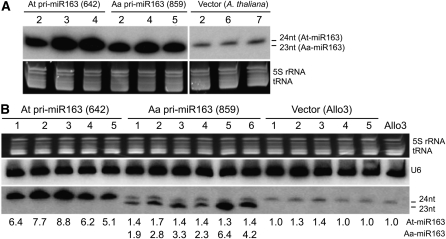
Can At MIR163 and Aa MIR163 pri-miRNAs that are transcribed in A. thaliana and allotetraploids be processed into mature miR163? Although At and Aa pri-miR163 share a high level of sequence identity (~86%), a 139-bp insertion was found at the 5′ region of Aa MIR163 transcribed region relative to At MIR163 (see Supplemental Figure 2 online). Sequence divergence between At and Aa pri-miR163 may lead to pri- and pre-miRNA structural changes (see Supplemental Figure 5 online), causing differences in miRNA processing efficiency and mature miR163 abundance. To test this, 35S:At-pri-MIR163 or 35S:Aa-pri-MIR163 was transformed into A. thaliana (Columbia [Col]) and allotetraploids (Allo3), respectively. Overexpression of At pri-miR163 (At642) and Aa pri-miR163 (Aa859) increased accumulation of mature At-miR163 (24 nucleotides) and Aa-miR163 (23 nucleotides), respectively (Figures 3A and 3B). In A. thaliana, transgenic At- and Aa-miRNA163 accumulated at higher levels than endogenous At-miR163 (vector control) (Figure 3A), confirming biogenesis of ectopic Aa MIR163 precursors. In allotetraploid overexpressors, mature At-miR163 accumulated 1- to 5-fold higher than Aa-miR163 and 2- to 4-fold higher than endogenous At-miR163. The data suggest that although At and Aa pri-miR163 are processed in the allotetraploids, there is a low processing efficiency of Aa pri-miR163 relative to At pri-miR163. However, since a high level of ectopic expression of Aa-miR163 was detected in A. thaliana, sequence divergence between At and Aa pri-miR163 precursors is probably not a major factor for the low accumulation of Aa-miR163 in A. arenosa or allotetraploids. These data suggest that the low abundance of mature Aa-miR163 is caused mainly by transcriptional repression of primary Aa MIR163 transcripts coupled with a relatively low efficiency of processing Aa pri-miR163 precursors in the allotetraploids. Repressive factor(s) that are present in allotetraploids and A. arenosa but absent in A. thaliana contribute to both transcriptional and posttranscriptional regulation of Aa-miR163.
Figure 3.
Posttranscriptional Regulation and miR163 Biogenesis of At and Aa pri-miR163 Precursors in A. thaliana and Allotetraploids.
(A) Small RNA gel blot analysis of total RNA from leaves of 4-week-old transgenic A. thaliana that overexpressed At pri-miR163 and Aa pri-miR163. The two separate blots had the same exposure time. Numbers correspond to different independent transgenic lines.
(B) Small RNA gel blot analysis of total RNA from leaves of 4-week-old transgenic allotetraploids overexpressing 24-nucleotide (nt) At or 23-nucleotide Aa miR163. Numbers below the blots indicate relative densitometric intensity ratios of miR163 abundance in the transgenic lines compared with the endogenous AtmiR163 level in Allo3, which were normalized to U6 and Allo3. The numbers above the gel correspond to individual transgenic plants.
At-miR163 and Aa-miR163 Mediate Target Gene Expression in A. thaliana
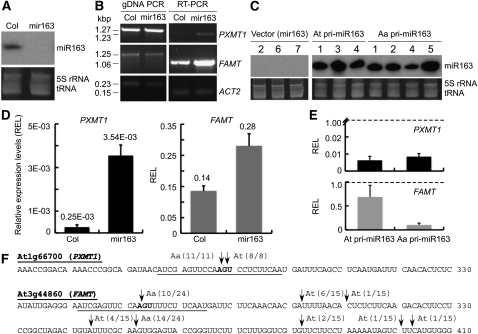
In the lines that overexpressed miR163, two target genes (PXMT1 and FAMT) were downregulated (see Supplemental Figure 6 online). However, the correlation between miR163 upregulation and target gene repression was not straightforward because endogenous miR163 is also present in the transgenic lines. The single nucleotide difference between the 24-nucleotide At-miR163 and the 23-nucleotide Aa-miR163 may also affect the target cleavage efficiency in the allotetraploids. To overcome these complications, we tested functional consequences of miR163 accumulation in a T-DNA insertion line for At MIR163, namely, the mir163 mutant. In the mutant, miR163 expression was undetectable (Figure 4A). As a result, both target genes (PXMT1 and FAMT) were upregulated (Figure 4B). Relative to the expression levels in A. thaliana (Col), PXMT1 was more induced than FAMT in the mir163 mutant, but FAMT had higher absolute levels than PXMT1 (Figure 4D). Overexpressing At pri-miR163 (At642) and Aa pri-miR163 (Aa859) in mir163 mutant plants increased ectopic accumulation of mature At and Aa-miR163, respectively (Figure 4C). As a result, expression of both PXMT1 and FAMT was reduced (Figure 4E; see Supplemental Figure 7 online). FAMT was more reduced by overexpressing Aa pri-miR163 than by overexpressing At pri-miR163. Compared with the At-miR163 cleavage site (Allen et al., 2004), the Aa-miR163 cleavage site was shifted one base in both targets (Figure 4F). This likely resulted from the single nucleotide difference between the two miR163 at the 20 nucleotide position, in which Aa-miR163 matches perfectly with the canonical binding site of the target, whereas At-miR163 has an extra U (Figures 1C, 1D, and 4F). An additional cleavage site was detected in FAMT transcripts when Aa MIR163 was overexpressed. Interestingly, At-miR163 did not cleave FAMT in the previously predicted site (Allen et al., 2004). Instead, in our assays, it cleaved six sites downstream from the predicted site, while Aa-miR163 cleaved two sites. It is possible that overexpression of At or Aa pri-miR163 resulted in the production of secondary small RNAs that could also target FAMT. Because of better sequence complementarity to Aa-miR163 than to At-miR163, FAMT is more downregulated in Aa-miR163 overexpressors than in At-miR163 overexpressors.
Figure 4.
Effects of miR163 Overexpression on Target Gene Regulation in the T-DNA Insertion Mutant Plants for mir163.
(A) Small RNA gel blot analysis showing presence of miR163 (24 nucleotides) in the wild type (Col) and absence of miR163 expression in the mir163 mutant.
(B) RT-PCR analysis of miR163 target genes, PXMT1 (At1g66700) and FAMT (At3g44860), in Col and mir163 lines. Corresponding genomic DNA was amplified as PCR controls, and Actin 2 was used as expression controls.
(C) Small RNA gel blot analysis of total RNA in mir163 mutant plants overexpressing At pri-miR163 or Aa pri-miR163. All RNA gel blots used the same probe labeling reaction for hybridization and had the same exposure time for image acquisition. Numbers above the gel correspond to different independent transgenic lines.
(D) qRT-PCR analysis of miR163 target gene expression in mir163 mutant plants. REL, relative expression level.
(E) qRT-PCR analysis of miR163 target gene expression (PXMT1 and FAMT) in mir163 plants overexpressing either At or Aa pri-miR163. The average R.E.L. was calculated between miR163 overexpressing lines and the vector control (dotted lines). Values are mean ± se, which are calculated from multiple transgenic lines as shown in (C). Except for PXMT1 in (E), the expression differences were statistically significant at P < 0.001.
(F) Analysis of miR163-mediated mRNA cleavage using 5′ rapid amplification of cDNA ends. Partial sequences of PXMT1 (At1g66700) and FAMT (At3g44860) are shown. Predicted target sites in PXMT1 and FAMT are underlined. The arrows indicate the detected cleavage sites in Aa transcripts (Aa) and At transcripts (At or without designation), respectively. The frequencies of cloned sequences are shown next to the arrows.
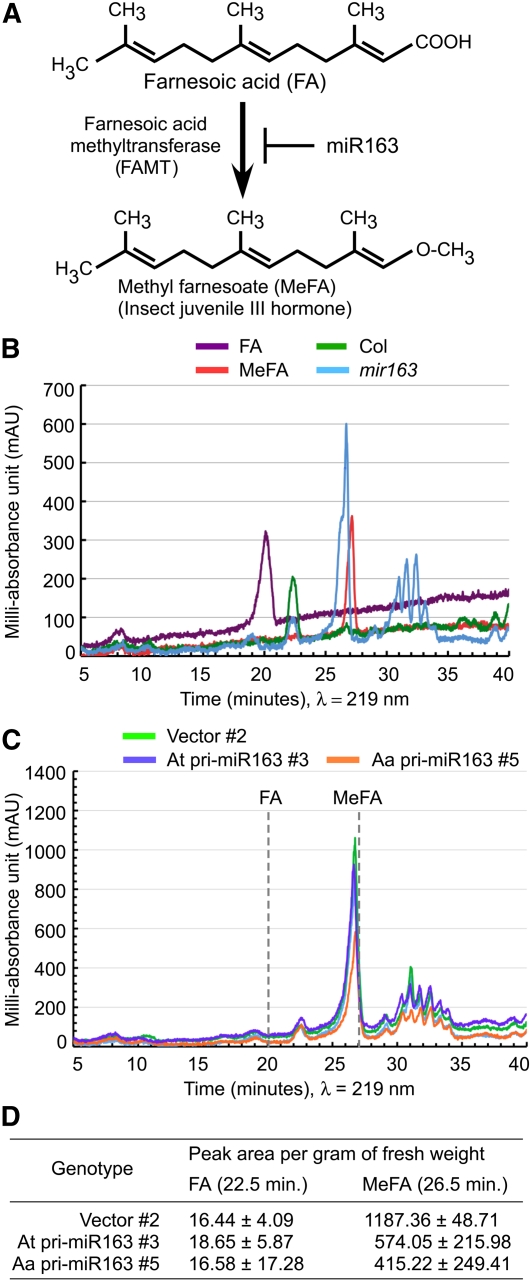
Altered At- and Aa-miR163 Abundance Changes Expression of Target Genes and Profiles of Metabolites
Products of miR163 target genes are members of the plant SABATH family of methyltransferases that have important biological functions by methylating hormones, signaling molecules, and other metabolites (D’Auria et al., 2003; Zhao et al., 2008). One of the miR163 target genes, FAMT (At3g44860), encodes farnesoic acid methyltransferase. The recombinant FAMT has enzymatic activity that converts FA into MeFA (Yang et al., 2006) (Figure 5A). Using HPLC coupled with UV detection (HPLC-UV) and chromatogram (219 nm) analysis, we compared crude metabolic profiles in wild-type and mir163 mutant plants. Synthetic FA and MeFA standards (with >95% all trans-stereochemistry) peaked at 20.2 and 27.2 min, respectively (Figure 5B). In chloroform extracts of rosette leaves, a prominent peak near the FA standard was detected in A. thaliana wild type (Col), whereas another prominent peak near the MeFA standard was found in the mir163 mutant. The data support the hypothesis that in the wild-type plants, expression of FAMT is negatively regulated by miR163, resulting in accumulation of FA. By contrast, loss of miR163 expression in the mir163 mutant leads to upregulation of FAMT and accumulation of MeFA through conversion of FA to MeFA. The slight difference in the retention time between leaf extracts and synthetic FA or MeFA standards may reflect different steric isoforms of biological materials. Additional peaks between 30 and 35 min were detected in the extracts from the mir163 mutant, which may suggest effects of miR163 loss on other target gene products.
Figure 5.
Effects of miR163 Mutation and Overexpression on Target Metabolites in the mir163 Mutant Plants.
(A) Conversion of FA to MeFA by FAMT is negatively regulated by miR163.
(B) HPLC chromatographs showing peaks of crude plant (leaf) extracts from Col (green) and mir163 (blue). Standards of FA (purple) and MeFA (red) are shown.
(C) HPLC chromatographs showing peaks of crude extracts in the vector control (green), At-miR163 transgenic lines (blue), and Aa-miR163 transgenic lines (orange), all in the mir163 mutant background.
(D) Quantification of peak areas near FA and MeFA (peak area per gram of fresh leaf weight) detected in the experiment in (C) with three biological replications. Values are mean ± sd.
In the mir163 mutant plants that overexpressed At or Aa-miR163, the peak near MeFA decreased compared with the vector transgenic control (Figure 5C). Interestingly, Aa-miR163 and At-miR163 overexpressors in the mir163 mutant resulted in significant reduction (~65 and ~48%) of characteristic peak values, respectively (Figure 5D), which is consistent with the severe reduction of FAMT expression in Aa-miR163 overexpressors relative to the At-miR163 overexpressors (Figure 4E). As noted previously, Aa-miR163 is probably a better modulator of MeFA production than At-miR163. The data suggest that the conversion of FA to MeFA is negatively regulated by miR163 in A. thaliana (Figure 5A).
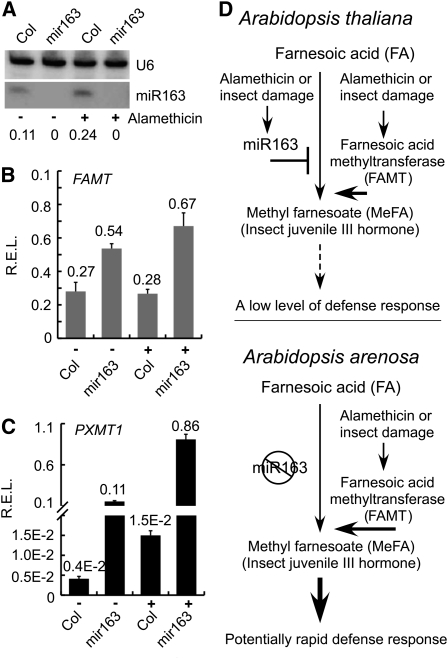
miR163 and Target Gene Expression Is Inducible
MeFA is an unepoxidized analog of insect juvenile hormone III made by some plants as a defense response against insect herbivores through interference with insect development (Toong et al., 1988; Shinoda and Itoyama, 2003). In A. thaliana, the endogenous FA and MeFA concentration is relatively low, but FAMT and PXMT1 expression is highly inducible (Yang et al., 2006). Among many inducing agents, including alamethicin, salicylic acid, methyl jasmonate, and wounding, the fungal elicitor alamethicin is the most effective inducer. Indeed, expression of both miR163 (Figure 6A) and its targets FAMT (Figure 6B) and PXMT1 (Figure 6C) was induced by alamethicin treatments. Expression levels of FAMT and PXMT1 were ~2- and ~25-fold higher, respectively, in Col-0 (wild type) than in the mir163 mutant. In alamethicin-treated A. thaliana plants, FAMT expression remained at relatively constant levels probably due to miR163 upregulation, whereas PXMT1 expression was induced ~3.6-fold by alamethicin. PXMT1 encodes a 1,7 paraxanthine methyltransferase, indicating that its substrate is likely to be a caffeine-related secondary metabolite (Zhao et al., 2008). The physiological relevance of PXMT1 needs further investigation because its absolute level was low. These different responses of miR163, PXMT1, and FAMT levels to alamethicin treatment may reflect a role for miR163 in mediating homeostasis of its target gene expression and potential defense response.
Figure 6.
Alamethicin Induces miR163 and Its Target Gene Expression in A. thaliana and a Model for miR163-Mediated Natural Variation of a Defense Mechanism in Related Species.
(A) Small RNA gel blot analysis of endogenous miR163 accumulation in 4- to 5-week-old leaves in A. thaliana wild type (Col) and miR163 T-DNA mutant (mir163) after 10-h treatments with 0.1% methanol (control) or alamethicin (10 μg/μL). Numbers indicate relative densitometric intensities of miR163 relative to the U6 loading control (n = 2).
(B) qRT-PCR of PXMT1 expression in Col and mir163 with or without alamethicin (10 μg/μL) treatment. The relative expression level (R.E.L.) was normalized to endogenous Ef1-α transcript levels (n = 3).
(C) qRT-PCR of FAMT expression in Col and mir163 with or without alamethicin (10 μg/μL) treatment (n = 3). Values are mean ± se. P = 0.01 (*), 0.001 (**), and 0.0001 (***), respectively.
(D) In inbreeding rapid cycling A. thaliana, both miR163 and its target gene (FAMT) are induced by wounding and/or fungal elicitors in response to insect bites and physical damages. As a result, FAMT expression is low because of miR163 accumulation, resulting in a low level of defense response. In outcrossing and late flowering A. arenosa, this defense response pathway is highly inducible due to loss of miR163 expression, leading to rapid and effective defense response. Constant maintenance of this defense pathway in an outcrossing and late flowering species (e.g., to prevent from insect bites and mechanical wounding) might have promoted the loss of miR163 expression.
We propose a model that explains natural variation of miR163-mediated defense responses in Arabidopsis-related species (Figure 6D). In A. thaliana, a high abundance of miR163 represses meFA production. In A. arenosa, the negative regulator (miR163) for MeFA production is suppressed, allowing expression of FAMT, which coverts FA to MeFA, a precursor of insect juvenile hormone III that affects insect growth and development (Toong et al., 1988; Shinoda and Itoyama, 2003). This miR163 repression is mediated by a relatively weak promoter in A. arenosa as well as species-specific repressors that are present in A. arenosa but absent in A. thaliana. In the allotetraploids, A. arenosa repressors may reduce expression of both A. thaliana and A. arenosa MIR163 loci. Over time, accumulation of mutations in the promoters as well as presence of repressors might have led to severe reduction of miR163 expression in A. arenosa. As a result, the MeFA-mediated defense pathway is more inducible and active in outcrossing and late flowering A. arenosa than in inbreeding and rapid cycling A. thaliana.
DISCUSSION
Expression of many miRNAs and their targets in plants is responsive to external stimuli (Sunkar and Zhu, 2004). miR395 is induced under depleted sulfur conditions (Kawashima et al., 2009), and miR399 is induced by phosphate starvation (Fujii et al., 2005). Both miR163 and its target genes (PXMT1 and FAMT) are induced by alamethicin treatments, suggesting their potential roles in defense response pathways. Expression induction of miR163 and its target genes (PXMT1 and FAMT) by alamethicin treatments and target cleavage preference between two miR163 homologs suggest a molecular mechanism for the evolution of a complex pathway through differential regulation of miRNA loci in closely related species.
A. arenosa is an outcrossing and natural strain living in wild conditions, in contrast with inbreeding A. thaliana (Clauss and Koch, 2006). Although both species have rapid life cycle, a relatively late flowering time means a longer vegetative development in A. arenosa than in A. thaliana (Wang et al., 2006). A combination of outcrossing and prolonged vegetative stage would subject A. arenosa to more pathogen/herbivore attacks. Over time, repression of miR163 expression in A. arenosa would provide an adaptive advantage for an outcrossing and late flowering species. The loss of negative regulator (e.g., miR163) in defense regulatory pathways could prepare A. arenosa to be more responsive to herbivory and wounding. Indeed, two miR163 targets in A. thaliana are inducible by herbivory and wounding and play a role in defense against pests (Chen et al., 2003). Moreover, miR163 also regulates another set of target genes encoding S-adenosylmethionine–dependent methyltransferases that methylate hormones, signaling molecules, and other secondary metabolites including volatiles (D’Auria et al., 2003; Zhao et al., 2008). The production of volatiles and secondary metabolites in large and pink flowers and the relationship between volatile production and obligated outcrossing in A. arenosa remain to be investigated. Plant defense against herbivory is an evolutionary arm race. Current data suggest that positive selection might have kept suppression of negative regulators such as miR163 by accumulation of mutations in the promoters as well as induction of trans-acting protein factors, resulting in epistatic effects on target gene regulation and potential downstream effects including defense responses.
Our finding of cis- and trans-regulation of a miRNA-mediated pathway provides new insights into the molecular basis for changes in gene expression and downstream phenotypic variation between species and in allopolyploids. Differential accumulation of miRNAs can be controlled by transcriptional and posttranscriptional regulation (Ha et al., 2009). At the transcriptional level, cis-promoter divergence between At MIR163 and Aa MIR163 upstream regions is the major factor contributing to the differential expression of MIR163 loci in A. thaliana and A. arenosa through chromatin modifications. The correlation of Aa MIR163 expression with H3K4me3 but not with H3K9ac in F1 allotetraploids suggests that it takes several generations in the allopolyploids to establish stable chromatin and expression patterns (Wang et al., 2004). In the allotetraploids, A. thaliana and A. arenosa genomes present within the same cellular environment has created a trans-acting background for homoeologous gene regulation. It is likely that trans-acting factors interact with chromatin modifiers, such as histone methyltransferases and deacetylases (Berger, 2007; Li et al., 2007), to remodel chromatin structure in the vicinity of MIR163 loci, leading to differential accumulation of At- and Aa-miR163.
At the posttranscriptional level, although At and Aa pri-miR163 are efficiently processed to yield mature miR163 in transgenic A. thaliana, their processing efficiencies are lower in transgenic allotetraploids than in A. thaliana. It has been shown that structure of the distal loop region of pri-miR172a is important for its efficient processing and accumulation of mature miR172a (Werner et al., 2010). The cleavage of pri-miRNA by DICER-LIKE1 (DCL1) is also affected by the structure of lower stem-loop region that is ~15 nucleotides from the miRNA/miRNA* duplex (Mateos et al., 2010; Song et al., 2010). Therefore, it is likely that cis-sequence divergence leads to secondary structural changes between the homoeologous miR163 precursors, affecting their biogenesis in the allotetraploids. Moreover, differential expression of species-specific miRNA biogenesis factor genes, such as DCL1, may confer additional trans-regulation of miRNA accumulation (Ha et al., 2009). We speculate that trans-interactions among homoeologous miRNA biogenesis factors, including DCL1, SERRATE, and HYPONASTIC LEAVES1, could modulate the level of homoeologous miR163 accumulation. Further studies in the allotetraploids will shed light on posttranscriptional regulation of homoeologous miRNA loci and their downstream effects on target gene expression, including epistatic regulation and potentially novel phenotypes.
In spite of a single-nucleotide difference between canonical At-miR163 and Aa-miR163, overexpression of either At or Aa-miR163 in A. thaliana or allotetraploids downregulates expression of the targets PXMT1 and FAMT. For FAMT transcripts, an additional and multiple cleavage sites were detected in Aa-miR163 and At-miR163 overexpressors, respectively. These cryptic cleavage sites may result from the production of secondary small RNAs that mediate additional cleavage of the targets. As a result, FAMT is more downregulated in Aa-miR163 overexpressors than in At-miR163 overexpressors. These data may reflect species-specific regulation of miR163 target homeostasis in Arabidopsis.
In the newly synthesized allotetraploids, trans-acting effects on homoeologous gene expression can occur immediately upon fertilization of A. thaliana ovules with A. arenosa pollen, providing a mechanism to reconcile the divergence between cis-regulatory elements and trans-acting factors in the related species. As examples, a subset of siRNAs whose biogenesis is dependent on RNA polymerase IV, known as p4-siRNAs, accumulates in the maternal gametophyte during seed development. This suggests a role for these siRNAs in trans-acting effects on RNA silencing and seed development (Mosher et al., 2009). In the pollen vegetative nucleus, reactivation of transposons generates 21-nucleotide siRNAs that accumulate in sperm cells (Slotkin et al., 2009). Although the functions of 21-nucleotide siRNAs in transposon regulation and DNA methylation are yet to be tested, these paternally derived siRNAs are predicted to act in trans during fertilization in the interspecific hybrids (Bourc’his and Voinnet, 2010). Similarly, genome-wide expression differences in miRNAs and siRNAs from homoeologous loci in the allotetraploids may generate trans-acting effects on the expression of a wide range of target genes (Ha et al., 2009). In conclusion, this example of miRNA-mediated natural variation between related species is predicted to provide a general mechanism for genome-wide cis- and trans-regulation of miRNA and protein-coding genes, leading to altered expression of target genes and pathways, epistatic effects, and transgressive phenotypes in interspecific hybrids and allopolyploids.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana autotetraploid (At4, CS3900), Arabidopsis arenosa (Aa, CS3901), and resynthesized allotetraploid lines (Allo3, aka Allo733, CS3895 and Allo8 aka Allo738, CS3896) were obtained as previously described (Ha et al., 2009). A. thaliana (CS879797) T-DNA insertion mutant for mir163 was obtained from the ABRC. All plants were grown at 22°C under a 16/8-h light/dark cycle. For plant transformation, A. thaliana (4 to 5 weeks old) or Allo3 (9 to 10 weeks old) plants were used for Agrobacterium tumefaciens–mediated transformation through floral dipping (Clough and Bent, 1998).
Alamethicin Treatment
A 10 mg/mL stock solution of alamethicin (Sigma-Aldrich) was prepared by dissolving the alamethicin in 100% methanol. Mature leaves from A. thaliana (4 to 5 weeks old) were cut at the petiole and placed in 10 μg/mL alamethicin (Sigma-Aldrich) solution (diluted 1000 times with water) and treated for 10 h. A control was included by placing the leaves in a beaker containing 0.1% methanol.
Cloning of the At and Aa MIR163 Loci and Plasmid Constructs
Genomic DNA from A. thaliana (At4) and A. arenosa (Aa) was used to amplify MIR163 upstream promoter fragments or transcribed miR163 precursors. Oligonucleotide primers were designed based on the A. thaliana genome or sequence information of Illumina reads of genomic DNA of A. arenosa (see Supplemental Table 3 online for primer sequences). The amplified fragments were cloned into pGEM-T vector (Promega) for sequence verification.
For GUS reporter constructs, a pAt-MIR163 (−1439/+150) or pAa-MIR163 (−1977/+150) promoter fragment and the GUS coding region were cloned into pEarleyGate103 (ABRC stock: CD3-685) by replacing the 35S-Gateway-GFP-His cassette in the vector through EcoRI and AvrII restriction sites. For 35S-driven overexpression constructs, At pri-miR163 (642 bp) or Aa pri-miR163 (859 bp) transcribed region was cloned into the pEarleyGate100 vector (ABRC stock: CD3-724) by replacing the gateway cassette in the vector through XhoI and AvrII restriction sites. All constructs were individually cloned into Agrobacterium strain GV3101 for plant transformation.
Because the T-DNA insertion mutant (mir163) is resistant to phosphinothricin, a binary vector, pHM301k (Chandrasekharan et al., 2003), containing a kanamycin plant selection marker was used for generating 35S-driven pri-miR163 overexpression constructs for plant transformation.
Histochemical and Fluorometric Assays for GUS Activity
Spatial expression of GUS in transgenic plants was visualized using histochemical GUS staining at 37°C for overnight (16 h) in the presence of 5-bromo-4-chloro-3-indoxyl-β-d-glucuronic acid (Gold Biotechnology) as a substrate (Jefferson et al., 1987). In the GUS fluorometric assay, 4-methylumbelliferyl-β-d-glucuronide and 4-methylumbelliferone (4-MU) (Gold Biotechnology) were used as a substrate and a standard, respectively. GUS activity (nmol 4-MU per hour per microgram protein) of T2 seedlings, mature leaves, and inflorescences were measured. A Student’s t test was used to determine the significance of promoter activity differences between GUS reporter and control transgenic plants in three independent assays per transgenic line.
RNA Isolation and cDNA Synthesis
Total RNA was isolated from mature leaves (3- to 4-week-old A. thaliana plants; 6- to 7-week-old A. arenosa or allotetraploids plants) or inflorescences using Plant RNA reagent (Invitrogen) and treated with RQ1 DNase (Promega) according to the manufacturers’ instructions. For cDNA synthesis, 1 μg of DNaseI-treated RNAs was incubated with Superscript III reverse transcriptase (Invitrogen) in the presence of 25 ng/μL oligo dT (12-18) primer (GeneLink) in a 20 μL reaction according to the manufacturers’ instructions.
RT-PCR and qRT-PCR
DNase-treated (500 ng) RNA was subjected to One-Step RT-PCR assays (Qiagen) using the primer pairs for PXMT1 or FAMT. Actin2 was used as an internal control for the amplification. For qPCR, FastStart Universal SYBR Green Master (Rox) (Roche Applied Science) was used for PCR in the presence of gene-specific primers and template cDNAs. Expression levels of target genes were normalized against transcript levels of either Elongation factor 1-α (EF1-α) or Ubiquitin10 (UBQ10) using oligonucleotide primers in RT-PCR and qRT-PCR assays (see Supplemental Table 2 online). Relative expression levels of transcripts were calculated. For each sample, three PCR reactions were performed with two biological replications. Primer sequences are shown in Supplemental Table 2 online.
Small RNA Gel Blot Analysis
Total leaf RNAs (10 μg) were used for small RNA gel blot analysis of miR163 accumulation as previously described (Lackey et al., 2010). An antisense 19-nucleotide oligonucleotide probe corresponding to mature At or Aa miR163 and an antisense U6 oligonucleotide probe (see Supplemental Table 2 online) were end-labeled with [γ-32P]ATP (6000 Ci/mmol) for hybridization. Small RNA gel blot signals for miR163 and U6 were detected using a phosphor imaging plate (Fuji Photo Film) and a Typhoon Trio variable mode imager (GE Healthcare) for image acquisition. Separate exposure times were used for miR163 and U6 signals to prevent saturation of target signals. Densitometric intensities of small RNA gel blot signals were obtained using ImageJ software (National Institutes of Health).
5′ Rapid Amplification of cDNA Ends for miR163 Target Cleavage Site Analysis
Total leaf RNAs from At642 and Aa859 transgenics in the mir163 mutant were used to determine the 5′ end of the 3′ cleavage product of At and Aa miR163 target transcripts, respectively. Total RNA (1 μg) was ligated to a RNA oligo-adapter, followed by cDNA synthesis with an oligo dT (12-18) primer (GeneLink) and gene-specific primers corresponding to PXMT1 and FAMT. Cleaved targets were then amplified using nested PCR with corresponding nested primers (see Supplemental Table 5 online). The PCR products were purified and cloned into a pGEM-T vector (Promega) for sequencing using the sp6 primer.
ChIP and PCR
ChIP was performed as previously described (Saleh et al., 2008) with the following modifications. Rosette leaves (3 g) of tetraploid A. thaliana (3 to 4 weeks old), A. arenosa (6 to 7 weeks old), and F1 and F8 generations of allotetraploids (Allo3, 6 to 7 weeks old) were harvested for ChIP analysis. Immunoprecipitation (IP) was performed using 750 μL sonicated chromatin with antibodies against H3-Cter (Abcam; Ab1791; 2 μg), H3K4me3 (Abcam; Ab8580; 4 μg), or H3K9ac (Abcam; Ab10812; 3.6 μg). For each sample, a mock (no antibody) and input DNA (no IP) controls are included in the IP reaction. Purified DNA from ChIP was resuspended in 50 μL TE, pH 7.5, and used for PCR amplification with oligonucleotide primers (see Supplemental Table 2 online). PCR products were resolved in 2.5% agarose gel and visualized by ethidium bromide staining.
For qPCR, purified DNA from ChIP (using αH3-Cter, αH3K4me3, or αH3K9ac) was diluted 10 times, and 1 μL diluted DNA was used for qPCR with the primer pairs (see Supplemental Table 2 online). For input DNA control, purified DNA was diluted 20 times, and 1 μL diluted-DNA was used for qPCR. Enrichment of DNA for a specific histone modification was normalized with the corresponding input DNA and the histone H3 level.
Organic Extraction of Crude Plant Extract and HPLC
Rosette leaves (~1.5 g) from 3- to 4-week-old wild-type or transgenic plants were ground in liquid nitrogen and extracted with chloroform at room temperature. Following solvent evaporation under nitrogen gas, each sample was resuspended in 500 μL HPLC-grade methanol. Crude extract was filtered using an Ultrafree-MC centrifugal filter with 0.2 μm low binding hydrophilic PTFE membrane (Millipore). Metabolite profile was determined using HPLC on a Varian Pro Star chromatography workstation (Varian). Separation of metabolites was achieved by injection of 50 μL crude extract into a Varian Pursuit C8 ChromSep Column (5 μm, 150 × 46 mm). Gradient separation of compounds was achieved in a 1 mL/min flow rate with water as solvent A and methanol as solvent B in the following parameters: 60% B (isocratic) from 0 to 5 min, 60 to 80% B (linear) from 5 to 30 min, 80% B (isocratic) from 30 to 40 min, 80 to 100% B (linear) from 40 to 41 min, 100% B (isocratic) from 41 to 46 min, 100 to 60% B (linear) from 46 to 47 min, and 60% B (isocratic) from 47 to 50 min. Eluted compounds were detected by UV absorbance at 219 nm. All trans (>95%) (E,E) FA (200 μM) and (E,E) MeFA (400 μM) (Echelon Biosciences) were used as reference standards. Quantification of peak at a specific retention time was done by integrating the peak area and normalization against the fresh weight of plant materials in three biological replications.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: MIR163 (At1g66725), PXMT1 (At1g66700), FAMT (At3g44860), ACTIN7 (At5g09810), ACTIN2 (At3g18780), UBQ10 (At4g05420), and EF1α (At1g07930).
Author Contributions
Z.J.C. and D.W-K.N. designed experiments. D.W-K.N. performed most experiments. C.Z. performed miR163 target cleavage assays. M.M. performed cloning and transformation. G.P. performed HPLC analysis. D.T. provided secondary metabolite and volatile analysis and data interpretation. M.W. designed HPLC experiments. D.W-K.N. and Z.J.C. analyzed the data and wrote the article.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment between A. thaliana and A. arenosa MIR163 Loci.
Supplemental Figure 2. Clustal V Alignment of the At and Aa pri-miR163.
Supplemental Figure 3. Phylogenetic Analysis of miR163 Target Genes.
Supplemental Figure 4. Pri-miR163 Expression in Arabidopsis-Related Species and Allotetraploids.
Supplemental Figure 5. Predicted Secondary Structure of At- and Aa-miR163 Precursors.
Supplemental Figure 6. Downregulation of miR163 Target Genes in Transgenic Arabidopsis Overexpressing At-miR163 or Aa-miR163.
Supplemental Figure 7. Downregulation of miR163 Target Genes in the mir163 Mutant Plants Overexpressing At-miR163 or Aa-miR163.
Supplemental Table 1. Putative cis-Elements Present in At and Aa MIR163 Upstream Regions.
Supplemental Table 2. Oligonucleotide Primers Used for Genotyping and Expression Analyses.
Supplemental Table 3. Oligonucleotide Primers Used for Cloning and Plasmid Construction.
Supplemental Table 4. pAt-MIR163 or pAa-MIR163 Driven GUS Expression in Arabidopsis thaliana and Allotetraploids (Allo3).
Supplemental Table 5. Oligonucleotide Adapters and Primers Used for Cleavage Analysis.
Acknowledgments
We thank Lu Tian for initial characterization of miR163 expression in A. thaliana and A. arenosa and Eric Bih and Colin Purmal for plant care and DNA analysis. The work is supported by a grant from the National Science Foundation (MCB1110957 to Z.J.C.).
References
- Allen E., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36: 1282–1290 [DOI] [PubMed] [Google Scholar]
- Ambros V. (2004). The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- Ason B., Darnell D.K., Wittbrodt B., Berezikov E., Kloosterman W.P., Wittbrodt J., Antin P.B., Plasterk R.H. (2006). Differences in vertebrate microRNA expression. Proc. Natl. Acad. Sci. USA 103: 14385–14389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Thuemmler F., van Laake L.W., Kondova I., Bontrop R., Cuppen E., Plasterk R.H. (2006). Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 38: 1375–1377 [DOI] [PubMed] [Google Scholar]
- Berger S.L. (2007). The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Bourc’his D., Voinnet O. (2010). A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science 330: 617–622 [DOI] [PubMed] [Google Scholar]
- Carlborg O., Haley C.S. (2004). Epistasis: Too often neglected in complex trait studies? Nat. Rev. Genet. 5: 618–625 [DOI] [PubMed] [Google Scholar]
- Carroll S.B. (2005). Evolution at two levels: On genes and form. PLoS Biol. 3: e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan M.B., Bishop K.J., Hall T.C. (2003). Module-specific regulation of the beta-phaseolin promoter during embryogenesis. Plant J. 33: 853–866 [DOI] [PubMed] [Google Scholar]
- Chen F., D’Auria J.C., Tholl D., Ross J.R., Gershenzon J., Noel J.P., Pichersky E. (2003). An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 36: 577–588 [DOI] [PubMed] [Google Scholar]
- Chen X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J. (2007). Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss M.J., Koch M.A. (2006). Poorly known relatives of Arabidopsis thaliana. Trends Plant Sci. 11: 449–459 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Comai L., Tyagi A.P., Winter K., Holmes-Davis R., Reynolds S.H., Stevens Y., Byers B. (2000). Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12: 1551–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria J.C., Chen F., Pichersky E. (2003). The SABATH family of MTS in Arabidopsis thaliana and other plant species. Recent Adv. Phytochem. 37: 253–283 [Google Scholar]
- Fujii H., Chiou T.J., Lin S.I., Aung K., Zhu J.K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Ha M., Lu J., Tian L., Ramachandran V., Kasschau K.D., Chapman E.J., Carrington J.C., Chen X., Wang X.J., Chen Z.J. (2009). Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. USA 106: 17835–17840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Pang M., Agarwal V., Chen Z.J. (2008). Interspecies regulation of microRNAs and their targets. Biochim. Biophys. Acta 1779: 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson C., Dilkes B., Comai L. (2006). Parent-dependent loss of gene silencing during interspecies hybridization. Curr. Biol. 16: 1322–1328 [DOI] [PubMed] [Google Scholar]
- Kawashima C.G., Yoshimoto N., Maruyama-Nakashita A., Tsuchiya Y.N., Saito K., Takahashi H., Dalmay T. (2009). Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 57: 313–321 [DOI] [PubMed] [Google Scholar]
- Koch M.A., Haubold B., Mitchell-Olds T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17: 1483–1498 [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Watanabe Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey E., Ng D.W., Chen Z.J. (2010). RNAi-mediated down-regulation of DCL1 and AGO1 induces developmental changes in resynthesized Arabidopsis allotetraploids. New Phytol. 186: 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V.N. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch A.R., Leitch I.J. (2008). Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483 [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. (2007). The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Mateos J.L., Bologna N.G., Chorostecki U., Palatnik J.F. (2010). Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr. Biol. 20: 49–54 [DOI] [PubMed] [Google Scholar]
- Mosher R.A., Melnyk C.W., Kelly K.A., Dunn R.M., Studholme D.J., Baulcombe D.C. (2009). Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286 [DOI] [PubMed] [Google Scholar]
- Ng D.W., Chandrasekharan M.B., Hall T.C. (2006). Ordered histone modifications are associated with transcriptional poising and activation of the phaseolin promoter. Plant Cell 18: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Kim E.D., Ha M., Lackey E., Liu J., Zhang Y., Sun Q., Chen Z.J. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R., Slack F.J. (2007). The evolution of animal microRNA function. Curr. Opin. Genet. Dev. 17: 145–150 [DOI] [PubMed] [Google Scholar]
- Ruthenburg A.J., Allis C.D., Wysocka J. (2007). Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol. Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T. (2002). Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Schübeler D., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., Bell S.P., Groudine M. (2004). The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Song J.T., Cheong J.J., Lee Y.H., Lee Y.W., Hwang I., Lee J.S., Choi Y.D. (2001). Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 98: 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda T., Itoyama K. (2003). Juvenile hormone acid methyltransferase: A key regulatory enzyme for insect metamorphosis. Proc. Natl. Acad. Sci. USA 100: 11986–11991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Axtell M.J., Fedoroff N.V. (2010). RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 20: 37–41 [DOI] [PubMed] [Google Scholar]
- Sterner D.E., Berger S.L. (2000). Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64: 435–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Zhu J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Reikhav S., Levy A.A., Barkai N. (2009). A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324: 659–662 [DOI] [PubMed] [Google Scholar]
- Toong Y.C., Schooley D.A., Baker F.C. (1988). Isolation of Insect Juvenile Hormone-Iii from a Plant. Nature 333: 170–171 [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H.S., Chen Z.J. (2006). Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics 173: 965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Madlung A., Lee H.S., Chen M., Lee J.J., Watson B., Kagochi T., Comai L., Chen Z.J. (2004). Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167: 1961–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S., Wollmann H., Schneeberger K., Weigel D. (2010). Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr. Biol. 20: 42–48 [DOI] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H. (2005). MicroRNA expression in zebrafish embryonic development. Science 309: 310–311 [DOI] [PubMed] [Google Scholar]
- Wittkopp P.J., Haerum B.K., Clark A.G. (2004). Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88 [DOI] [PubMed] [Google Scholar]
- Wray G.A. (2007). The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8: 206–216 [DOI] [PubMed] [Google Scholar]
- Yang Y., Yuan J.S., Ross J., Noel J.P., Pichersky E., Chen F. (2006). An Arabidopsis thaliana methyltransferase capable of methylating farnesoic acid. Arch. Biochem. Biophys. 448: 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N., Ferrer J.L., Ross J., Guan J., Yang Y., Pichersky E., Noel J.P., Chen F. (2008). Structural, biochemical, and phylogenetic analyses suggest that indole-3-acetic acid methyltransferase is an evolutionarily ancient member of the SABATH family. Plant Physiol. 146: 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]