Abstract
This work identifies LOW QUANTUM YIELD OF PHOTOSYSTEM II1 (LQY1), a Zn finger protein that shows disulfide isomerase activity, interacts with the photosystem II (PSII) core complex, and may act in repair of photodamaged PSII complexes. Two mutants of an unannotated small Zn finger containing a thylakoid membrane protein of Arabidopsis thaliana (At1g75690; LQY1) were found to have a lower quantum yield of PSII photochemistry and reduced PSII electron transport rate following high-light treatment. The mutants dissipate more excess excitation energy via nonphotochemical pathways than wild type, and they also display elevated accumulation of reactive oxygen species under high light. After high-light treatment, the mutants have less PSII–light-harvesting complex II supercomplex than wild-type plants. Analysis of thylakoid membrane protein complexes showed that wild-type LQY1 protein comigrates with the PSII core monomer and the CP43-less PSII monomer (a marker for ongoing PSII repair and reassembly). PSII repair and reassembly involve the breakage and formation of disulfide bonds among PSII proteins. Interestingly, the recombinant LQY1 protein demonstrates a protein disulfide isomerase activity. LQY1 is more abundant in stroma-exposed thylakoids, where key steps of PSII repair and reassembly take place. The absence of the LQY1 protein accelerates turnover and synthesis of PSII reaction center protein D1. These results suggest that the LQY1 protein may be involved in maintaining PSII activity under high light by regulating repair and reassembly of PSII complexes.
INTRODUCTION
Harnessing the tremendous energy of photons by photosynthesis is both essential and risky for green plants; this energy powers plant metabolism but can also damage the photosynthetic apparatus. Despite the importance of carbon fixation in agriculture, key mechanistic details remain to be elucidated for some aspects of photosynthesis, such as energy dissipation, cyclic electron transport, and formation, repair, and degradation of the photosynthetic apparatus (Baena-González and Aro, 2002; Tanaka and Makino, 2009). Under high irradiance conditions, photosystem II (PSII) undergoes rapid damage and repair cycles, resulting in the degradation and replacement of damaged D1 protein in the reaction center (Demmig-Adams and Adams, 1992; Yokthongwattana et al., 2001; Baena-González and Aro, 2002). PSII repair is a complex process that involves disassembly and reassembly of a thylakoid membrane protein complex composed of dozens of proteins and hundreds of cofactors (Baena-González and Aro, 2002; Mulo et al., 2008). The repair cycle of PSII requires the participation of more than 25 auxiliary proteins (Mulo et al., 2008), including heat shock protein 70 family members, PSII 22 kDa protein (PSB29), LOW PSII ACCUMULATION protein (LPA1), which are essential for repair of PSII, and small J-domain proteins AtJ8, AtJ11, and AtJ20 (Schroda et al., 1999; Yokthongwattana et al., 2001; Keren et al., 2005; Peng et al., 2006; Chen et al., 2010).
The continued identification of new proteins associated with PSII indicates that there is more to learn about its structure and function. This is challenging for a variety of reasons, including low protein abundance, labile interactions with core and antenna complexes, and functional redundancy (Mulo et al., 2008). Analysis of in vivo chlorophyll fluorescence is a powerful, noninvasive technique to identify mutations affecting photosynthesis (Maxwell and Johnson, 2000; Müller et al., 2001; Kramer et al., 2004). In an attempt to identify functions for novel chloroplast proteins, thousands of Arabidopsis thaliana T-DNA mutants with insertions in nuclear genes for chloroplast-targeted proteins are being analyzed for a variety of mutant phenotypes, including chlorophyll fluorescence (Lu et al., 2008, 2011; Ajjawi et al., 2010).
This screen identified two mutants of the At1g75690 gene, LOW QUANTUM YIELD OF PSII1 (LQY1), with high-light–induced defects in PSII photochemistry. The LQY1 protein is predicted to have a chloroplast transit peptide, a transmembrane domain, and a putative Zn finger homologous to Escherichia coli DnaJ. The results presented in this article are consistent with the hypothesis that LQY1 is involved in the repair and reassembly cycle of the PSII–light-harvesting complex II (LHCII) supercomplex under high irradiance.
RESULTS
Identification of lqy1 Mutants
As part of a reverse genetics project designed to discover novel functions for chloroplast-targeted proteins, ~5200 Arabidopsis homozygous T-DNA lines were analyzed for alterations in PSII photosynthetic electron transport (Lu and Last, 2008; Lu et al., 2008; Ajjawi et al., 2010). Chlorophyll fluorescence measurements of the maximum photochemical efficiency of PSII (i.e., variable fluorescence/maximal fluorescence of dark-adapted leaves [Fv/Fm]) and nonphotochemical quenching (NPQ) were performed on plants grown under 100 μmol photons m−2 s−1. To look for mutants that failed to acclimate to photoinhibitory light conditions, Fv/Fm was measured immediately following a 3-h high-light treatment (1500–1700 μmol photons m−2 s−1). Finally, recovery from photoinhibition was assessed by measuring Fv/Fm after a 48-h recovery under 100 μmol photons m−2 s−1 (Lu et al., 2008). More than 300 putative mutants were identified based on these parameters, and the data are publicly available at http://bioinfo.bch.msu.edu/2010_LIMS.
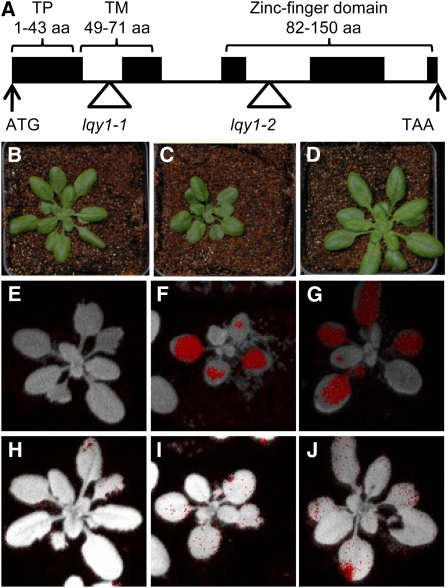
Two mutants of the nuclear-encoded gene At1g75690 (SALK_021824C and WiscDsLox376F05) were found to have a lower maximum photochemical efficiency of PSII (Fv/Fm) after high-light treatment, as displayed in red pixels (Figures 1F and 1G). Fv/Fm returned to normal following a 2-d recovery under growth light conditions (Figures 1H to 1J). These results were confirmed with twice-backcrossed mutants. Based on the mutant phenotype, we named SALK_021824C and WiscDsLox376F05 as lqy1-1 and lqy1-2, respectively. The lqy1-1 and lqy1-2 mutants carry T-DNA insertions in the first and third introns of At1g75690, respectively (Figure 1A).
Figure 1.
Mutations of the LQY1 Gene Cause Altered Chlorophyll Fluorescence Parameters.
(A) Gene structure and domains of LQY1 inferred by DNA sequence analysis. Black boxes represent exons and lines represent introns. The start and stop codons are indicated, and the T-DNA insertions in lqy1-1 and lqy1-2 are represented by triangles. aa, amino acids; TM, transmembrane domain; TP, chloroplast transit peptide.
(B) to (D) Images of 4-week-old Col wild-type (B), lqy1-1 (C), and lqy1-2 (D) plants under a 12-/12-h photoperiod.
(E) to (G) False-color images representing Fv/Fm after a 3-h high-light treatment in 3-week-old Col wild-type (E), lqy1-1 (F), and lqy1-2 (G) plants. Red pixels in (E) to (G) indicate that Fv/Fm is below the cutoff value (0.482).
(H) to (J) False-color images representing Fv/Fm after a 3-h high-light treatment and a 2-d recovery period in 3-week-old Col wild-type (H), lqy1-1 (I), and lqy1-2 (J) plants. Red pixels indicate that Fv/Fm is below the cutoff value (0.725).
The LQY1 gene is predicted to encode a 154-amino acid protein based on The Arabidopsis Information Resource 9 annotation (www.Arabidopsis.org). As illustrated in Figure 1A, the coding region includes an N-terminal chloroplast transit peptide (1–43 amino acids) based on TargetP prediction (Emanuelsson et al., 2000), a transmembrane domain (49–71 amino acids) by the transmembrane domain prediction program TMHMM (Krogh et al., 2001), and a Zn finger domain (82–150 amino acids) by Pfam (Bateman et al., 2004). Prediction of transmembrane helices by TMHMM also suggests that the C-terminal Zn finger is located on the lumen side of thylakoid membrane (Krogh et al., 2001). Consistent with the in silico analysis, proteomics studies showed that this protein is located in the thylakoid membrane (Kleffmann et al., 2004; Peltier et al., 2004; Zybailov et al., 2008).
PSII Activity in lqy1 Mutants Is More Susceptible to High-Light Stress
Comparing false-color images from Fv/Fm measurements provides a quick and robust screen for mutants. To further analyze photosynthetic defects in the mutants, minimum fluorescence of dark-adapted leaves (Fo), maximum fluorescence of dark-adapted leaves (Fm), Fv/Fm, and different components of NPQ were quantitatively determined. After a 3-h high-light treatment, 4-week-old lqy1 mutants had much higher Fo and NPQ and much lower Fv/Fm than wild-type control plants (Table 1). Interestingly, Fm values between the mutants and the wild type are not significantly different (Table 1), indicating that the reduction of Fv/Fm in lqy1 mutants is primarily caused by the increase in Fo. NPQ can be divided into energy-dependent quenching (qE), state-transition quenching, and photoinhibitory quenching (qI) according to their relaxation kinetics (Müller et al., 2001; Baker et al., 2007). Only qE and qI were calculated in this study because state-transition quenching is only significant under very low light in most plants (Müller et al., 2001; Krause and Jahns, 2003). After a 3-h high-light treatment, lqy1 mutants had higher qI than wild-type control plants (Table 1). The substantial increases in qI indicate that the mutants experience more photoinhibition than wild type under high-light stress (Dodd et al., 1998).
Table 1.
Chlorophyll Contents and Chlorophyll Fluorescence Parameters in 4-Week-Old Wild-Type and lqy1 Mutants
| Measured Parameter | Growth Light |
After 3-h High Light |
||||
| Col WT | lqy1-1 | lqy1-2 | Col WT | lqy1-1 | lqy1-2 | |
| Chl a (mg/g FW) | 1.414 ± 0.059 | 1.413 ± 0.028 | 1.489 ± 0.069 | 1.370 ± 0.016 | 1.385 ± 0.067 | 1.415 ± 0.034 |
| Chl b (mg/g FW) | 0.490 ± 0.025 | 0.486 ± 0.009 | 0.514 ± 0.024 | 0.491 ± 0.008 | 0.503 ± 0.024 | 0.512 ± 0.012 |
| Chl a/b | 2.893 ± 0.029 | 2.907 ± 0.016 | 2.894 ± 0.006 | 0.265 ± 0.024 | 2.754 ± 0.007 | 2.764 ± 0.009 |
| Fo | 0.091 ± 0.006 | 0.139 ± 0.006*** | 0.113 ± 0.005* | 0.106 ± 0.006 | 0.170 ± 0.006*** | 0.140 ± 0.006** |
| Fm | 0.508 ± 0.028 | 0.692 ± 0.023*** | 0.570 ± 0.018 | 0.297 ± 0.018 | 0.334 ± 0.010 | 0.277 ± 0.007 |
| Fv/Fm | 0.821 ± 0.005 | 0.800 ± 0.005** | 0.803 ± 0.005* | 0.644 ± 0.006 | 0.491 ± 0.009*** | 0.497 ± 0.013*** |
| NPQ | 1.832 ± 0.011 | 1.815 ± 0.015 | 1.835 ± 0.022 | 2.529 ± 0.038 | 2.936 ± 0.050*** | 2.933 ± 0.052*** |
| qE | 1.388 ± 0.009 | 1.304 ± 0.011*** | 1.365 ± 0.016 | 1.611 ± 0.040 | 1.637 ± 0.038 | 1.661 ± 0.027 |
| qI | 0.444 ± 0.007 | 0.511 ± 0.010*** | 0.470 ± 0.010* | 0.918 ± 0.017 | 1.299 ± 0.026*** | 1.272 ± 0.036*** |
Measurements of chlorophyll fluorescence parameters were done on plants after 20 min of dark adaption. For NPQ, qE, and qI measurements, an actinic light treatment (531 μmol photons m−2 s−1) was performed for 715 s. During actinic illumination, 36 saturation pulses (2800 μmol photons m−2 s−1) were applied at 20-s intervals. After termination of actinic light, recovery of Fm′ was monitored for 14 min as described by Müller et al. (2001). During the recovery, 16 saturation pulses were applied. After a 3-h high-light treatment, NPQ, qE , and qI were calculated with Fm determined before the high-light treatment. Data are presented as means ± se (n = 4 for chlorophyll contents and n = 8 for chlorophyll fluorescence parameters). The asterisk indicates a significant difference between the mutant and Col wild type (WT) (Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001). FW, fresh weight.
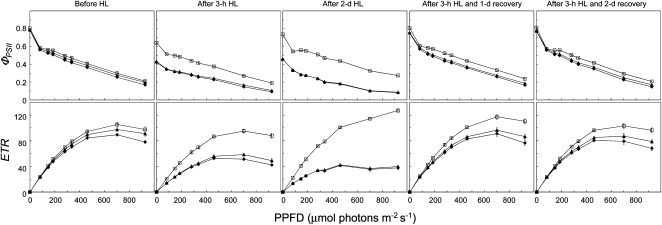
Light–response curves, which assess PSII quantum yield at a range of actinic light, are another standard tool for assessment of photosynthetic parameters. To further characterize the photosynthetic apparatus in lqy1 mutants, light–response curves of PSII quantum yield (ΦPSII) and electron transport rate (ETR) were determined before and after a 3-h high-light treatment. Before high-light treatment, there was a slight reduction in ΦPSII and ETR in lqy1 mutants at most actinic light intensities used in the assay (Figure 2). After a short-term high-light treatment (3 h), ΦPSII in lqy1 mutants is much lower than that in wild type; this is consistent with the earlier finding that Fv/Fm is much lower in lqy1 mutants than in wild type after high-light treatment (Table 1). The reduction in ΦPSII and ETR can be partially reversed by moving the plants back to permissive light condition for 1 to 2 d (Figure 2).
Figure 2.
Light–Response Curves of PSII Quantum Yield and ETR in Wild-Type and lqy1 Mutants.
The measurements were performed at the following light intensities: 0, 81, 145, 186, 281, 335, 461, 701, and 926 μmol photons m−2 s−1. Data for Col wild-type (open squares), lqy1-1 (filled diamonds), and lqy1-2 (filled triangles) plants are presented as mean ± se (n = 8). HL, high light. Note that lqy1-1 and 1-2 response curves are superimposed in the after 2-d HL ΦPSII and PSII ETR graphs due to their similar phenotypes.
In addition to a short-term high-light treatment and recovery under permissive light conditions, we also tested the effect of extended high-light treatment on lqy1 mutants (2 d). After an extended high-light treatment, the difference between wild-type and lqy1 mutants became more obvious than after the 3-h treatment (Figure 2). The 2-d high-light treatment led to small but statistically significant reductions of ΦPSII and ETR in lqy1 mutants at most actinic light intensities tested (145, 186, 281, 335, 461, and 701 μmol photons m−2 s−1, P = 0.05). By contrast, extended high-light treatment had the opposite effects on ΦPSII and ETR in wild-type plants (Figure 2). Compared with the 3-h high-light treatment, the 2-d high-light treatment led to a small but statistically significant increase of ΦPSII and ETR in wild-type plants at all actinic light intensities tested (81, 145, 186, 281, 335, 461, 701, and 926 μmol photons m−2 s−1, P = 0.05). Taken together, detailed chlorophyll fluorescence analyses showed that PSII in lqy1 mutants is more susceptible to high-light stress.
lqy1 Mutants Accumulate More Reactive Oxygen Species Than Wild Type after High-Light Treatment
Taken together, the chlorophyll fluorescence data show that lqy1 mutant plants are more sensitive to high light. Because high-light intensity can cause oxidative damage to plants (Demmig-Adams and Adams, 1992; Triantaphylidès and Havaux, 2009), we stained for superoxide (O2−), hydrogen peroxide (H2O2), and singlet oxygen (1O2) following a 2-d high-light treatment. Nitro blue tetrazolium (NBT) staining for O2− shows that that lqy1 mutants produce more of the formazan product than wild type after high-light treatment, consistent with increased accumulation of O2− (Figures 3A to 3F). In addition, high-light–treated lqy1 mutant plants also stain more intensely with diaminobenzidine (DAB), consistent with the hypothesis that the plants accumulate more H2O2 in their leaves (Figures 3G to 3L). In high-light–treated lqy1 mutants, NBT staining is stronger in younger leaves, whereas DAB staining is more pronounced in older leaves. The detection of 1O2 was performed on detached leaves using the Invitrogen singlet oxygen sensor green (SOSG) fluorescence indicator (Figures 3M to 3R). The stronger fluorescence signal in high-light–treated lqy1 mutants than in wild type is consistent with the hypothesis that the mutants accumulate more 1O2 in their leaves than the corresponding wild type (Figures 3P to 3R). In summary, the results from O2−H2O2, and 1O2 detection are consistent with the hypothesis that lqy1 mutants accumulate more reactive oxygen species (ROS) than wild type following high-light treatment.
Figure 3.
Analysis of ROS in Wild-Type and lqy1 Mutants.
(A) to (C) NBT staining of O2− in Col wild-type (A), lqy1-1 (B), and lqy1-2 (C) plants under growth light.
(D) to (F) NBT staining of O2− in Col wild-type (D), lqy1-1 (E), and lqy1-2 (F) plants after growing under high light for 2 d.
(G) to (I) DAB staining of H2O2 in Col wild-type (G), lqy1-1 (H), and lqy1-2 (I) plants under growth light.
(J) to (L) DAB staining of H2O2 in Col wild-type (G), lqy1-1 (H), and lqy1-2 (I) plants after growing under high light for 2 d.
(M) to (O) Detection of 1O2 with SOSG in Col wild-type (M), lqy1-1 (N), and lqy1-2 (O) plants under growth light.
(P) to (R) Detection of 1O2 with SOSG in Col wild-type (P), lqy1-1 (Q), and lqy1-2 (R) plants after growing under high light for 2 d.
High-Light Effects on Transcript and Protein Levels for LQY1 and PSII Core Proteins
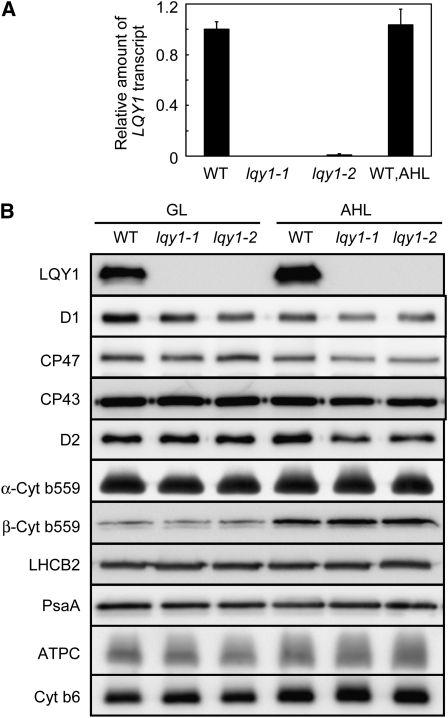
The analysis of the lqy1 mutants described above shows that the mutants have reduced photosynthetic efficiency, especially after high-light treatment. This suggests that the wild-type LQY1 gene product may have a photoprotective role under high-light stress. To investigate the influence of light on mRNA and protein expression, the abundance of LQY1 transcript was measured using quantitative RT-PCR. The relative abundance of LQY1 transcript in wild-type leaves treated in high light (1500–1700 μmol photons m−2 s−1) is not significantly different from wild-type plants grown in low light (100 μmol photons m−2 s−1) (Figure 4A). LQY1 transcript was undetectable in the lqy1 mutants, confirming that they are null alleles. Because LQY1 protein is in the thylakoid membrane (Kleffmann et al., 2004; Peltier et al., 2004; Zybailov et al., 2008), total thylakoid membrane proteins were analyzed for LQY1 protein and PSII core subunits by immunoblotting. When normalized to chlorophyll, the amount of LQY1 protein in wild-type leaves increased ~45% (mean value from five independent biological replicates) after a 2-d high-light treatment (Figure 4B; see Supplemental Figure 1 online). This might be an overestimation of LQY1 protein increase because of the ~30% reduction of total chlorophyll content in wild-type leaves after a 2-d high-light treatment (see Supplemental Table 1 online). Even accounting for this possibility, the abundance of LQY1 protein relative to PSII core proteins is higher after a 2-d high-light treatment, consistent with LQY1 protein having a role in photoprotection. The lack of detectable cross-reacting material in thylakoid membranes from lqy1-1 and 1-2 mutants indicates that the antibody is specific for the target protein (Figure 4B; see Supplemental Figure 1 online). After a 2-d high-light treatment, the relative abundances of PSII reaction center proteins D1, PsbB, PsbC, and D2 are slightly lower in lqy1 mutant plants than in wild type (also calculated on an equal chlorophyll basis; Figure 4B; see Supplemental Figure 1 online). This reduction was not observed in the relative abundance of LHCII chlorophyll a/b binding protein LHCB2, PSI reaction center protein PsaA, ATP synthase subunit C, or cytochrome b6 (Figure 4B; see Supplemental Figure 1 online).
Figure 4.
Analysis of LQY1 Transcript and Thylakoid Membrane Protein Accumulation.
(A) Relative amount of the LQY1 transcript determined by quantitative RT-PCR. The amount of LQY1 transcript was normalized by that of the ACT2 transcript (At3g18780). Values (mean ± se, n = 3) are given as the ratio to the amount of LQY1 transcript in wild type (WT) under growth light. For LQY1 transcript levels after a 2-d high-light treatment, only the wild-type value is shown because the transcript was not detectable in either lqy1 mutant. AHL, after 2 d of high light.
(B) Representative immunoblots of LQY1 and PSII proteins in wild-type and lqy1 mutants. Thylakoid membrane proteins were separated by SDS-urea-PAGE, electroblotted to a PVDF membrane, and probed with affinity-purified anti-LQY1 or antisera against known thylakoid membrane proteins obtained from Eva-Mari Aro or Agrisera Co. The lanes on each gel were loaded on an equal chlorophyll basis. Cyt, cytochrome; GL, growth light.
Effects of LQY1 Reduction on PSII Complexes
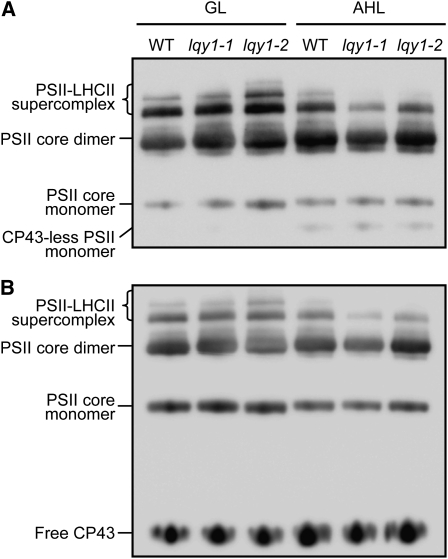
To further investigate the effects of the lack of functional LQY1 protein on PSII structure and function, we analyzed the relative proportions of various PSII complexes in wild type and lqy1 mutants. Thylakoid membranes were solubilized in dodecyl β-d-maltoside, membrane protein complexes were separated by blue native (BN)-PAGE, and the complexes were analyzed by immunoblotting with antibodies against PSII core proteins. PSII goes through rapid damage, repair, and reassembly cycles under high light (Demmig-Adams and Adams, 1992; Yokthongwattana et al., 2001; Baena-González and Aro, 2002), and the CP43-less PSII core monomer is a marker for ongoing PSII repair cycles (Lundin et al., 2008). The immunoblot using anti-D1 antiserum shows that after a 2-d high-light treatment, both wild-type and lqy1 mutants accumulate the CP43-less PSII core monomer (Figure 5A), indicating that PSII repair is occurring under these conditions. More importantly, results with anti-D1 and anti-CP43 antisera show that lqy1 mutants have less PSII-LHCII supercomplex than corresponding wild-type thylakoid membranes (Figure 5; see Supplemental Figure 2 online).
Figure 5.
Immunological Analysis of PSII Complexes Separated by BN-PAGE.
(A) A representative immunoblot with anti-D1 antiserum of a BN-PAGE gel. Thylakoid membrane proteins were solubilized with 2% dodecyl β-d-maltoside and separated by BN-PAGE. A sample with an equal amount of chlorophyll (1.5 μg) was loaded in each lane. GL, growth light; AHL, after 2 d of high light; WT, wild type.
(B) A representative immunoblot with anti-CP43 antiserum of a BN-PAGE gel as in (A).
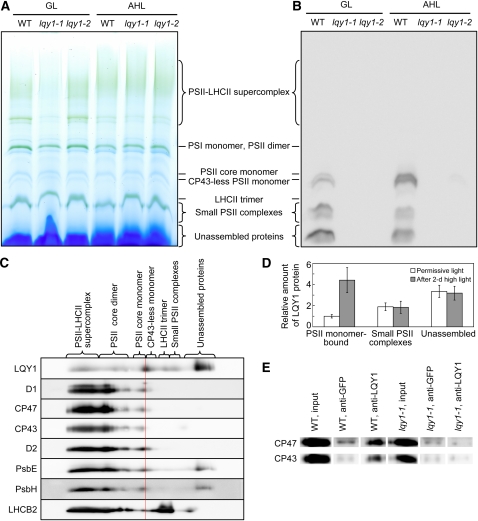
Interaction between LQY1 and PSII Complexes
The observed reduction in the abundance of PSII-LHCII supercomplex in high-light–treated lqy1 mutants suggests a role for LQY1 in repair and reassembly of photodamaged PSII complexes. To test this hypothesis, we asked whether LQY1 protein is associated with PSII complexes. Although LQY1 did not comigrate with PSII complexes solubilized with 2% dodecyl β-d-maltoside (see Supplemental Figure 3 online; LQY1 protein is found in the region of the gel associated with unassembled protein), positive results were obtained following treatment with the milder detergent 1.5% digitonin and BN-PAGE (Figure 6). Interestingly, a substantial proportion of LQY1 protein binds to the PSII core monomer and reaction center, repair-associated CP43-less PSII monomer, and this fraction of LQY1 protein increases threefold after high-light treatment (Figures 6B and 6D).
Figure 6.
Evidence for Interaction of LQY1 Protein with PSII Protein Complexes.
(A) A representative unstained BN-PAGE gel. Thylakoid membranes were solubilized with 1.5% digitonin and separated on a NativePAGE 3 to 12% Bis-Tris mini gel. A sample with an equal amount of chlorophyll (7 μg) was loaded in each lane. GL, growth light; AHL, after 2 d of high light; WT, wild type.
(B) A representative immunoblot with anti-LQY1 antiserum of a gel as in (A). The positions of various PSII protein complexes were determined by immunoblotting with antisera against PSII core proteins D1 and CP43. Little signal is observed in lqy1 mutants, validating the specificity of the antibody.
(C) 2-D electrophoresis and immunoblots of PSII protein complexes from Col wild-type leaves after a 2-d high-light treatment. Protein complexes were separated as in (A) by BN-PAGE (first dimension), and proteins in the complexes were separated by SDS-PAGE (second dimension). The 2-D gel was electroblotted and probed with antisera against LQY1 and various PSII proteins. The positions of detected PSII protein complexes are indicated. The red line separates the CP43-less monomer from the PSII core monomer.
(D) Relative abundance of LQY1 protein in Col wild-type plants as detected in (B). The values (mean ± se, n = 3) are given as the ratio to PSII monomer-bound LQY1 in Col wild-type plants under growth light. Note that in Figure 6B, the relative intensity of LQY1 in small PSII complexes from wild-type plants decreased after a 2-d high-light treatment. However, this is not seen in Figure 6D, which represents the mean values from three independent BN-PAGE/immunoblots.
(E) Immunoprecipitation of CP47 and CP43 by anti-LQY1 antibody. Thylakoid membranes were extracted from Col wild-type and lqy1-1 leaves after a 2-d high-light treatment.
To verify the association of LQY1 with PSII protein complexes, the lane from high-light–treated wild-type leaf extract was excised from a BN-PAGE gel. The proteins of the complexes were separated by SDS-PAGE in the second dimension and were immunoblotted with antibodies or antisera against LQY1 and PSII core proteins (Figure 6C). These two-dimensional (2-D) gel results confirmed that LQY1 protein binds to the PSII core monomer and CP43-less core monomer. Based on these results, we cannot rule out the possibility that LQY1 interacts with larger PSII complexes. Coimmunoprecipitation of CP47 and CP43 by anti-LQY1 antibody further supports that there is interaction between LQY1 and PSII core subunits (Figure 6E).
LQY1 Homologs Are Found in Other Land Plants
To ask whether LQY1 is evolutionarily conserved, as might be expected for a protein involved in maintenance of PSII function, we looked for LQY1 homologs in other photosynthetic species. As shown in Supplemental Figure 4A online, a BLASTP search with LQY1, a full-length protein sequence, revealed the presence of homologs across land plants (e.g., Physcomitrella patens, Selaginella moellendorffii, sitka spruce [Picea stichensis], rice [Oryza sativa], sorghum [Sorghum bicolor], tobacco [Nicotiana tabacum], tomato [Solanum lycopersicum], potato [Solanum tubersosum], soybean [Glycine max], grape vine [Vitis vinifera], castor bean [Ricinus communis], and poplar [Populus trichocarpa]). However, homologs were not found in cyanobacteria or algae. Nearly all of the LQY1 homologs appear to have all three motifs predicted from the Arabidopsis sequence: transit peptide, single-pass transmembrane domain, and a C-terminal region homologous to the Zn finger in the molecular chaperone protein DnaJ (Figures 7A to 7D; see Supplemental Figure 4A online). The presence of a LQY1 homolog in tobacco chloroplasts was demonstrated by immunodetection in thylakoid membranes isolated from wild-type tobacco leaves (see Supplemental Figure 4B online).
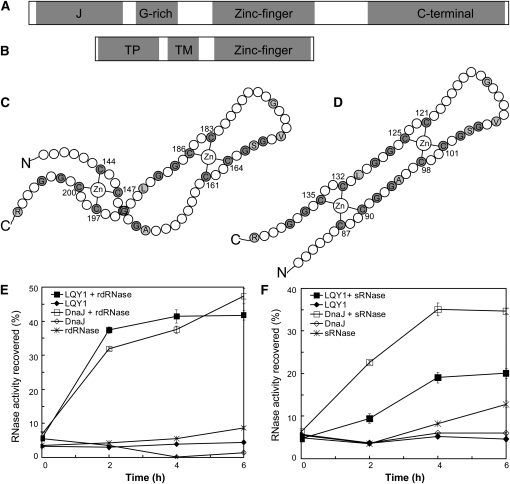
Figure 7.
The LQY1 Zn Finger Domain Has Protein Disulfide Isomerase Activity.
(A) Domains in E. coli DnaJ. The protein contains a J-domain, a Gly-rich motif, a Zn finger domain, and a C-terminal region.
(B) Domains inferred from the Arabidopsis LQY1 sequence. The protein contains a transit peptide (TP), a transmembrane region (TM), and a Zn finger domain according to in silico analysis.
(C) The topology of the E. coli DnaJ protein (Shi et al., 2005). Cys and Gly residues are shown in gray. Other residues conserved between LQY1 and DnaJ Zn finger are shown in single-letter amino acid codes.
(D) Putative topology of Arabidopsis LQY1 protein based on its homology to E. coli DnaJ.
(E) Assays for oxidative renaturation of rdRNase by recombinant LQY1 Zn finger. Restoration of activity to inactive reduced and denatured RNase A was measured for LQY1 and positive-control E. coli DnaJ. The values (mean ± se, n = 4) are normalized by the activity of native RNase A. Filled squares, LQY1 and rdRNase; filled diamonds, LQY1, no rdRNase; open squares, DnaJ and rdRNase; open diamonds, DnaJ, no rdRNase; asterisks, rdRNase, no LQY1 or DnaJ.
(F) Reductive renaturation of sRNase by recombinant LQY1 Zn finger. The values (mean ± se, n = 4) are normalized as in (E). Filled squares, LQY1 and sRNase; filled diamonds, LQY1, no sRNase; open squares, DnaJ and sRNase; open diamonds, DnaJ, no sRNase; asterisks, sRNase, no LQY1 or DnaJ.
Enzymatic Activity of LQY1 Protein
Although LQY1 has sequence similarity to DnaJ proteins in the conserved Zn finger, it does not have the other domains found in a typical DnaJ protein: the J-domain, Gly-rich region, or C-terminal motif (Figures 7A and 7B; see Supplemental Figure 4A online). To test the hypothesis that this putative Zn finger binds Zn2+, inductively coupled plasma-mass spectrometry was used to analyze metals in the protein. Recombinant LQY1 preparations obtained from E. coli cells grown in complete Luria-Bertani medium without Zn supplementation contained 1.8 Zn equivalents per protein subunit. This is close to the expected Zn:protein ratio of two based on similarity between the Zn finger domains in LQY1 and DnaJ (Figures 7C and 7D).
Full-length DnaJ is capable of reactivating reduced and denatured RNase A (rdRNase A), which contains eight sulfhydryl groups. This reactivation is mediated by the Zn finger domain, and it involves oxidation of thiol groups and subsequent reformation of disulfides to create a native and enzymatically active conformation (Shimada et al., 2007). To test the hypothesis that the LQY1 Zn finger has similar activity, we measured oxidative renaturation of rdRNase A and reductive renaturation of scrambled RNase (sRNase) A. To improve protein solubility, the transmembrane domain in LQY1 was removed along with the transit peptide to produce LQY172-154. After a 2-h incubation, ~40% of maximal RNase activity was recovered by incubation of rdRNase A with recombinant LQY1 Zn finger (Figure 7E). This oxidase activity of LQY1 Zn finger was comparable to that of E. coli DnaJ (Figure 7E). Reactivation of sRNase A involves the reduction of mispaired disulfides and subsequent rearrangement to an active conformation (Pigiet and Schuster, 1986; Hasegawa et al., 2003). The LQY1 Zn finger has measurable reductase activity with this substrate, although it is lower than that of the control E. coli DnaJ (Figure 7F). Taken together, the measurements of rdRNase and sRNase renaturation by LQY1 Zn finger show that this protein has a protein disulfide isomerase activity analogous to the DnaJ Zn finger. This is consistent with the hypothesis that LQY1 is involved in the repair and reassembly of photodamaged PSII. In fact, the PSII core proteins D1, D2, CP47, and CP43 each have two or more Cys residues (Shimada et al., 2007) and are possible substrates for LQY1.
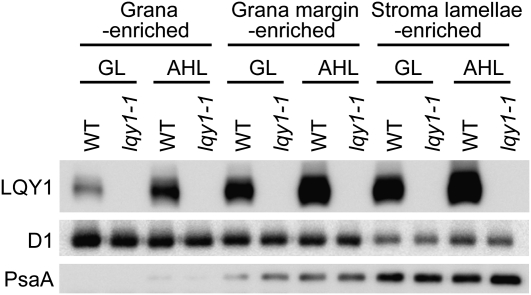
LQY1 Protein Is More Abundant in Stroma-Exposed Thylakoids
A number of key steps of PSII repair and reassembly take place in stroma-exposed thylakoids (Mulo et al., 2008) . If LQY1 protein participates in repair and assembly, we hypothesized that the majority of LQY1 would be localized in stroma-exposed thylakoids. To further analyze the role of LQY1 in PSII repair and reassembly, thylakoid grana core, grana margin, and stroma lamellae were subfractionated by differential ultracentrifugation. As shown in Figure 8, the relatively lower amount of PSII core protein D1 in stroma lamellae and the lack of detectable PSI core protein PsaA in grana core indicate that the subfractionation method was successful (Figure 8). The distribution of representative proteins of the PSII and PSI complexes does not change due to the absence of the functional LQY1 protein (lqy1-1), under either light condition. Interestingly, the content of LQY1 in grana core-, grana margin-, and stroma lamellae-enriched wild-type thylakoid fractions increases substantially after a 2-d high-light treatment (Figure 8), consistent with the relatively higher level of total LQY1 upon high-light treatment (Figure 4; see Supplemental Figure 1 online). In wild-type plants growing at the same light intensity, LQY1 protein is more abundant in grana margin than in grana core and even more abundant in stroma lamellae (Figure 8). This is consistent with the hypothesis that LQY1 plays a role in the dynamic repair and reassembly of photodamaged PSII complexes.
Figure 8.
Analysis of LQY1 Protein in Grana-, Grana Margin-, and Stroma Lamellae-Enriched Thylakoid Membranes.
Thylakoid membranes were solubilized by 0.4% digitonin and fractionated by ultracentrifugation. Fractionated thylakoid membrane proteins were separated by SDS-Urea-PAGE and analyzed by immunoblots. The lanes on each gel were loaded on an equal chlorophyll basis. GL, growth light; AHL, after 2-d high light; WT, wild type.
Absence of Functional LQY1 Accelerates Turnover of the D1 Protein
Pulse-chase labeling experiments were used to investigate the impact of LQY1 deficiency on D1 protein synthesis and degradation. Detached leaves were incubated with [35S]methionine in the dark for 16 h, pulse-labeled under 500 μmol photons m−2 s−1 light for 20 to 60 min, and chased in the light for 1 to 3 h in the presence of unlabeled Met (see Supplemental Figure 5 online). The D1 turnover rate is ~30% higher in lqy1 mutants than in wild type (Table 2), and the half-time for D1 turnover is ~30% lower in lqy1 mutants than in wild type (Table 2). Interestingly, the relative rate of D1 synthesis is also faster in lqy1 mutants than in wild type (Table 2). Taken together, the pulse-chase experiments indicate that the absence of functional LQY1 protein results in accelerated turnover and synthesis of D1 protein (Table 2).
Table 2.
Comparison of the Rate Constant and Half-Time for D1 Turnover and the Relative Rate for D1 Synthesis in Wild-Type and lqy1 Mutants
| Degradation |
Synthesis | ||
| Genotype | kPI x 103, min−1 | t1/2, min | Relative Synthesis Rate |
| WT | 5.87 ± 0.58 | 120 ± 11 | 1.00 ± 0.00 |
| lqy1-1 | 7.95 ± 1.36 | 94 ± 16 | 1.37 ± 0.10 |
| lqy1-2 | 7.84 ± 0.76 | 90 ± 9 | 1.29 ± 0.05 |
The degradation rate constant (kPI) and degradation half-time (t1/2) for D1 degradation were calculated from data fitted to first order reaction kinetics (A = A0e−kt) according to Sirpiö et al. (2007). Relative rate of D1 synthesis was calculated during a 1-h pulse under high light. Data are presented as means ± se (n = 3). WT, wild type.
DISCUSSION
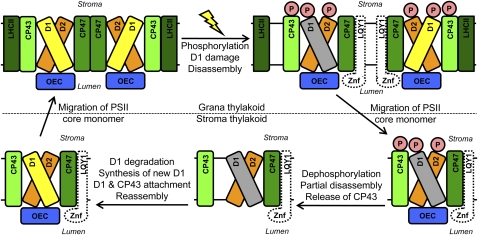
Under high irradiance, proteins in the PSII reaction center are damaged and undergo rapid repair and reassembly to enable photosynthetic electron transport to continue (Demmig-Adams and Adams, 1992; Yokthongwattana et al., 2001; Baena-González and Aro, 2002). As schematically illustrated in Figure 9, this is a complex multistep process involving: (1) high-light induced phosphorylation and damage to PSII-LHCII supercomplexes and PSII core dimers in grana stacks, (2) disassembly of photodamaged PSII-LHCII supercomplexes and PSII core dimers into PSII core monomers, (3) migration of PSII core monomer to stroma-exposed thylakoid membranes, (4) partial disassembly of the PSII core monomer and the release of CP43, (5) degradation of photodamaged D1 and synthesis of new D1, (6) reassembly of D1 and CP43 to form the PSII core monomer, (7) migration of PSII core monomers back to the grana stacks, and (8) dimerization into PSII core dimers and reformation of PSII-LHCII supercomplexes (Aro et al., 2005; Mulo et al., 2008).
Figure 9.
PSII Repair and Reassembly and Hypothetical Role of LQY1.
This schematic diagram for high-light induced PSII repair and reassembly is modified from Mulo et al. (2008). Undamaged D1 protein is yellow-filled and damaged D1 protein is gray-filled. The thylakoid membrane-localized LQY1 protein, schematically drawn as an unfilled and dashed shape, comigrates with PSII core monomer and CP43-less core monomer on BN-PAGE gels. The Zn finger domain (Znf) in LQY1 is predicted to locate at the lumen side of thylakoid membrane. The presence of LQY1 in these complexes, its mutant phenotype, and protein disulfide isomerase activity are consistent with a role in PSII photodamage repair. The specific proteins with which it interacts and the substrates for its disulfide isomerase activity remain to be determined. OEC, oxygen evolving complex.
Several lines of evidence presented in this study indicate that the lqy1 mutants are compromised in the maintenance of PSII activity during light stress. Under high-light treatment, mutants lacking functional LQY1 protein are defective in PSII photosynthetic electron transport (Table 1, Figures 1, 2, and 5). After prolonged high-light treatment, these mutants accumulate more ROS than wild type (Figure 3). High irradiance treatment leads to a consistent increase in immunologically detectable LQY1 protein in wild-type plants (Figure 4; see Supplemental Figure 1 online) and a decrease of PSII-LHCII supercomplex in lqy1 mutants (Figure 5; see Supplemental Figure 2 online). Taken together, the data indicate that LQY1 protein is important in maintaining normal levels of PSII-LHCII supercomplex under light stress. The data also lead us to hypothesize that LQY1 is involved in the repair and reassembly of photodamaged PSII (Figure 9). Interestingly, the difference between lqy1 mutants and Columbia (Col) wild type appears to be less obvious on older plants or leaves (Table 1; see Supplemental Tables 2 and 3 online). The effect of age on photosynthetic phenotype is also seen in our NPQ data in that the 3-week-old plants tend to have unusually high NPQ (Table 1; see Supplemental Tables 2 and 3 online).
The hypothesis that LQY1 plays a role in PSII repair and reassembly is supported by multiple lines of evidence; for example, the interaction of LQY1 with PSII complexes and the protein disulfide isomerase activity of LQY1 Zn finger. LQY1 was found to comigrate with the PSII core monomer and CP43-less PSII monomer (a marker for ongoing PSII repair cycle) on BN-PAGE gels, and more LQY1 was found to associate with PSII monomer forms under high light (Figures 6B and 6D). The interaction between LQY1 and PSII monomer forms was further demonstrated by 2-D PAGE immunoblotting and immunoprecipitation of CP47 and CP43 with anti-LQY1 antibody (Figures 6C and 6E). Disassembly and reassembly of PSII involves breakage and reformation of disulfide bonds between Cys residues (Shimada et al., 2007), and recombinant LQY1 Zn finger domain demonstrated protein disulfide isomerase activity in vitro (Figure 7). Based on LQY1 topology predictions by TMHMM (Krogh et al., 2001), the C-terminal Zn finger is hypothesized to be located to the lumen side of thylakoid membrane, where the PSII oxygen evolving complex is located (Figure 9). PSII core subunits D1, D2, CP47, and CP43 and oxygen-evolving complex proteins PsbO and PsbP contain one or more Cys residues (Shimada et al., 2007). These PSII proteins are possible targets for LQY1. Therefore, it is plausible that the protein disulfide isomerase activity of LQY1 is involved in the repair and reassembly of PSII.
Additional lines of evidence supporting the role of LQY1 in PSII repair and reassembly include LQY1 protein enrichment in stroma lamellae and increased D1 turnover and synthesis in the lqy1 mutant. It is well established that key steps of PSII repair and reassembly (for example, partial disassembly of PSII core monomer into the CP43-less monomer, degradation of photodamaged D1, assembly of newly synthesized D1, and reformation of PSII core monomer) take place in stroma-exposed thylakoids (Mulo et al., 2008). Our thylakoid subfractionation and subsequent immunoblotting analysis show that LQY1 is more abundant in stroma lamellae than in grana core (Figure 8). The pulse-chase experiments show that the degradation and synthesis of D1 protein are accelerated in the absence of functional LQY1 protein (Table 2; see Supplemental Figure 5 online). These four lines of evidence strongly support the hypothesis that LQY1 is involved in the repair and reassembly of photodamaged PSII.
It is interesting to consider which step(s) of PSII repair and reassembly might involve LQY1. The distribution of D1 protein among grana core, grana margin, and stroma lamellae is not altered in the lqy1 mutant (Figure 8). This suggests that the migration of the PSII core monomer between grana core- and stroma-exposed thylakoids is not affected in lqy1 mutants (see Figure 9 for PSII repair cycle). Based on the relatively faster D1 turnover and synthesis in lqy1 mutant plants, it is likely that LQY1 plays a role in PSII repair and reassembly by influencing D1 turnover and synthesis. Despite the acceleration of D1 turnover and synthesis, the overall amounts of D1 protein and PSII supercomplex are reduced in lqy1 mutants (Figures 4 and 5; see Supplemental Figures 1 and 2 online), indicating that the effectiveness of PSII repair might be lower in lqy1 mutants. It is not uncommon for plants lacking auxiliary PSII repair and reassembly proteins to have higher rates of D1 turnover. For example, the absence of PSII peripheral protein PsbI accelerates D1 degradation (Dobáková et al., 2007). Dobáková et al. (2007) postulated that newly synthesized and replaced D1 in the PsbI deletion mutant is unstable and undergoes further rounds of degradation and reinsertion.
Despite evidence supporting the role of LQY1 in efficient PSII repair and reassembly of photodamaged PSII proteins, the protein appears not to be absolutely essential for PSII core repair. One line of evidence is that lqy1 mutants recover from short-term high-light treatment after a shift back to 100 μmol photons m−2 s−1 light conditions for 1 to 2 d (Figure 2). The labile interaction of LQY1 with PSII complexes is another noteworthy feature of this protein. In contrast to the situation with 1.5% digitonin, when thylakoid membrane was solubilized in 2% dodecyl β-d-maltoside, LQY1 was found in the region containing unassembled proteins on a BN-PAGE gel (see Supplemental Figure 3 online). The fact that LQY1 accumulation does not depend on strong interaction with PSII is suggested by studies in tobacco: LQY1 protein accumulates in the tobacco ΔpsbE cytochrome b559 α subunit mutant (Swiatek et al., 2003; see Supplemental Figure 4B online). These plants, which are compromised in PSII photosynthetic activity, have reduced levels of core proteins D1, D2, CP43, and CP47 (Swiatek et al., 2003; Suorsa et al., 2004). The presence of the LQY1 protein in PSII-deficient tobacco plants indicates that the LQY1 accumulation does not depend on functional PSII.
Functional redundancy could contribute to the relatively mild and reversible phenotype of lqy1 mutants. LQY1 has similarities to proteins involved in PSII repair and reassembly in addition to PsbI (Lima et al., 2006; Fu et al., 2007; Ma et al., 2007; Dominguez-Solis et al., 2008; Mulo et al., 2008). For example, the cyclophilin proteins CYP20 and CYP38 are reported to play important roles in assembly and stabilization of PSII (Rokka et al., 2000; Fu et al., 2007; Dominguez-Solis et al., 2008). Similar to the lqy1 mutants, Arabidopsis plants lacking CYP20 or CYP38 have reduced PSII quantum yield and less PSII-LHCII supercomplexes under high light (Fu et al., 2007; Dominguez-Solis et al., 2008; Sirpiö et al., 2008). Most notably, the CYP38 cyclophilin protein has a peptidyl–prolyl cis-trans isomerase protein folding activity, and D1 and D2 proteins in Arabidopsis plants lacking CYP38 have a shorter half-life under high light (Fulgosi et al., 1998; Fu et al., 2007), similar to the lqy1 mutants.
CONCLUSION
Taken together, our results provide evidence that the LQY1 protein is involved in the repair and reassembly of photodamaged PSII proteins. PSII repair and reassembly involves isomerization of disulfide bonds, and recombinant LQY1 protein has the ability to break and reform disulfide bonds of substrate proteins. The location of the LQY1 protein on stroma-exposed thylakoids and the acceleration of D1 turnover and synthesis in the lqy1 mutants offer additional support for this hypothesis. The absence of functional LQY1 protein causes reduced amounts of PSII supercomplexes, increased sensitivity to high light, and increased ROS accumulation. Although we cannot rule out the possibility that LQY1 is directly involved in ROS detoxification, the comigration of LQY1 with CP43-less PSII monomer is consistent with the role of LQY1 in PSII repair and reassembly. Further studies of LQY1 and other thylakoid accessory proteins should lead to a more detailed understanding of the dynamics of PSII core protein repair and assembly (Yokthongwattana et al., 2001; Mulo et al., 2008; Chen et al., 2010).
METHODS
Plant Materials and Growth Conditions
T-DNA and transgenic Arabidopsis thaliana lines used in this study were in the Col background (Alonso et al., 2003; Woody et al., 2007). The lqy1-1, lqy1-2 mutants were obtained from the Arabidopsis Biological Resource Center (stock nos. SALK_021824C and CS853351) and were backcrossed twice to Col, and the backcrossed lines were redeposited at the Arabidopsis Biological Resource Center as CS66008 (lqy1-1) and CS66009 (lqy1-2).
Seeds were sown on soil, stratified in the dark at 4°C for 3 d, and grown in a growth chamber on a 12-h/12-h photoperiod. The irradiance was 100 μmol photons m−2 s−1 using a mixture of cool white fluorescent and incandescent bulbs, the temperature was 21°C, and the relative humidity was set to 50%. Plants for chlorophyll fluorescence assays and protein analysis were 3, 4 and 5 weeks old, respectively.
To study the effect of high light, plants were placed in a high-light growth chamber (1500–1700 μmol photons m−2 s−1) for 3 h or 2 d while maintaining the 12-h-light/12-h-dark cycle.
Chlorophyll Measurement and Chlorophyll Fluorescence
Chlorophyll was extracted with 80% acetone in 2.5 mM Hepes-KOH, pH 7.5), and the amount of chlorophyll was determined as described by Wellburn (1994). Chlorophyll fluorescence parameters were measured on dark-adapted plants with the MAXI version of the IMAGING-PAM M-Series chlorophyll fluorescence system (Heinz-Walz Instruments), as described by Lu et al. (2008). The plants were dark-adapted for 20 min before making measurements.
For NPQ measurements, after initial determination of Fo and Fm and a 40-s delay, an actinic light treatment (531 μmol photons m−2 s−1) was performed for 715 s. During actinic illumination, 36 saturation pulses (2800 μmol photons m−2 s−1) were applied at 20-s intervals. After termination of actinic light, recovery of Fm′ was monitored for 14 min as described by Müller et al. (2001). During this time period, 16 saturation pulses were applied, with the interval increasing exponentially.
For measurements of light–response curves, plants were illuminated at the following light intensities: 0, 81, 145, 186, 281, 335, 461, 701, and 926 μmol photons m−2 s−1. The duration of illumination at individual light intensities was 3 min. A saturation pulse was applied at the end of the 3-min illumination.
In Situ Detection of ROS
In situ detection of O2− was performed by treating plants with NBT as previously described by Kawai-Yamada et al. (2004). Aerial portions of the plants were detached from the roots, vacuum-infiltrated with 10 mM NaN3 in 10 mM potassium phosphate buffer, pH 7.8, for 2 min, and incubated in 0.1% NBT (in 10 mM potassium phosphate buffer, pH 7.8) for 2 h at room temperature. Stained leaves were boiled in acetic acid:glycerol:ethanol (1:1:3 [v/v/v]) for 5 min and photographed.
In situ detection of H2O2 was performed by treating plants with DAB-HCl as previously described by Thordal-Christensen et al. (1997). Aerial portions of the plants were detached from the roots, vacuum-infiltrated with 5 mM DAB-HCl, pH 3.8, for 3 min, and incubated in the same solution under growth or high light for 3 h. Stained leaves were boiled in an acetic acid:glycerol:ethanol solution and photographed (see above).
In vivo detection of 1O2 was performed by treating detached leaves with SOSG (Invitrogen) as previously described by Flors et al. (2006). The petioles of freshly detached leaves were submerged in a solution containing 260 mM SOSG and 50 mM phosphate buffer, pH 7.5, in the dark for 2.5 h and then under growth light (100 μmol photons m−2 s−1) or high light (1500–1700 μmol photons m−2 s−1) for 3 h. After a brief rinse in distilled water, the fluorescence from SOSG in the leaves was imaged with a luminescent image analyzer LAS3000 (Fuji Film).
Quantitative RT-PCR
Total RNA was extracted from Arabidopsis rosette leaves using the RNeasy plant mini kit (QIAGEN), digested with RQ1 RNase-free DNase (Promega), and reverse-transcribed with random primers and Moloney murine leukemia virus RT (Promega). Quantitative PCR was performed on a 7500 Real-Time PCR system with Power SYBR Green PCR master mix (Applied Biosystems; Ajjawi et al., 2010). Primers LQY1_FL and LQY1_FR (see Supplemental Table 4 online) were used to quantify the steady-state LQY1 transcript level. Primers ACT2_L and ACT2_R (see Supplemental Table 4 online) were used to quantify the control ACT2 (At3g18780) transcript level.
Isolation of Thylakoid Membranes
Thylakoid membranes were isolated as described in Suorsa et al. (2006) with minor modifications. Mature leaves were excised and blended with an Omni-Mixer homogenizer (Omni) in ice-cold 50 mM HEPES-KOH, pH 7.5, containing 330 mM sorbitol, 2 mM EDTA, 1 mM MgCl2, 5 mM ascorbate, 0.05% bovine serum albumin, 10 mM NaF, and 0.25 mg/mL Pefabloc SC protease inhibitor (Roche). Ascorbic acid and Pefabloc SC were added to the buffer immediately before blending. The resulting lysate was filtered through four layers of cheesecloth and centrifuged for 4 min at 2500g at 4°C using a swing-bucket rotor. The pellet was resuspended and centrifuged in 50 mM HEPES-KOH, pH 7.5, containing 5 mM sorbitol, 10 mM NaF, and 0.25 mg/mL Pefabloc SC. The thylakoid pellet was resuspended and centrifuged twice in 50 mM HEPES-KOH, pH 7.5, containing 100 mM sorbitol, 10 mM MgCl2, 10 mM NaF, and 0.25 mg/mL Pefabloc SC. The final pellet was resuspended in a small volume of the same buffer.
Protein Analysis
Proteins were separated with SDS-PAGE (15% polyacrylamide, 6 M urea; Laemmli, 1970), using a Hoefer SE 250 mini-vertical gel electrophoresis unit or a Hoefer SE 400 sturdier vertical slab gel electrophoresis unit (Hoefer). After electrophoresis, the proteins were electroblotted onto a polyvinylidene difluoride (PVDF) membrane (Millipore) using Trans-Blot electrophoretic transfer cell (Bio-Rad).
BN-PAGE was performed as described by Rokka et al. (2005) with minor modifications. Thylakoid preparations were either solubilized with 1 to 2% (w/v) dodecyl β-d-maltoside (Sigma) on ice for 2 min or with 1.5% digitonin at room temperature for 10 min. Electrophoresis was performed using NativePAGE 3-12% Bis-Tris mini gel and XCell SureLock mini-cell (Invitrogen) according to the manufacturer’s protocols at 4°C. After electrophoresis, the proteins were transferred to a PVDF membrane as described above.
For 2-D electrophoresis, the lanes on the BN-PAGE gel were cut out and incubated in SDS-PAGE denaturing buffer (50 mM Tris, pH 6.8, 10% glycerol, 2% SDS, 0.002% bromophenol blue, and 50 mM dithiothreitol at room temperature for 1 h. Protein subunits of the complexes were separated using SDS-PAGE using a NuPAGE 4-12% Bis-Tris mini gel with 2-D well at 4°C. After electrophoresis, the proteins were visualized by silver staining, or they were transferred to a PVDF membrane as described above.
Immunodetection of proteins on a PVDF membrane was performed using the SuperSignal west pico rabbit immunoglobulin G detecting kit (Thermo Scientific) and analyzed by LAS3000 imaging system (Fuji). For quantification of thylakoid proteins, gels were loaded on an equivalent chlorophyll basis, in amounts allowing the immunodetection to be in the linear range. Unless otherwise indicated, all primary antibodies and antisera were raised in rabbits. Antisera against D1 and D2 proteins were a kind gift from Dr. Eva-Mari Aro (University of Turku, Finland). Anti-phosphothreonine antibody was purchased from Invitrogen. Antisera against other photosynthesis proteins were purchased from Agrisera.
Production of Anti-LQY1 Polyclonal Antibodies
Affinity-purified anti-LQY1 polyclonal antibodies were made by Open Biosystems. An 18-amino acid peptide (corresponding to amino acids 137–154 of LQY1) with an additional N terminal Cys residue, CGSGVQPRYLDRREFKDDD, was synthesized, conjugated with keyhole limpet hemocyanin, and used to raise antibody against LQY1 protein.
Detection of LQY1 Homolog in psbE Tobacco Plants
To analyze LQY1 homologs in plants lacking PSII activity, wild-type tobacco plants and ΔpsbE tobacco plants (kindly provided by Eva-Marie Aro, University of Turku; psbE encodes the α subunit of cytochrome b559 of PSII) were grown in Murashige-Skoog medium supplemented with 3% (w/v) Suc under low light (10 μmol photons m−2 s−1; Swiatek et al., 2003; Suorsa et al., 2004). Mature green leaves were used for thylakoid membrane extraction and immune detection of LQY1 homolog, avoiding senescent leaves.
Expression and Purification of Recombinant LQY1 and DnaJ proteins in Escherichia coli
Total RNA was extracted, digested with DNase, and reverse-transcribed with oligo(dT)15 primers as described above. Full-length LQY1 cDNA (LQY11-154), LQY1 cDNA lacking a transit peptide (LQY144-154), LQY1 cDNA lacking the transit peptide, and the transmembrane domain (LQY172-154) was amplified using the mRNA:cDNA hybrid, Pfu DNA polymerase (Promega) with forward primers LQY1_BamH1_ATG, LQY1_BamH1_noTP, LQY1_BamH1_noTM, and a reverse primer LQY1_Xho1_TAA (see Supplemental Table 4 online). The DnaJ gene in E. coli was amplified by PCR using primers DnaJ_BamH1_ATG and DnaJ_Xho1_TAA (see Supplemental Table 4 online). The resulting PCR products were AT-cloned into pGEM-T Easy Vector and sequenced to confirm the absence of PCR errors. Xho1/BamH1-digested LQY1 fragments were subcloned into pET28a expression vector (Novagen) and were expressed in E. coli strain BL21 (DE3) (Stratagene).
An overnight culture of BL21 (DE3) harboring the LQY172-154 or DnaJ gene was diluted 1:20 and grown at 37°C for 1 h as described by Shimada et al. (2007). Expression of recombinant proteins was induced with 1 mM isopropyl β-d-thiogalactoside and cells were grown at 30°C overnight. Recombinant proteins were affinity-purified with Ni-NTA resins under native conditions according to the QIAexpressionist protocol (QIAGEN). Protein concentration was determined using bicinchoninic acid assay.
Inductively Coupled Plasma-Mass Spectrometry
To assay the number of Zn ions per subunit of protein, affinity-purified recombinant LQY1 protein was diluted by 500-fold with 2% HNO3, and 1 mL of diluted sample was used for inductively coupled plasma-mass spectrometry analysis at the Department of Geological Sciences at Michigan State University. For quantification, a procedure blank and a dilution series of Zn standards (0, 4, 10, 20, 40, 50, 100, and 200 precipitate) were used.
Assay of Protein Disulfide Isomerase Activity
Protein disulfide oxidase activity was assayed by the oxidative reactivation of rdRNase (Pigiet and Schuster, 1986; Hasegawa et al., 2003). rdRNase A was prepared from native RNase A (Roche) using 0.15 M dithiothreitol and 6 M guanidine hydrochloride (in 0.1 M Tris-HCl, pH 8.6) according to Pigiet and Schuster (1986). Reactivation of rdRNase was initiated by incubating rdRNase with affinity-purified LQY172-154 at 37°C. The final concentrations of rdRNase and LQY172-154 were 2.9 and 10 mM, respectively. To measure RNase activity, yeast RNA (Sigma-Aldrich) was precipitated with ethanol and sodium acetate at −20°C to remove the unprecipitable fraction; precipitable yeast RNA was dissolved in 50 mM Tris-HCl, pH 7.5, and incubated with an aliquot of reactivated rdRNase at 37°C for 10 min (Hasegawa et al., 2003). The final concentrations of RNase and RNA were 36 nM and 1 mg/mL, respectively. After the reaction, yeast RNA was precipitated with ethanol and sodium acetate at −20°C overnight. The absorption of the supernatant, which contained degraded RNA, was determined at 260 nm.
Protein disulfide reductase activity was assayed by measuring the reductive reactivation of scrambled RNase (Pigiet and Schuster, 1986; Hasegawa et al., 2003). To make scrambled RNase, rdRNase A was sparged with oxygen and then incubated at room temperature in the dark for 4 d (Pigiet and Schuster, 1986). Reactivation of scrambled RNase was initiated by incubating scrambled RNase with affinity-purified LQY172-154 at 37°C as described above but in the presence of 1 mM glutathione and 10 nM glutathione disulfide.
Immunoprecipitation
Immunoprecipitation of PSII core protein CP47 and CP43 by anti-LQY1 antibody was performed as described in Klostermann et al. (2002) with minor modifications. Thylakoid membrane proteins were solubilized in 10 mM Hepes-KOH, pH 8.0, 200 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, and 1.5% digitonin on ice for 45 min. After centrifugation, the supernatant was diluted by an equal volume of the same buffer without digitonin and preincubated with control agarose resin at 4°C for 2 h. Preincubated thylakoid membrane proteins (200 μL, 0.5 mg chlorophyll/mL) were incubated at 4°C overnight with 10 μg of affinity-purified anti-LQY1 antibody or anti-green fluorescent protein antibody cross-linked to 10 μg Protein A/G Plus Agarose in Pierce spin columns. After removing excess fluid by centrifugation, the resin was washed three times with 20 mM HEPES-KOH, pH 8.0, and 80 mM NaCl, and the proteins were eluted with 50 μL of 8 M urea in sample buffer.
In Vivo Pulse Labeling of Thylakoid Proteins
In vivo protein labeling was performed essentially according to Sirpiö et al. (2007). Mature leaves were detached, and each leaf petiole was submerged in 0.2 mL of labeling solution containing 20 μCi/mL [35S]methionine, 0.1% Tween 20, 1 mM KH2PO4, pH 6.3, and 20 μg/mL cycloheximide in the dark for 16 h. The leaves were then pulse labeled under 500 μmol photons m−2 s−1 for 20 and 60 min and briefly rinsed with unlabeled Met solution (10 mM Met in 0.1% Tween 20 and 1 mM KH2PO4, pH 6.3) to remove unincorporated [35S]methionine. Chase experiments with unlabeled Met were performed under 500 μmol photons m−2 s−1 light and samples were collected after 1, 2, and 3 h. The amount of radioactivity incorporated into D1 protein was normalized to that incorporated into ATP synthase α and β subunits (CF1) according to Sirpiö et al. (2007).
Subfractionation of Thylakoid Membranes
Total thylakoid membranes were isolated as described above. Fractionation of grana core-enriched, grana margin-enriched, and stroma lamellae-enriched thylakoid was performed as described by Kyle et al. (1984). Thylakoids were diluted in resuspension buffer containing 15 mM Tricine, pH 7.8, 100 mM sorbitol, 10 mM NaCl, and 5 mM MgCl to a chlorophyll concentration of 1 mg/mL, and an equal volume of 0.8% digitonin in resuspension buffer was added. After incubation at room temperature for 2 min, the reactions were stopped by the addition of 8× volume of ice-cold resuspension buffer. After centrifugation at 1000g at 4°C for 3 min, the supernatant was collected and centrifuged at 10,000g at 4°C for 30 min to pellet grana core-enriched membranes. To collect grana margin-enriched membranes, the supernatant was centrifuged at 40,000g at 4°C for 30 min. To collect stroma lamellae-enriched membranes, the supernatant was centrifuged at 144,000g at 4°C for 60 min.
DNA Sequence Analysis
LQY1 homologs were obtained by BLASTP. The 5′-end protein sequence for the LQY1 homolog in Physcomitrella patens (XP_001781381.1) was incomplete and lacked its transit peptide. Therefore, the partial coding region sequence was used to compare with P. patens sequences at the U.S. Department of Energy Joint Genome Institute (http://genome.jgi-psf.org/cgi-bin/runAlignment?db=Phypa1_1&advanced=1), and scaffold_301 was retrieved. Complete genomic sequence (including 500 bp upstream and downstream of the original partial sequence) was used to predict new gene structures with the FGENESH program (a Hidden Markov Model-based gene prediction program; http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind). The DNA sequence data for LQY1 homologs in tobacco (Nicotiana tabacum), tomato (Solanum lycopersicum), and Solanum tuberosum were obtained from the Solanaceae Genome Browser (http://solanaceae.plantbiology.msu.edu/). The sequence identification numbers are PUT-173a-Nicotiana_tabacum-76569, PUT-171a-Solanum_lycopersicum-29725, and PUT-157a-Solanum_tuberosum-44449, respectively. The consensus expressed sequence tag sequences were used to predict gene structures with the FGENESH program. Multiple sequence alignment was done using the MultAlin software (http://bioinfo.genotoul.fr/multalin/multalin.html; Corpet, 1988).
Prediction of subcellular localization of proteins was performed with the TargetP server (http://www.cbs.dtu.dk/services/TargetP/; Emanuelsson et al., 2000). Prediction of transmembrane domains was performed using the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/; Krogh et al., 2001). The search for the DnaJ-related Zn finger domain was done with Pfam 24.0 (http://pfam.sanger.ac.uk/; Bateman et al., 2004).
Accession Numbers
Amino acid sequence data for LQY1 and its homologs in other species are in the GenBank/EMBL databases under the following accession numbers: Arabidopsis, NP_177698; soybean (Glycine max), ACU13537.1; rice (Oryza sativa) cv japonica, NP_001049532.1; sitka spruce (Picea stichensis), ADE77026.1; poplar (Populus trichocarpa), XP_002300713.1; castor bean (Ricinus communis), EEF52706.1; sorghum (Sorghum bicolor), XP_002447952.1; Selaginella moellendorffii, XP_002978765.1; and grape vine (Vitis vinifera), XP_002276528.1. The original protein sequence for the LQY1 homolog in P. patens (XP_001781381.1) was incomplete and replaced by a new prediction as described above. The accession number for E. coli DnaJ is AAA00009.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Relative Abundance of LQY1 and PSII Proteins.
Supplemental Figure 2. A Representative Immunoblot with Anti-CP43 Antiserum of a BN-PAGE Gel.
Supplemental Figure 3. LQY1 Protein Does Not Copurify with PSII Protein Complexes in 2% Dodecyl β-d-Maltoside.
Supplemental Figure 4. LQY1 Homologs in Other Land Plants.
Supplemental Figure 5. Pulse-Chase Analysis of D1 Turnover and Synthesis.
Supplemental Table 1. Chlorophyll Contents in 4-Week-Old Wild-Type and lqy1 Mutants after 2-d High-Light Treatment.
Supplemental Table 2. Chlorophyll Fluorescence Parameters in 3-Week-Old Wild-Type and lqy1 Mutants.
Supplemental Table 3. Chlorophyll Fluorescence Parameters in 5-Week-Old Wild-Type and lqy1 Mutants.
Supplemental Table 4. Primers Used in This Study.
Acknowledgments
We thank Eva-Mari Aro, Maija Holmström, and Sari Sirpiö (University of Turku) for generously providing anti-sera for D1 and D2 proteins, for sharing ΔpsbE tobacco plants, and for technical advice on pulse-chase experiments. We thank Peter Jahns (Heinrich-Heine-University) and Thomas D. Sharkey and Sean E. Weise (Michigan State University [MSU]) for technical advice on photosynthesis measurements. Thanks to David M. Kramer and Kaori Kohzuma (MSU) for providing results leading to the observation that the lqy1 phenotype is dependent on plant age. We thank Christoph Benning and Rebecca Roston (MSU) for providing facilities for pulse-chase experiments. We are grateful to Krishna K. Niyogi (University of California, Berkeley) for discussion and critical review of the manuscript. We thank Kathleen M. Imre (MSU), who performed the chlorophyll fluorescence screens that led to identification of lqy1-1, Björn Lundin (MSU), who provided technical advice on thylakoid membrane subfractionation, and Linda J. Savage (MSU), who managed the Chloroplast 2010 Project. This work was supported by U.S. National Science Foundation 2010 Project Grant MCB-0519740.
References
- Ajjawi I., Lu Y., Savage L.J., Bell S.M., Last R.L. (2010). Large-scale reverse genetics in Arabidopsis: Case studies from the Chloroplast 2010 Project. Plant Physiol. 152: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aro E.-M., Suorsa M., Rokka A., Allahverdiyeva Y., Paakkarinen V., Saleem A., Battchikova N., Rintamäki E. (2005). Dynamics of photosystem II: A proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56: 347–356 [DOI] [PubMed] [Google Scholar]
- Baena-González E., Aro E.M. (2002). Biogenesis, assembly and turnover of photosystem II units. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 1451–1459, discussion 1459–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.R., Harbinson J., Kramer D.M. (2007). Determining the limitations and regulation of photosynthetic energy transduction in leaves. Plant Cell Environ. 30: 1107–1125 [DOI] [PubMed] [Google Scholar]
- Bateman A., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32(Database issue): D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.M., Holmström M., Raksajit W., Suorsa M., Piippo M., Aro E.M. (2010). Small chloroplast-targeted DnaJ proteins are involved in optimization of photosynthetic reactions in Arabidopsis thaliana. BMC Plant Biol. 10: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16: 10881/–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B., Adams, III W.W. (1992). Photoprotection and other responses of plants to high light stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43: 599–626 [Google Scholar]
- Dobáková M., Tichý M., Komenda J. (2007). Role of the PsbI protein in photosystem II assembly and repair in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 145: 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I., Critchley C., Woodall G., Stewart G. (1998). Photoinhibition in differently coloured juvenile leaves of Syzygium species. J. Exp. Bot. 49: 1437–1445 [Google Scholar]
- Dominguez-Solis J.R., He Z.Y., Lima A., Ting J., Buchanan B.B., Luan S. (2008). A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc. Natl. Acad. Sci. USA 105: 16386–16391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Flors C., Fryer M.J., Waring J., Reeder B., Bechtold U., Mullineaux P.M., Nonell S., Wilson M.T., Baker N.R. (2006). Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J. Exp. Bot. 57: 1725–1734 [DOI] [PubMed] [Google Scholar]
- Fu A., He Z., Cho H.S., Lima A., Buchanan B.B., Luan S. (2007). A chloroplast cyclophilin and maintenance of functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulgosi H., Vener A.V., Altschmied L., Herrmann R.G., Andersson B. (1998). A novel multi-functional chloroplast protein: identification of a 40-kDa immunophilin-like protein located in the thylakoid lumen. EMBO J. 17: 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa G., Suwa M., Ichikawa Y., Ohtsuka T., Kumagai S., Kikuchi M., Sato Y., Saito Y. (2003). A novel function of tissue-type transglutaminase: Protein disulphide isomerase. Biochem. J. 373: 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai-Yamada M., Ohori Y., Uchimiya H. (2004). Dissection of Arabidopsis Bax inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell 16: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N., Ohkawa H., Welsh E.A., Liberton M., Pakrasi H.B. (2005). Psb29, a conserved 22-kD protein, functions in the biogenesis of Photosystem II complexes in Synechocystis and Arabidopsis. Plant Cell 17: 2768–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann T., Russenberger D., von Zychlinski A., Christopher W., Sjölander K., Gruissem W., Baginsky S. (2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14: 354–362 [DOI] [PubMed] [Google Scholar]
- Klostermann E., Droste Gen Helling I., Carde J.P., Schünemann D. (2002). The thylakoid membrane protein ALB3 associates with the cpSecY-translocase in Arabidopsis thaliana. Biochem. J. 368: 777–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer D.M., Johnson G., Kiirats O., Edwards G.E. (2004). New fluorescence parameters for the determination of q(a) redox state and excitation energy fluxes. Photosynth. Res. 79: 209–218 [DOI] [PubMed] [Google Scholar]
- Krause G.H., Jahns P. (2003). Pulse amplitude modulated chlorophyll fluorometry and its application in plant science. In Light-Harvesting Antennas in Photosynthesis, Green B.R., Parson W.W., (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 373–399 [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kyle D.J., Kuang T.Y., Watson J.L., Arntzen C.J. (1984). Movement of a sub-population of the light harvesting complex (LHCII) from grana to stroma lamellae as a consequence of its phosphorylation. Biochim. Biophys. Acta 765: 89–96 [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lima A., Lima S., Wong J.H., Phillips R.S., Buchanan B.B., Luan S. (2006). A redox-active FKBP-type immunophilin functions in accumulation of the photosystem II supercomplex in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 103: 12631–12636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Last R.L. (2008). Web-based Arabidopsis functional and structural genomics resources. The Arabidopsis Book 6: e0118, doi/10.1199/tab.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Savage L.J., Ajjawi I., Imre K.M., Yoder D.W., Benning C., Dellapenna D., Ohlrogge J.B., Osteryoung K.W., Weber A.P.M., Wilkerson C.G., Last R.L. (2008). New connections across pathways and cellular processes: industrialized mutant screening reveals novel associations between diverse phenotypes in Arabidopsis. Plant Physiol. 146: 1482–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Savage L.J., Larson M.D., Wilkerson C.G., Last R.L. (2011). Chloroplast 2010: A database for large-scale phenotypic screening of Arabidopsis mutants. Plant Physiol. 155: 1589–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin B., Nurmi M., Rojas-Stuetz M., Aro E.M., Adamska I., Spetea C. (2008). Towards understanding the functional difference between the two PsbO isoforms in Arabidopsis thaliana: Insights from phenotypic analyses of psbo knockout mutants. Photosynth. Res. 98: 405–414 [DOI] [PubMed] [Google Scholar]
- Ma J., Peng L., Guo J., Lu Q., Lu C., Zhang L. (2007). LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19: 1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Maxwell K., Johnson G.N. (2000). Chlorophyll fluorescence: A practical guide. J. Exp. Bot. 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Müller P., Li X.P., Niyogi K.K. (2001). Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125: 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo P., Sirpiö S., Suorsa M., Aro E.M. (2008). Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynth. Res. 98: 489–501 [DOI] [PubMed] [Google Scholar]
- Peltier J.-B., Ytterberg A.J., Sun Q., van Wijk K.J. (2004). New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 279: 49367–49383 [DOI] [PubMed] [Google Scholar]
- Peng L.W., Ma J.F., Chi W., Guo J.K., Zhu S.Y., Lu Q.T., Lu C.M., Zhang L.X. (2006). LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigiet V.P., Schuster B.J. (1986). Thioredoxin-catalyzed refolding of disulfide-containing proteins. Proc. Natl. Acad. Sci. USA 83: 7643–7647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokka A., Aro E.-M., Herrmann R.G., Andersson B., Vener A.V. (2000). Dephosphorylation of photosystem II reaction center proteins in plant photosynthetic membranes as an immediate response to abrupt elevation of temperature. Plant Physiol. 123: 1525–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokka A., Suorsa M., Saleem A., Battchikova N., Aro E.M. (2005). Synthesis and assembly of thylakoid protein complexes: multiple assembly steps of photosystem II. Biochem. J. 388: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M., Vallon O., Wollman F.A., Beck C.F. (1999). A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.Y., Tang W., Hao S.F., Wang C.C. (2005). Contributions of cysteine residues in Zn2 to zinc fingers and thiol-disulfide oxidoreductase activities of chaperone DnaJ. Biochemistry 44: 1683–1689 [DOI] [PubMed] [Google Scholar]
- Shimada H., Mochizuki M., Ogura K., Froehlich J.E., Osteryoung K.W., Shirano Y., Shibata D., Masuda S., Mori K., Takamiya K.I. (2007). Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell 19: 3157–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirpiö S., Allahverdiyeva Y., Suorsa M., Paakkarinen V., Vainonen J., Battchikova N., Aro E.M. (2007). TLP18.3, a novel thylakoid lumen protein regulating photosystem II repair cycle. Biochem. J. 406: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirpiö S., Khrouchtchova A., Allahverdiyeva Y., Hansson M., Fristedt R., Vener A.V., Scheller H.V., Jensen P.E., Haldrup A., Aro E.M. (2008). AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 55: 639–651 [DOI] [PubMed] [Google Scholar]
- Suorsa M., Regel R.E., Paakkarinen V., Battchikova N., Herrmann R.G., Aro E.M. (2004). Protein assembly of photosystem II and accumulation of subcomplexes in the absence of low molecular mass subunits PsbL and PsbJ. Eur. J. Biochem. 271: 96–107 [DOI] [PubMed] [Google Scholar]
- Suorsa M., Sirpiö S., Allahverdiyeva Y., Paakkarinen V., Mamedov F., Styring S., Aro E.M. (2006). PsbR, a missing link in the assembly of the oxygen-evolving complex of plant photosystem II. J. Biol. Chem. 281: 145–150 [DOI] [PubMed] [Google Scholar]
- Swiatek M., Regel R.E., Meurer J., Wanner G., Pakrasi H.B., Ohad I., Herrmann R.G. (2003). Effects of selective inactivation of individual genes for low-molecular-mass subunits on the assembly of photosystem II, as revealed by chloroplast transformation: The psbEFLJoperon in Nicotiana tabacum. Mol. Genet. Genomics 268: 699–710 [DOI] [PubMed] [Google Scholar]
- Tanaka A., Makino A. (2009). Photosynthetic research in plant science. Plant Cell Physiol. 50: 681–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zhang Z.G., Wei Y.D., Collinge D.B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11: 1187–1194 [Google Scholar]
- Triantaphylidès C., Havaux M. (2009). Singlet oxygen in plants: production, detoxification and signaling. Trends Plant Sci. 14: 219–228 [DOI] [PubMed] [Google Scholar]
- Wellburn A.R. (1994). The spectral determination of chlorophyll a and chlorophyll b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144: 307–313 [Google Scholar]
- Woody S.T., Austin-Phillips S., Amasino R.M., Krysan P.J. (2007). The WiscDsLox T-DNA collection: An Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant Res. 120: 157–165 [DOI] [PubMed] [Google Scholar]
- Yokthongwattana K., Chrost B., Behrman S., Casper-Lindley C., Melis A. (2001). Photosystem II damage and repair cycle in the green alga Dunaliella salina: Involvement of a chloroplast-localized HSP70. Plant Cell Physiol. 42: 1389–1397 [DOI] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]