Abstract
The development of refractive error is mediated by both environmental and genetic factors. We performed regression-based quantitative trait locus (QTL) linkage analysis on Ashkenazi Jewish families to identify regions in the genome responsible for ocular refraction. We measured refractive error on individuals in 49 multi-generational American families of Ashkenazi Jewish descent. The average family size was 11.1 individuals and was composed of 2.7 generations. Recruitment criteria specified that each family contain at least two myopic members. The mean spherical equivalent refractive error in the sample was −3.46D (SD=3.29) and 87% of individuals were myopic. Microsatellite genotyping with 387 markers was performed on 411 individuals. We performed multipoint regression-based linkage analysis for ocular refraction and a log transformation of the trait using the statistical package Merlin-Regress. Empirical genomewide significance levels were estimated through gene-dropping simulations by generating random genotypes at each of the 387 markers in 200 replicates of our pedigrees. Maximum LOD scores of 9.5 for ocular refraction and 8.7 for log-transformed refraction (LTR) were observed at 49.1 cM on chromosome 1p36 between markers D1S552 and D1S1622. The empirical genomewide significance levels were P=0.065 for ocular refraction and P<0.005 for LTR, providing strong evidence for linkage of refraction to this locus. The inter-marker region containing the peak spans 11 Mb and contains approximately 189 genes. Conclusion: We found genomewide significant evidence for linkage of refractive error to a novel QTL on chromosome 1p36 in an Ashkenazi Jewish population.
Introduction
Refractive errors are a set of optical abnormalities in which the focal point of the eye does not coincide with the sensory retina, causing optical defocus and resulting in reduced vision. Myopia, or nearsightedness, results when the image of distant objects is focused in front of the retinal plane, causing blurred distance vision. Myopia of -1 diopter (D) or worse affects over 30 million (25.4%) Americans and 49.6 million (26.6%) Europeans aged 40 and above. The highest rates of myopia have been observed in East Asian populations. In Singapore, Japan and Taiwan, the prevalence of myopia among teenagers and young adults exceeds 50% (Au Eong et al. 1993; Lin et al. 1999; Matsumura and Hirai 1999; Saw 2003; He et al. 2004; Fan et al. 2004).
Although proper optical correction can usually improve vision, uncorrected and inadequately corrected refractive error are major causes of visual impairment worldwide (Tielsch et al. 1990; Attebo et al. 1999; Munoz et al. 2000; VanNewkirk et al. 2001). Myopia has also been implicated in a number of potentially blinding eye conditions including: peripheral retinal abnormalities (Pierro et al. 1992), age related cataracts (Lim et al. 1999; Leske et al. 2002), glaucoma (Mitchell et al. 1999), myopic retinal degeneration (Vongphanit et al. 2002), and choroidal neovascularization (Cohen et al. 1996). Furthermore, common modes of refractive correction such as contact lenses or refractive surgery can lead to adverse effects (Schein and Poggio 1990; Stulting et al. 1999).
There is convincing evidence that the development of refractive error is mediated by both environmental and genetic influences. An increase in the prevalence of myopia, as seen in Asia over the last few decades (Matsumura and Hirai 1999), strongly suggests that secular changes in environmental exposures and/or behavioral factors influence the population distribution of ocular refraction. Additionally, significant geographical differences in the prevalence of refractive errors have been noted within ethnic groups, pointing to environmental mediation of refractive development (Au Eong et al. 1993; Saw et al. 2001). The intensity of upclose visual activities during childhood and adolescence appears to play an important role in myopisation (Zylbermann et al. 1993; Ben Simon et al. 2004). Although the prevalence of myopia amongst young Israeli adults appears to be similar to that of Western populations (Rosner and Belkin 1991), Zylbermann et al. (1993) found that Israeli Jewish boys who attended Orthodox school were, on average, more myopic than similarly aged boys who were educated in secular schools. The authors postulated that the intensive visual demand associated with a religious education is likely responsible for high rates of myopia in the Jewish Orthodox community.
Although individual behavior and environmental influences can affect refractive development, the contribution of genetics is also thought to be important. Epidemiological studies in various populations have consistently shown strong familial patterns for ocular refraction (Alsbirk 1979; Bear et al. 1981; Hammond et al. 2001; Lee et al. 2001; Lyhne et al. 2001; Wojciechowski et al. 2005). Twin studies by Hammond et al. (2001) and Lynhe et al. (2001) both estimated the heritability (h2) of ocular refraction to be 0.85 or above. High heritability estimates have also been reported for the well-characterized Beaver Dam, Wisconsin (Lee et al. 2001) (h2=0.74) and Salisbury, Maryland (Wojciechowski et al. 2005) (h2=0.61) cohorts.
Linkage studies of ocular refraction have generally focused on highly penetrant, severe forms of myopia (Young et al. 1998a; b; Naiglin et al. 1999; Paluru et al. 2003; Young et al. 2004). Young et al. (1998b) first found evidence for linkage to high, or so-called “pathological” myopia on chromosome 18p11. Subsequent studies have identified areas of linkage to high myopia on chromosomal regions 12q22-23 (Young et al. 1998a), 7q36 (Naiglin et al. 1999) and 17q21-22 (Paluru et al. 2003). In a study of Ashkenazi Jewish and Amish families reported by Ibay et al. (2004), these loci were not found to play a major role in susceptibility to common myopia. In a study of 51 families in the United Kingdom, Farbrother et al. (2004) confirmed linkage of the MYP3 locus on chromosome 12q to high myopia. The authors also found weak evidence for linkage to the 17q locus. Among the same Ashkenazi Jewish population as in the present study, Stambolian et al. (2004) reported significant evidence for linkage of more moderate degrees of myopia to chromosome 22q12 (HLOD=3.54).
While ocular refraction is an inherently quantitative measure, these linkage studies characterized myopia as a dichotomous trait. To our knowledge, only one study has reported results from a genomewide linkage scan of ocular refraction characterized as a quantitative trait (Hammond et al. 2004). By taking into account the entire distribution of the continuous outcome, quantitative trait locus (QTL) linkage analyses may be more suitable to isolate the genomic loci that contribute to the whole range of ocular refraction than penetrance-based, or parametric, linkage methods. In a genomewide scan of 221 dizygotic twin pairs, Hammond et al. (2004) reported significant linkage (LOD>3.2) to refractive error at four loci, with a maximum LOD score of 6.1 at 40 cM on chromosome 11p13. They found additional linkage peaks at chromosomes 3q26 (LOD 3.7), 8p23 (LOD 4.1), and 4q12 (LOD 3.3). A recent parametric analysis in the Old Order Amish independently confirmed linkage of myopia to a locus on chromosome 8p23 (Stambolian et al. 2005). In the current study, we report results of QTL linkage analyses to refractive error in multigenerational American Ashkenazi Jewish families. Our results are the first report of significant linkage of myopia to a novel QTL on chromosome 1.
Materials and methods
Family recruitment and evaluation
The study protocol adhered to the tenet of the Declaration of Helsinki and was approved by the University of Pennsylvania and the National Human Genome Research Institute, National Institutes of Health institutional review boards. The family ascertainment procedure and examination protocol has been described in detail elsewhere (Stambolian et al. 2004). Briefly, participants were invited to take part in the study through a mass mailing of 3,900 letters that included questionnaires requesting contact information that were sent to all known Orthodox Jewish families residing in Lakewood, NJ. Second and third mailings were sent to individuals who did not respond to the previous mailings. A total of 1,310 questionnaires were returned. These individuals were called and family histories of spectacle wear and ocular refraction status were obtained by telephone. All participants included in the study were of Ashkenazi Jewish heritage. In order to be eligible to participate in the study, families had to satisfy the following criteria: (1) the proband must be myopic and have either an affected parent or child; (2) only one parent of the proband should be myopic; and (3) the family must be willing to participate. These recruitment conditions were established to preferentially select families that are consistent with a dominant mode of inheritance of a susceptibility allele to myopia (Durner et al. 1992). Medical records were obtained for each member of selected families. Data were gathered for all eligible parents, cousins, grandparents, siblings, children, aunts and uncles of each proband.
The final sample consisted of 542 individuals in 49 pedigrees with an average of 11.1 members per family. Families were, on average, composed of 2.7 generations. Blood was drawn and genotyping was carried out on 411 (75.7%) individuals. Four hundred-and-seventy-one (86.9%) participants were myopic with a mean spherical equivalent refractive error of −1.0 or less. Population characteristics are presented in Table 1.
Table 1.
Sample characteristics
| Individuals
|
Pedigrees
|
Refraction
|
|||||
|---|---|---|---|---|---|---|---|
| Number | Number genotyped (%) | Number | Mean size | Mean number of generations | Number myopic (%) | Mean SE refraction (D) | Variance |
| 542 | 410 (75.7) | 49 | 11.1 | 2.7 | 471 (86.9) | −3.5 | 11.2 |
DNA extraction and genotyping
High-molecular-weight DNA was isolated from buffy coats with a kit (Puregene; Gentra Systems, Inc.; Minneapolis, MN, USA). Samples were stored in a DNA repository under a unique code. A genomewide scan was performed at the Center for Inherited Disease Research (CIDR) by use of automated fluorescent microsatellite analysis. PCR products were sized on an ABI 3700 sequencer. The marker set used was a modification of the Cooperative Human Linkage Center marker set, version 9 (387 markers; average spacing 9 cM; average heterozygosity 0.76). The error rate, which was based on paired genotypes from blind duplicate samples, was 0.06%. The overall missing-data rate was 3.6%. All genotyping was performed blind to clinical status.
Phenotypic evaluation
Eligibility requirements for entry of a family into the study required an index case that met the following criteria: (1) cycloplegic refraction of −1.00 spherical equivalent (as long as there was −1.00 D or higher in each meridian if astigmatism was present) or worse in those under age 50, (2) manifest refraction of −1 spherical equivalent (as long as there was −1 D or higher in each meridian if astigmatism was present) or worse in those over age 50, and (3) no history of systemic or ocular disease which might predispose to myopia including premature birth. This same classification scheme was used to determine affection status for all individuals in the pedigrees, and subjects who did not meet this standard for affection were considered unaffected. If a subject was reported to have been myopic, but this diagnosis could not be confirmed with either medical records, measurement of the prescription of an old pair of eyeglasses, or current physical examination, this person was treated as being of unknown phenotype.
Our ascertainment strategy for the multiply affected pedigrees was to obtain eye records of all affected individuals, the parents of those individuals, and any other family members connecting affected pairs. We also sought unaffected siblings as well as affected cousins, grandparents, uncles and aunts. Eye records of subjects were reviewed to determine if cycloplegia was properly utilized for those under age 50. Subjects with eye records older than 2 years and subjects with improper exams were re-examined by their local eye doctor or one of the authors (D.S.). While most exams occurred on the side of the family with the history of myopia, family history of myopia was collected for the parents and siblings of both parents of the proband (i.e., all grandparents and aunts/uncles of the proband). Sometimes, members of the family of the unaffected parent of the proband were examined so that we could be certain of their clinical status.
We defined the quantitative trait ocular refraction (REF) as the spherical equivalent refractive error, averaged between the eyes; where the spherical equivalent is the spherical spectacle correction plus one-half the cylindrical correction.
Statistical methods
Error and relationship testing
Mendelian inconsistencies and potential relationship errors were assessed and corrected prior to data analysis by use of SIBPAIR (Duffy 1997), GAS (Young 1995) and RELCHECK (Boehnke and Cox 1997; Broman and Weber 1998) software.
Quantitative trait locus (QTL) linkage analysis
QTL linkage analyses were performed using the software package MERLIN, version 0.10.2 (Abecasis et al. 2002). The statistical package implements an extended Haseman-Elston regression-based linkage method through its MERLIN-REGRESS subroutine (Sham et al. 2002). The method is applicable to extended pedigrees and to pedigrees selected based on trait values, provided that correct values for population parameters of the trait distribution are specified (Sham et al. 2002). The estimates of the regression coefficients are unbiased by sample selection because pairwise identity-by-descent (IBD) sharing proportions are regressed on functions of trait values. However, for the method to be applicable to selected samples, both independent and dependent variables are centered on their population means (Sham et al. 2002). Consequently, this regression approach requires the specification of the population mean, variance and heritability of the trait.
While the underlying distribution of ocular refraction is unknown in this population, epidemiological data are available from a number of other source populations. The mean spherical equivalent ocular refraction in our sample was −3.46 D (SD=3.29 D). Since our ascertainment scheme selected for multiplex myopic families, and our sample mean is likely to be biased towards myopic values relative to the population mean, we specified a mean refractive error of −2.50 D for the source population while the trait variance was set to 10. These parameter values are consistent with those reported for an Israeli Orthodox Jewish population by Zylbermann et al. (1993). Numerous studies have reported heritability estimates for refractive error ranging from 0.5 to 0.9 (Alsbirk 1979; Bear et al. 1981; Teikari et al. 1991, 1992; Hammond et al. 2001; Lee et al. 2001; Lyhne et al. 2001; Wojciechowski et al. 2005). Hence, the trait heritability was set to a conservative value of 0.6 in our statistical model.
The distribution of ocular refraction was slightly left skewed (skewness=−0.48) and slightly leptokurtotic (kurtosis=3.47) in our sample. Some pedigrees contained highly myopic individuals causing extreme intra-family deviations from normality and highly discordant relative pairs. We therefore applied a logarithmic transformation (transformed refraction = ln[20.23 − spherical equivalent refraction]) to the data in order to minimize the effect of departures from normal distribution assumptions as well as extreme phenotypic discordance on our linkage statistics and their associated significance levels. The skewness and kurtosis of our sample’s log-transformed trait distribution were 0 and 3.37, respectively. Linkage analyses were subsequently carried out on both the untransformed ocular refraction (REF) and the log-transformed refraction (LTR).
Since the distribution of ocular refraction is unknown in our source population, we specified the underlying population mean of the LTR as the direct transformation of the population’s putative mean ocular refraction of −2.50 D and used the sample variance of the transformed distribution as the population variance parameter in our statistical model. Hence, the population mean and variance of LTR were correspondingly set to 3.12 and 0.02.
We performed genomewide multipoint linkage analyses at 371 autosomal microsatellite markers using MERLIN-REGRESS. The average marker heterozygosity was 0.77 and the sex-averaged mean intermarker spacing was 9.3 cM. In addition, we carried out interval mapping analyses at two evenly spaced locations between consecutive markers. MERLIN uses the Lander-Green algorithm (Lander and Green 1987) to estimate inter-marker allele sharing between family members.
We explored the sensitivity of the linkage statistic to differing parameter specifications by varying the population parameters in the statistical models for LTR, and repeating linkage analyses. The model parameters were individually varied from the initial model as follows: mean=3.01, 3.06, 3.19 and 3.29; variance=0.0067, 0.01, 0.04 and 0.06; and heritability=0.1, 0.3, 0.45, 0.75, 0.9 and 1. Furthermore, we compared linkage statistics of the regression-based procedure to the variance components method, which is also implemented in MERLIN.
Simulations and genomewide significance levels
The MERLIN-REGRESS procedure provides point-wise, nominal, LOD scores and significance levels based on the χ2 distribution and asymptotic theory (Sham et al. 2002). Genomewide significance levels can be assessed through a gene-dropping simulation procedure implemented in the software package. Briefly, MERLIN generates random datasets of markers (unlinked to the trait) within pedigrees while preserving family structure, population marker allele frequencies, marker spacing and individual phenotypic information. These simulations can provide empirical type-1 error rates given sample-specific pedigree structures, phenotypic distribution and marker information.
We estimated genomewide significance levels by generating 200 whole-genome replicates of unlinked markers in our families and assessing the distribution of maximum LOD scores obtained from each simulation. Since there were 371 markers in each scan, a total of 74,200 markers that were unlinked to the trait (but not all independent of each other) were simulated. Individual gene-dropping simulations required, on average, 15 h of computing time for one whole-genome scan replicate and the number of replicates performed were thus limited by the availability of computational resources. Nevertheless, 200 whole-genome replicates are sufficient to establish genomewide significance levels with a high degree of confidence. Simulations under different model specifications were accomplished in the same manner using 100 wholegenome replicates (i.e., 37,100 markers unlinked to the trait were simulated). The empiric genomewide significance level of our observed maximum LOD score was estimated as the proportion of simulated replicates in which the maximum LOD score exceeded the value of our observed maximum LOD score. Genomewide P values of secondary peaks were calculated as the proportion of replicates that contained one or more LOD scores greater than the LOD score at that position. In addition, we defined the empirical locus-specific p-value as the proportion of simulations in which the LOD score at a given locus exceeded the observed test LOD score at the same locus.
Results
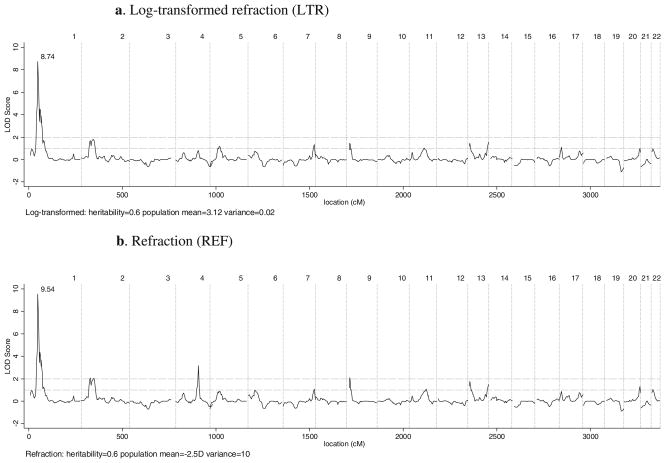
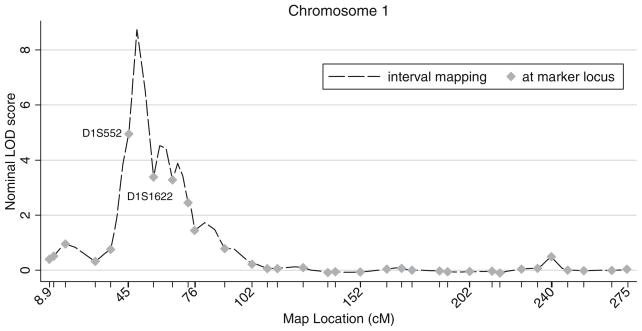
The maximum at-marker multipoint LOD score for LTR was 5.09 (nominal pointwise P<0.0001) at marker D1S552 at 45.33 cM on chromosome 1. The same marker showed a LOD score of 4.59 (nominal pointwise P<0.0001) for REF. QTL linkage results for multipoint scans of REF and LTR are presented in Table 2. Interval mapping yielded multipoint LOD score peaks of 8.74 and 9.54 at 49.1 cM on chromosome 1 for LTR and REF respectively (Table 2). Figure 1 shows multipoint LOD scores for LTR (panel a) and REF (panel b). The linkage statistics follow a similar pattern for both traits but the peaks are attenuated in the analysis of LTR compared to REF. Figure 2 shows LOD scores for multipoint linkage of LTR to chromosome 1. The LOD score profile for linkage of REF to chromosome 1 was similar and is not presented.
Table 2.
QTL linkage LOD scores for refraction (REF) and log-transformed refraction (LTR)
| Chromosome | Marker | Position (cM) | Refraction (REF)
|
Log-transformed refraction (LTR)
|
||||
|---|---|---|---|---|---|---|---|---|
| LOD | Nominal pointwise | Empirical genomewide | LOD | Nominal pointwise | Empirical genomewide | |||
| P value | P value | P value | P value | |||||
| 1 | D1S3669 | 37.1 | 0.50 | 0.07 | 1 | 0.90 | 0.02 | 0.975 |
| – | 39.8 | 1.41 | 0.005 | 0.945 | 2.10 | 0.0009 | 0.415 | |
| – | 42.6 | 3.33 | 0.00005 | 0.39 | 4.09 | 0.00001 | 0.08 | |
| D1S552 | 45.3 | 4.59 | <0.00001 | 0.23 | 5.09 | <0.00001 | 0.035 | |
| – | 49.1 | 9.54 | <0.00001 | 0.065 | 8.74 | <0.00001 | <0.005 | |
| – | 52.9 | 7.08 | <0.00001 | 0.085 | 6.52 | <0.00001 | <0.005 | |
| D1S1622 | 56.7 | 3.43 | 0.00004 | 0.38 | 3.38 | 0.00003 | 0.10 | |
| – | 59.7 | 4.35 | <0.00001 | 0.25 | 4.55 | <0.00001 | 0.055 | |
| – | 62.6 | 4.13 | 0.00001 | 0.28 | 4.41 | <0.00001 | 0.06 | |
| D1S255 | 65.5 | 3.14 | 0.00007 | 0.445 | 3.25 | 0.00005 | 0.125 | |
| – | 67.8 | 3.64 | 0.00002 | 0.345 | 3.78 | 0.00002 | 0.085 | |
| – | 7.2 | 3.19 | 0.00006 | 0.435 | 3.32 | 0.00005 | 0.115 | |
| D1S3721 | 72.6 | 2.32 | 0.0005 | 0.69 | 2.40 | 0.0004 | 0.28 | |
| – | 73.6 | 2.20 | 0.0007 | 0.755 | 2.35 | 0.0005 | 0.305 | |
| – | 74.6 | 1.65 | 0.003 | 0.905 | 1.95 | 0.0014 | 0.51 | |
| D1S2134 | 75.7 | 1.13 | 0.011 | 0.985 | 1.47 | 0.005 | 0.755 | |
| – | 8.3 | 1.26 | 0.008 | 0.96 | 1.77 | 0.002 | 0.58 | |
| – | 84.9 | 0.99 | 0.02 | 0.995 | 1.51 | 0.004 | 0.735 | |
| 2 | – | 44.8 | 1.36 | 0.006 | 0.95 | 1.04 | 0.014 | 0.945 |
| D2S405 | 48.0 | 2.01 | 0.0012 | 0.81 | 1.65 | 0.003 | 0.665 | |
| – | 5.5 | 2.04 | 0.0011 | 0.805 | 1.70 | 0.003 | 0.625 | |
| – | 53.0 | 1.68 | 0.003 | 0.895 | 1.45 | 0.005 | 0.77 | |
| 4 | D4S2394 | 129.9 | 1.40 | 0.006 | 0.945 | 0.67 | 0.09 | 1 |
| – | 134.4 | 3.17 | 0.00007 | 0.435 | 0.80 | 0.04 | 0.99 | |
| – | 138.9 | 1.05 | 0.014 | 0.985 | 0.33 | 0.03 | 1 | |
| 9 | D9S2169 | 14.2 | 1.19 | 0.01 | 0.985 | 0.90 | 0.02 | 0.975 |
| – | 16.8 | 2.09 | 0.001 | 0.79 | 1.46 | 0.005 | 0.765 | |
| – | 19.33 | 1.82 | 0.002 | 0.865 | 1.38 | 0.006 | 0.80 | |
Regions containing LOD scores above 2.0 for either REF or LTR are shown. Dashes in the marker column represent inter-marker regions. Nominal, pointwise, P values were output by MERLIN-REGRESS and are based on the χ2 distribution and asymptotic theory. Empirical P values were estimated via whole-genome gene-dropping simulations of unlinked markers using the simulate procedure in MERLIN-REGRESS. Genomewide significance for the maximum LOD score at 49.1 cM on chromosome 1 represents the proportion of replicates in which the maximum LOD score exceeded the threshold at that marker. Genomewide significance levels of secondary peaks were calculated as the proportion of replicates that contained one or more LOD scores greater than the position-specific LOD score
Fig. 1.
Whole-genome, multipoint, QTL linkage scan of log-transformed refraction (a) and refraction (b) in multigenerational Ashkenazi Jewish families. For the log-transformed trait (LTR), the mean, variance and heritability were set to 3.12, 0.02 and 0.6. For refraction (REF), the model mean, variance and heritability were fixed to −2.5D, 10 and 0.6, respectively. Vertical dashed lines represent boundaries between chromosomes. Horizontal dashed lines mark LOD scores of 1 and 2
Fig. 2.
Multipoint QTL linkage scores for log-transformed refraction (LTR) on chromosome 1. Marker locations and their corresponding LOD scores are marked with diamonds. The connecting line represents interval mapping scores. Marker locations are also shown as ticks on the horizontal axis. The peak multipoint inter-marker LOD score was 8.7 at 49.1 cM
Interval mapping also yielded local LOD score peaks above two on chromosomes 2, 4 and 9 for the REF. For LTR, only the peak on chromosome 1 had a LOD score above two (Table 2).
Empirical genomewide significance levels were obtained separately for the maximum LOD scores of LTR and REF at 49.1 cM on chromosome 1. The heritability, population mean and variance parameters in our simulations were identical to those specified in the QTL linkage analysis. None of the LTR replicates had maximum LOD scores greater than 8.7 and 13 REF replicates had maximum LOD scores above 9.5, representing genomewide significance levels of P<0.005 and P=0.065 for LTR and REF, respectively. Genomewide P values for LTR at markers D1S552 (LOD=5.09) and D1S1622 (LOD=3.44) were 0.03 and 0.10, respectively.
We assessed the sensitivity of the LOD score profiles on chromosome 1 to misspecification of the trait heritability, trait mean and variance parameters. Individually varying the heritability, the mean or the variance from the assumed population values of 0.6, 3.12 and 0.02, respectively, had negligible effects on the maximum LOD score within a wide range of parameter specifications (Table 3). The LOD score at 49.1 cM on chromosome 1 remained above 7 for heritabilities between 0.3 and 0.75, variances ranging from 0.0067 to 0.06 and population means between 3.01 and 3.29. These mean values for LTR correspond to mean ocular refractions between 0 and −6.5 D on the non-transformed scale. For variance components linkage analysis, maximum multipoint LOD scores of 1.80 (nominal P=0.002) and 2.23 (nominal P=0.0007) were obtained at marker D1S552 for REF and LTR, respectively.
Table 3.
Maximum linkage LOD scores and empirical genomewide (GW) p-values at 49.1 cM on chromosome 1 for LTR using different parameter specifications in MERLIN-REGRESS
| Mean | LOD | GW P value* | Variance | LOD | GW P value* | Heritability | LOD | GW P value* |
|---|---|---|---|---|---|---|---|---|
| 0.1 | 5.05 | 0.01 | ||||||
| 3.01 | 7.47 | 0.03 | 0.0067 | 8.75 | 0.14 | 0.3 | 7.11 | <0.01 |
| 3.06 | 8.01 | <0.01 | 0.01 | 8.78 | 0.09 | 0.45 | 8.41 | <0.01 |
| 3.12** | 8.74 | <0.01 | 0.02 | 8.74 | <0.01 | 0.6 | 8.74 | <0.01 |
| 3.19 | 8.29 | <0.01 | 0.04 | 8.2 | <0.01 | 0.75 | 7.12 | 0.08 |
| 3.29 | 3.59 | <0.01 | 0.06 | 7.42 | <0.01 | 0.9 | 4.36 | 0.19 |
| 1 | 2.66 | 0.12 |
Parameter values for the mean, variance and heritability were, respectively, 3.12, 0.02 and 0.6, when not otherwise indicated
Empirical genomewide p-values for each statistical model were calculated using gene-dropping simulations implemented in the simulate option of MERLIN-REGRESS. P-values were based on 100 replicates of 371 randomly generated markers
The “presumed correct” model (bold) assumed a population mean for LTR of 3.12, a variance of 0.02 and a heritability of 0.6. Values of 3.01, 3.06, 3.12, 3.19 and 3.29 for mean LTR correspond to refractions of 0, −1, −2.5, −4 and −6.5 D, respectively, on the non-transformed scale
Discussion
We have found evidence for a novel QTL locus for ocular refraction on chromosome 1p36 in an Ashkenazi Jewish population. The linkage peak on chromosome 1 has not previously been reported for either qualitative (i.e., myopia) or quantitative ocular refraction phenotypes.
The statistical procedure implemented in MERLIN-REGRESS estimates the test statistic of individual linkage signals based on asymptotic properties and large-sample central limit theorem. Sham et al. found their test statistic to be unbiased and that it provided correct type I error rates for simulated sibships ranging in size from 2 to 6 (Sham et al. 2002). The authors maintain that the method is applicable to general pedigrees of all types although they limited their simulation studies of non-nuclear families to cousin pedigrees only. Our gene-dropping simulations suggest that the magnitude of the linkage statistics were elevated in our study, yielding inflated type-1 error rates if P values had not been determined empirically. This may have been the result of extreme between-relative discordance in refraction having a large influence on the regression slope. In addition, this upward bias in LOD scores under the null hypothesis of no linkage was greater for REF than for LTR even though the sample’s marginal trait distributions were similar for the untransformed (skewness=− 0.48, kurtosis=3.47) and transformed (skewness= 0, kurtosis=3.37) data, with the transformed trait being slightly closer to normality. Sham et al. (2002) suggest that liberal results can occur when a small number of families contribute inordinately to the linkage statistic. MERLIN-REGRESS provides a measure of family informativeness, conditional on the trait values of the family members, to assess each family’s expected contribution to the test statistic. For the untransformed trait, a single family accounted for 19.3% of the total informativeness and the five most informative families contributed 45.4% to the informativeness index. After transformation, the distribution of family informativeness was much less skewed. For LTR, the most informative and five most informative families accounted for 7.5% and 30.8% of the total informativeness, respectively. This demonstrates the importance of appropriate data transformation and of calculation of empirical significance levels for this statistic. The empirical genomewide P value (a family-wise error rate or FWER) associated with our maximum LOD score of 9.5 for REF was 0.065 whereas our maximum LOD of 8.7 for LTR yielded a genomewide significance smaller than 0.005. Lander and Kruglyak (1995) suggested that allele-sharing LOD scores between 3.3 and 3.8 were needed to establish genomewide significance for non-parametric linkage scans at a genomewide P value (FWER) of 0.05. However, these guidelines were established for allele sharing methods and relative pair designs and are unlikely to be applicable in general pedigree situations. In our sample of extended pedigrees, the simulations of LTR indicate that a MERLIN-REGRESS LOD score of 4.69 or larger is required to exhibit a genomewide significant p-value of 0.05 or less.
The discrepancy between empirical LOD scores obtained through simulation of unlinked markers and theoretical expectations emphasizes the importance of performing computer simulation studies to determine study significance, especially when the applicability of asymptotic theory is in question, such as with small sample sizes and extended, and unbalanced, pedigree structures. In addition, careful consideration must be given to choosing an appropriate transformation of the data since departures from distributional assumptions can have a significant effect on study error rates, as illustrated by the differences in empirical type-1 error rates between REF and LTR.
We compared the results from the regression-based approach to standard variance components linkage analysis, which yielded LOD scores of 1.8 (P=0.002) for refraction and 2.23 (P=0.0007) for LTR at D1S552. These results support the evidence for linkage to this locus and suggest that the regression-based approach may be more powerful than the variance components method in highly ascertained samples.
We carried out linkage analyses while varying the statistical model parameters in order to determine the sensitivity of the MERLIN-REGRESS linkage statistic to possible parameter misspecifications. Our sensitivity results indicate that alternate specifications of the statistical model parameters within a plausible range have little effect on the magnitude of the maximum linkage signal in our data (Table 3). Specifically, the linkage peak on chromosome 1 decreased substantially only for extreme population mean and heritability specifications. Empirical P values associated with the maximum linkage peak on chromosome 1 were below a genomewide significance level of 0.05 for all models except those that specified either very low variances or high heritabilities (Table 3). Even using our sample mean (which was most likely skewed towards myopia due to our ascertainment scheme) as the underlying population mean would have yielded a LOD score profile similar to that of our putative model. Therefore, assuming that our linkage signal is true, it appears that the regression method implemented in MERLIN-REGRESS is robust to misspecifications of the statistical model parameters.
Using the same population, we have previously reported linkage of myopia, characterized as a binary trait, to a locus on chromosome 22 (Stambolian et al. 2004). The current QTL analysis shows only suggestive evidence for linkage of ocular refraction to this region (peak LOD score on chromosome 22=0.93 at 9.1 cM for LTR, empiric locus-specific P=0.03). In addition, the prior parametric analysis did not reveal linkage of the dichotomous trait, myopia, to chromosome 1. While these results may appear incongruous, several differences may account for this discrepancy. First, QTL linkage analyses take account of the entire sample distribution of the trait. In particular, the QTL regression method implemented in MERLIN-REGRESS capitalizes on between-relative trait differences and sums to infer statistical linkage. Hence, individuals who are highly discordant for a quantitative trait (for example, a −1 D myope and a −15 D myope) may be in the same outcome group in binary trait analyses. In our sample, ocular refractions ranged from −15.875 to +6.75 D and 87% of individuals had myopia of 1 D or more. Therefore, quantitative analyses provided additional resolution of the phenotype within broad categorical classes. Second, ocular refraction is determined by various anatomical components that modulate the eye’s refractive power as well as its size. Myopia results primarily from a relative axial elongation of the posterior chamber, a component of the axial length, which has been shown to be a highly heritable trait in a variety of human populations (Alsbirk 1979; Hammond et al. 2001; Lyhne et al. 2001; Biino et al. 2005). A “myopia” gene may affect the sign of optical defocus whereas QTLs affecting ocular component growth may be responsible for regulating differences in the degree of refractive error. Hence, binary trait analyses may allow for the detection of “myopia” genes but be unable to discriminate between varying degrees of myopia whereas QTL methods would be more useful in identifying genes that regulate refraction across the entire phenotypic spectrum.
Using variance components linkage analyses, Biino et al. (2005) recently reported suggestive linkage (LOD>2) of ocular axial length to a locus at 2p24 (marker D2S1360 at 38.3 cM) in Sardinian families. They also reported suggestive evidence for linkage of anterior chamber depth to chromosome 1 and of radius of corneal curvature to chromosomes 7, 2 and 3. This group did not report linkage results for ocular refraction although the values of these biometric components, and their relation to one-another, fully determine refraction. We found mild evidence for linkage of refraction (maximum LOD for LTR on chromosome 2=1.70, empiric locus-specific P=0.01) to a locus near marker D2S405 at 50.4 cM, or approximately 12 cM from Biino et al.’s signal for axial length. This may implicate ocular axial length, which is the primary determinant of myopia, as the underlying cause for our linkage signal on chromosome 2.
Previous studies have mapped high (or “pathological”) myopia susceptibility loci to chromosomal regions 7q (Naiglin et al. 2002), 12q (Young et al. 1998a), 17q (Paluru et al. 2003) and 18p (Young et al. 1998b). We found no evidence for linkage of refractive error to loci on chromosomes 12 or 18. However, we found a small linkage peak (LOD=1.13 for LTR, empiric locusspecific P=0.01) at 10.7 cM on chromosome 17. Paluru and collaborators (2003) have previously mapped a locus for autosomal dominant high myopia to a region flanked by markers D17S5787 and D17S1811 located at 75.0 and 82.6 cM on chromosome 17. Given the distance of more than 60 cM between our weak linkage signal on chromosome 17 from this previously reported linkage peak, it is unlikely that the two studies are giving indication of linkage to the same locus.
Using a parametric statistical approach, Naiglin et al. (2002) reported suggestive evidence for linkage of familial high myopia (multipoint LOD score of 2.81) to a locus at 7q36 flanked by markers D7S798 (169.0 cM) and D7S2546 (173.0 cM). We found mild evidence for linkage of refraction to marker D7S3058 at 173.7 cM on chromosome 7 (LOD=1.33 for LTR, empiric locus-specific P value=0.02). Although our peak was comparatively shallow, the signal is intriguing in view of its close proximity to Naiglin et al.’s locus and represents support for a myopia susceptibility locus on chromosome 7.
One previous genomewide QTL analysis has been carried out for ocular refraction. In a study of 221 dizygotic British twin pairs, Hammond et al. (2004) reported significant (i.e., LOD>3.2) linkage peaks at 40 cM (LOD=6.1) on chromosome 11p13 as well as to loci at 3q26 (LOD=3.7), 8p23 (LOD=4.1), and 4q12 (LOD=3.3). Interestingly, the linkage region on chromosome 11 contains the PAX6 gene at 39–42 cM, which is fundamentally related to eye development in a number of species (Wawersik and Maas 2000; Dominguez et al. 2004) and in which mutations are known to cause a variety of ocular abnormalities in humans (Hanson et al. 1999). Our analyses revealed a peak LOD score of 1.02 (empiric locus-specific P value=0.03) at 90.6 cM on chromosome 11 for LTR. Since our peak is about 50 cM away from the previously mapped locus, our results should not be viewed as confirmatory of an ocular refraction QTL on chromosome 11. However, they are interesting in light of Hammond et al’s linkage results and should be followed-up with a denser marker map and/or the typing of additional families.
The area of most significant linkage, with a maximum LOD of 8.7 (empirical genome-wide P<0.005) observed between markers D1S552 (LOD=5.09) and D1S1622 (LOD=3.44), maps roughly to a region between 19 Mb and 30 Mb on chromosome 1. In our simulations for LTR, a genomewide significance of P<0.05 corresponded to a LOD score of 4.69, marking this as the most likely region to contain a QTL for ocular refraction. This region is predicted to contain approximately 189 genes. However, the linkage region on chromosome 1 that contains LOD scores of two or greater is considerably wider and spans a genetic distance of approximately 35 cM (or more than 20 Mb) from D1S552 to D1S3721. This large region includes several genes that are expressed in human eye tissue (Table 4). While none of these genes are obvious candidates for refractive regulation, they will be carefully examined in our follow-up studies of this region. Our future investigations will involve replicating this linkage signal with an independent sample of Ashkenazi Jewish families. Furthermore, we will attempt to refine the area of linkage by using a dense SNP map in the region, which will also permit us to perform family-based association studies.
Table 4.
Genes expressed in human eye tissue in chromosome 1 linkage region
| Gene symbol | Gene name | Cytogenic location | Representative clone |
|---|---|---|---|
| RCC1 | Regulator of chromosome condensation 1 | p35.3 | BC007300 |
| EPB41 | Erythrocyte membrane protein band 4.1 | p35.3 | BC081539 |
| NBL1 | Neuroblastoma, suppression of tumorigenicity 1 | p36.12 | BC012037 |
| KIF17 | Kinesin family member 17 | p36.12 | BC065927 |
| FAM43B | Family with sequence similarity 43, member B | p36.12 | BC015675 |
| HP1BP3 | Heterochromatin protein 1, binding protein 3 | p36.12 | BC046170 |
| EPHA8 | EPH receptor A8 | p36.12 | BC072417 |
| EPHB2 | EPH receptor B2 | p36.12 | BC041107 |
| HTR1D | 5-hydroxytryptamine (serotonin) receptor 1D | p36.12 | BC00720 |
| E2F2 | E2F transcription factor 2 | p36.12 | BC053676 |
| TCEB3 | Transcription elongation factor B (SIII), polypeptide 3 | p36.11 | BC020448 |
| HMGCL | 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase | p36.11 | BC021145 |
| GRHL3 | Grainyhead-like 3 (Drosophila) | p36.11 | BC036890 |
| PAFAH2 | Platelet-activating factor acetylhydrolase 2, | p36.11 | BC001158 |
| DHDDS | Dehydrodolichyl diphosphate synthase | p36.11 | BC014644 |
| NUDC | Nuclear distribution gene C homolog (A. nidulans) | p36.11 | BC007280 |
Since the eye can be decomposed into individual components that contribute to refraction, additional linkage studies should begin to focus on these constituents. Given the multiplicity of loci identified to date for high myopia in various populations, it is possible that considerable locus heterogeneity also exists for ocular refraction. Our use of a relatively isolated population may have led to the identification of a population-specific locus for refraction. Nevertheless, considering the ubiquity of less severe forms of myopia worldwide it is likely that a common underlying genetic mechanism(s) is responsible for refractive development among humans.
Acknowledgments
This study was supported in part by U.S. Public Health National Eye Institute grant EY12226 (DS) and in part by funds from the intramural program of the National Human Genome Research Institute, NIH (JEB-W, GI, RW). RW also received a William C. Ezell-CIBA Vision Fellowship from the American Optometric Foundation. The authors thank the families for their participation in the study and Dr. Reuvain Shanik and Rabbi Yitzchok Rozsansky for their enthusiasm and effort on the project. Genotyping services were provided by CIDR. CIDR is fully funded through a federal contract from the National Institutes of Health to Johns Hopkins University (contract number N01-HG-65403).
Contributor Information
Robert Wojciechowski, Email: kassaq@mail.nih.gov, Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA. Inherited Disease Research Branch, National Human Genome Research Institute (NIH), 333 Cassell Dr., Suite 1200, Baltimore, MD 21224, USA, Tel.: +1-410-5504627, Fax: +1-410-5507513.
Chris Moy, Department of Ophthalmology, University of Pennsylvania, Philadelphia, PA, USA.
Elise Ciner, Pennsylvania College of Optometry, Philadelphia, PA, USA.
Grace Ibay, Inherited Disease Research Branch, National Human Genome Research Institute (NIH), 333 Cassell Dr., Suite 1200, Baltimore, MD 21224, USA.
Lauren Reider, Department of Ophthalmology, University of Pennsylvania, Philadelphia, PA, USA.
Joan E. Bailey-Wilson, Inherited Disease Research Branch, National Human Genome Research Institute (NIH), 333 Cassell Dr., Suite 1200, Baltimore, MD 21224, USA
Dwight Stambolian, Department of Ophthalmology, University of Pennsylvania, Philadelphia, PA, USA.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Alsbirk PH. Refraction in adult West Greenland Eskimos. A population study of spherical refractive errors, including oculometric and familial correlations. Acta Ophthalmol (Copenh) 1979;57(1):84–95. doi: 10.1111/j.1755-3768.1979.tb06663.x. [DOI] [PubMed] [Google Scholar]
- Attebo K, Ivers RQ, Mitchell P. Refractive errors in an older population: the Blue Mountains Eye Study. Ophthalmology. 1999;106(6):1066–1072. doi: 10.1016/S0161-6420(99)90251-8. [DOI] [PubMed] [Google Scholar]
- Au Eong KG, Tay TH, Lim MK. Race, culture and Myopia in 110,236 young Singaporean males. Singapore Med J. 1993;34(1):29–32. [PubMed] [Google Scholar]
- Bear JC, Richler A, Burke G. Nearwork and familial resemblances in ocular refraction: a population study in Newfoundland. Clin Genet. 1981;19(6):462–472. doi: 10.1111/j.1399-0004.1981.tb02065.x. [DOI] [PubMed] [Google Scholar]
- Ben Simon GJ, Peiss M, Anis E, Nakra T, Luski A, Spierer A. Spectacle use and reduced unaided vision in third grade students: a comparative study in different educational settings. Clin Exp Optom. 2004;87(3):175–179. doi: 10.1111/j.1444-0938.2004.tb03171.x. [DOI] [PubMed] [Google Scholar]
- Biino G, Palmas MA, Corona C, Prodi D, Fanciulli M, Sulis R, Serra A, Fossarello M, Pirastu M. Ocular refraction: heritability and genome-wide search for eye morphometry traits in an isolated Sardinian population. Hum Genet. 2005;116(3):152–159. doi: 10.1007/s00439-004-1231-6. [DOI] [PubMed] [Google Scholar]
- Boehnke M, Cox NJ. Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet. 1997;61(2):423–429. doi: 10.1086/514862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Weber JL. Estimation of pairwise relationships in the presence of genotyping errors. Am J Hum Genet. 1998;63(5):1563–1564. doi: 10.1086/302112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SY, Laroche A, Leguen Y, Soubrane G, Coscas GJ. Etiology of choroidal neovascularization in young patients. Ophthalmology. 1996;103(8):1241–1244. doi: 10.1016/s0161-6420(96)30515-0. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Ferres-Marco D, Gutierrez-Avino FJ, Speicher SA, Beneyto M. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat Genet. 2004;36(1):31–39. doi: 10.1038/ng1281. [DOI] [PubMed] [Google Scholar]
- Duffy DL. Sibpair: a program for nonparametric linkage/association analysis. Am J Hum Genet. 1997;61(Suppl):A197. [Google Scholar]
- Durner M, Greenberg DA, Hodge SE. Inter- and intrafamilial heterogeneity: effective sampling strategies and comparison of analysis methods. Am J Hum Genet. 1992;51(4):859–870. [PMC free article] [PubMed] [Google Scholar]
- Fan DS, Lam DS, Lam RF, Lau JT, Chong KS, Cheung EY, Lai RY, Chew SJ. Prevalence, incidence, and progression of myopia of school children in Hong Kong. Invest Ophthalmol Vis Sci. 2004;45(4):1071–1075. doi: 10.1167/iovs.03-1151. [DOI] [PubMed] [Google Scholar]
- Farbrother JE, Kirov G, Owen MJ, Pong-Wong R, Haley CS, Guggenheim JA. Linkage analysis of the genetic loci for high myopia on 18p, 12q, and 17q in 51 U.K. families. Invest Ophthalmol Vis Sci. 2004;45(9):2879–2885. doi: 10.1167/iovs.03-1156. [DOI] [PubMed] [Google Scholar]
- Hammond CJ, Andrew T, Tat MY, Spector TD. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet. 2004;75(2):294–304. doi: 10.1086/423148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond CJ, Snieder H, Gilbert CE, Spector TD. Genes and environment in refractive error: the twin eye study. Invest Ophthalmol Vis Sci. 2001;42(6):1232–1236. [PubMed] [Google Scholar]
- Hanson I, Churchill A, Love J, Axton R, Moore T, Clarke M, Meire F, van Heyningen V. Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum Mol Genet. 1999;8(2):165–172. doi: 10.1093/hmg/8.2.165. [DOI] [PubMed] [Google Scholar]
- He M, Zeng J, Liu Y, Xu J, Pokharel GP, Ellwein LB. Refractive error and visual impairment in urban children in southern china. Invest Ophthalmol Vis Sci. 2004;45(3):793–799. doi: 10.1167/iovs.03-1051. [DOI] [PubMed] [Google Scholar]
- Ibay G, Doan B, Reider L, Dana D, Schlifka M, Hu H, Holmes T, O’Neill J, Owens R, Ciner E, Bailey-Wilson JE, Stambolian D. Candidate high myopia loci on chromosomes 18p and 12q do not play a major role in susceptibility to common myopia. BMC Med Genet. 2004;5(1):20. doi: 10.1186/1471-2350-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA. 1987;84(8):2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KE, Klein BE, Klein R, Fine JP. Aggregation of refractive error and 5-year changes in refractive error among families in the Beaver Dam Eye Study. Arch Ophthalmol. 2001;119(11):1679–1685. doi: 10.1001/archopht.119.11.1679. [DOI] [PubMed] [Google Scholar]
- Leske MC, Wu SY, Nemesure B, Hennis A. Risk factors for incident nuclear opacities. Ophthalmology. 2002;109(7):1303–1308. doi: 10.1016/s0161-6420(02)01094-1. [DOI] [PubMed] [Google Scholar]
- Lim R, Mitchell P, Cumming RG. Refractive associations with cataract: the Blue Mountains Eye Study. Invest Ophthalmol Vis Sci. 1999;40(12):3021–3026. [PubMed] [Google Scholar]
- Lin LL, Shih YF, Tsai CB, Chen CJ, Lee LA, Hung PT, Hou PK. Epidemiologic study of ocular refraction among schoolchildren in Taiwan in 1995. Optom Vis Sci. 1999;76(5):275–281. doi: 10.1097/00006324-199905000-00013. [DOI] [PubMed] [Google Scholar]
- Lyhne N, Sjolie AK, Kyvik KO, Green A. The importance of genes and environment for ocular refraction and its determiners: a population based study among 20–45 year old twins. Br J Ophthalmol. 2001;85(12):1470–1476. doi: 10.1136/bjo.85.12.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H, Hirai H. Prevalence of myopia and refractive changes in students from 3 to 17 years of age. Surv Ophthalmol. 1999;44(Suppl 1):S109–S115. doi: 10.1016/s0039-6257(99)00094-6. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Hourihan F, Sandbach J, Wang JJ. The relationship between glaucoma and myopia: the Blue Mountains Eye Study. Ophthalmology. 1999;106(10):2010–2015. doi: 10.1016/s0161-6420(99)90416-5. [DOI] [PubMed] [Google Scholar]
- Munoz B, West SK, Rubin GS, Schein OD, Quigley HA, Bressler SB, Bandeen-Roche K. Causes of blindness and visual impairment in a population of older Americans: the Salisbury Eye Evaluation Study. Arch Ophthalmol. 2000;118(6):819–825. doi: 10.1001/archopht.118.6.819. [DOI] [PubMed] [Google Scholar]
- Naiglin L, Clayton J, Gazagne C, Dallongeville F, Malecaze F, Calvas P. Familial high myopia: evidence of an autosomal dominant mode of inheritance and genetic heterogeneity. Ann Genet. 1999;42(3):140–146. [PubMed] [Google Scholar]
- Naiglin L, Gazagne C, Dallongeville F, Thalamas C, Idder A, Rascol O, Malecaze F, Calvas P. A genome wide scan for familial high myopia suggests a novel locus on chromosome 7q36. J Med Genet. 2002;39(2):118–124. doi: 10.1136/jmg.39.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluru P, Ronan SM, Heon E, Devoto M, Wildenberg SC, Scavello G, Holleschau A, Makitie O, Cole WG, King RA, Young TL. New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci. 2003;44(5):1830–1836. doi: 10.1167/iovs.02-0697. [DOI] [PubMed] [Google Scholar]
- Pierro L, Camesasca FI, Mischi M, Brancato R. Peripheral retinal changes and axial myopia. Retina. 1992;12(1):12–17. doi: 10.1097/00006982-199212010-00003. [DOI] [PubMed] [Google Scholar]
- Rosner M, Belkin M. A nation-wide study of myopia prevalence in Israel. Findings in a population of 312,149 young adults. Metab Pediatr Syst Ophthalmol. 1991;14(2):37–41. [PubMed] [Google Scholar]
- Saw SM. A synopsis of the prevalence rates and environmental risk factors for myopia. Clin Exp Optom. 2003;86(5):289–294. doi: 10.1111/j.1444-0938.2003.tb03124.x. [DOI] [PubMed] [Google Scholar]
- Saw SM, Hong RZ, Zhang MZ, Fu ZF, Ye M, Tan D, Chew SJ. Near-work activity and myopia in rural and urban schoolchildren in China. J Pediatr Ophthalmol Strabismus. 2001;38(3):149–155. doi: 10.3928/0191-3913-20010501-08. [DOI] [PubMed] [Google Scholar]
- Schein OD, Poggio EC. Ulcerative keratitis in contact lens wearers. Incidence and risk factors. Cornea. 1990;9(suppl 1):S55–S58. doi: 10.1097/00003226-199010001-00023. [DOI] [PubMed] [Google Scholar]
- Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet. 2002;71(2):238–253. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolian D, Ciner EB, Reider LC, Moy C, Dana D, Owens R, Schlifka M, Holmes T, Ibay G, Bailey-Wilson JE. Genome-wide scan for myopia in the Old Order Amish. Am J Ophthalmol. 2005;140(3):469–476. doi: 10.1016/j.ajo.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Stambolian D, Ibay G, Reider L, Dana D, Moy C, Schlifka M, Holmes T, Ciner E, Bailey-Wilson JE. Genomewide linkage scan for myopia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 22q12. Am J Hum Genet. 2004;75(3):448–459. doi: 10.1086/423789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stulting RD, Carr JD, Thompson KP, Waring GO, III, Wiley WM, Walker JG. Complications of laser in situ keratomileusis for the correction of myopia. Ophthalmology. 1999;106(1):13–20. doi: 10.1016/S0161-6420(99)90000-3. [DOI] [PubMed] [Google Scholar]
- Teikari JM, Kaprio J, Koskenvuo M, O’Donnell J. Heritability of defects of far vision in young adults—a twin study. Scand J Soc Med. 1992;20(2):73–78. [PubMed] [Google Scholar]
- Teikari JM, O’Donnell J, Kaprio J, Koskenvuo M. Impact of heredity in myopia. Hum Hered. 1991;41(3):151–156. doi: 10.1159/000153994. [DOI] [PubMed] [Google Scholar]
- Tielsch JM, Sommer A, Witt K, Katz J, Royall RM. Blindness and visual impairment in an American urban population. The Baltimore Eye Survey. Arch Ophthalmol. 1990;108(2):286–290. doi: 10.1001/archopht.1990.01070040138048. [DOI] [PubMed] [Google Scholar]
- VanNewkirk MR, Weih L, McCarty CA, Taylor HR. Cause-specific prevalence of bilateral visual impairment in Victoria, Australia: the Visual Impairment Project. Ophthalmology. 2001;108(5):960–967. doi: 10.1016/s0161-6420(01)00554-1. [DOI] [PubMed] [Google Scholar]
- Vongphanit J, Mitchell P, Wang JJ. Prevalence and progression of myopic retinopathy in an older population. Ophthalmology. 2002;109(4):704–711. doi: 10.1016/s0161-6420(01)01024-7. [DOI] [PubMed] [Google Scholar]
- Wawersik S, Maas RL. Vertebrate eye development as modeled in Drosophila. Hum Mol Genet. 2000;9(6):917–925. doi: 10.1093/hmg/9.6.917. [DOI] [PubMed] [Google Scholar]
- Wojciechowski R, Congdon N, Bowie H, Munoz B, Gilbert D, West SK. Heritability of refractive error and familial aggregation of myopia in an elderly American population. Invest Ophthalmol Vis Sci. 2005;46(5):1588–1592. doi: 10.1167/iovs.04-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A. GAS (Genetic Analysis System) Version 2.0. Oxford, United Kingdom: 1995. [Google Scholar]
- Young TL, Deeb SS, Ronan SM, Dewan AT, Alvear AB, Scavello GS, Paluru PC, Brott MS, Hayashi T, Holleschau AM, Benegas N, Schwartz M, Atwood LD, Oetting WS, Rosenberg T, Motulsky AG, King RA. X-linked high myopia associated with cone dysfunction. Arch Ophthalmol. 2004;122(6):897–908. doi: 10.1001/archopht.122.6.897. [DOI] [PubMed] [Google Scholar]
- Young TL, Ronan SM, Alvear AB, Wildenberg SC, Oetting WS, Atwood LD, Wilkin DJ, King RA. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998a;63(5):1419–1424. doi: 10.1086/302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TL, Ronan SM, Drahozal LA, Wildenberg SC, Alvear AB, Oetting WS, Atwood LD, Wilkin DJ, King RA. Evidence that a locus for familial high myopia maps to chromosome 18p. Am J Hum Genet. 1998b;63(1):109–119. doi: 10.1086/301907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylbermann R, Landau D, Berson D. The influence of study habits on myopia in Jewish teenagers. J Pediatr Ophthalmol Strabismus. 1993;30(5):319–322. doi: 10.3928/0191-3913-19930901-12. [DOI] [PubMed] [Google Scholar]