Abstract
The transcription factor REST silences neuronal gene expression in non-neuronal cells. In neurons, the protein is sequestered in the cytoplasm in part through binding to huntingtin. Polyglutamine expansions in huntingtin, which causes Huntington's disease (HD), abrogates REST-huntingtin binding. Consequently, REST translocates to the nucleus, occupies RE1 repressor sequences and decreases neuronal gene expression. In this work, we found that levels of several microRNAs (miRNAs) with upstream RE1 sites are decreased in HD patient cortices relative to healthy controls. Interestingly, one of these, the bifunctional brain enriched miR-9/miR-9*, targets two components of the REST complex: miR-9 targets REST and miR-9* targets CoREST. These data provide evidence for a double negative feedback loop between the REST silencing complex and the miRNAs it regulates.
Keywords: miRNA, REST, Huntington's disease, cortex, coordination, RNA interference
Introduction
Huntington's disease (HD) is an autosomal dominant neurodegenerative disease caused by CAG trinucleotide repeat expansion in huntingtin, which encodes Huntingtin (Htt) (The Huntington's Disease Collaborative Research Group, 1993). Patients with HD experience abnormal motor movements, cognitive decline and psychiatric disturbances that frequently result in premature death. Although Htt is ubiquitously expressed, patients with HD show predominantly CNS manifestations. One of the molecular phenotypes in HD patients is transcriptional misregulation in striatum and distinct cortical regions (Hodges et al., 2006). One putative mechanism underlying the transcriptional changes is aberrant cellular distribution of the transcriptional repressor RE1-silencing transcription factor (REST, also known as neuron-restrictive silencer factor, NRSF) (Lunyak et al., 2002; Zuccato et al., 2003, 2007).
REST expression is highest in pluripotent stem cells and decreases upon restriction to neural progenitor cells and subsequently to neurons (Ballas et al., 2005). In mature, healthy neurons, REST is expressed at low levels and primarily sequestered in the cytoplasm in part through interaction with Htt (Zuccato et al., 2003). However, in patients with HD, mutant Htt fails to bind REST, enabling its nuclear translocation (Zuccato et al., 2003). Once in the nucleus, REST can bind RE1 consensus sequences and recruit corepressors including mSin3, REST corepressor 1 (CoREST, also known as RCOR1), and methyl CpG binding protein 2 (MeCP2) (Andrés et al., 1999) to inactivate neuron-specific genes (Zuccato et al., 2003; Conaco et al., 2006). Similarly, REST repressor complex binding to RE1 sequences upstream of miRNA transcripts may alter miRNA expression profiles (Conaco et al., 2006; Klein et al., 2007).
MiRNAs are small noncoding RNAs that participate in post- transcriptional regulation through sequence complementarity to the 3′ untranslated regions (UTRs) of mRNAs. Binding of a miRNA to its target mRNA typically results in its translational repression through mRNA degradation or translational inhibition (Bartel, 2004). Given their essential role in modulation of cellular processes such as cell fate, identity, and function (Sempere et al., 2004; Kim et al., 2007; Makeyev et al., 2007), we screened a panel of predicted REST-regulated miRNAs (Jothi et al., 2008) in samples collected from healthy control and HD patient brains (HD grades 1–4). Our data show miRNA misregulation during HD progression, and moreover, evidence that miR-9/miR-9*, a REST-regulated miRNA, targets two components of the REST silencing complex.
Materials and Methods
Human tissues.
Postmortem brain samples were obtained from the New York Brain Bank (Columbia University, New York, NY), the Harvard Brain Bank (Harvard University, Cambridge, MA), and the New Zealand Neurological Foundation Brain Bank (University of Auckland, Auckland, New Zealand) with informed consent from families. Tissues were collected from Brodmann's area 4 (BA4) cortex of healthy control (n = 7; ages 46–62) and HD grade 1 (n = 7; ages 42–70), 2 (n = 6; ages 45–78), 3 (n = 2; ages 60–65), and 4 (n = 4; 53–60). CAG repeat length for controls was 16–21; HD grade 1–4 was 37–46. Postmortem time for brain samples was 2–20 h.
miRNA expression profiling.
RNA was collected from cortex using TRIzoL (Invitrogen). Expression profiles for the mature miRNAs were obtained by quantitative PCR (QPCR) using TaqMan Array Human MiRNA Panel v1.0 (TLDA) or TaqMan MiRNA Assays (Applied Biosystems; ABI) according to the manufacture on an ABI Prism 7900HT. Mature miRNAs were normalized to RNU48 and are shown as fold change relative to controls.
Cloning 3′ UTRs and endogenous miRNAs.
The 3′ UTR of REST (NCBI 36, Oct 2005) (supplemental Fig. S2, available at www.jneurosci.org as supplemental material) was verified in NT2, Neuroblastoma, A549, HEK 293 and HeLa cells by sequencing of cDNAs prepared from TRIzoL (Invitrogen) isolated RNA, then PCR amplified using Expand High Fidelity DNA polymerase (RocheApplied Science) and 3′ UTR specific primers (see supplemental methods, available at www.jneurosci.org). Both REST and CoREST 3′ UTRs were cloned from HEK 293 cDNAs into psiCHECK-2 (Promega) downstream of Renilla luciferase. Human miR-9–3 loci were amplified from HEK 293 genomic DNA and cloned into a Zero Blunt TOPO (Invitrogen) containing a Pol III mouse U6 promoter. mU6 miR-124a-1 and miR-132 hairpins were cloned as described above.
Perfect target controls.
Perfect target (PT) controls for miR-9, 9* and 132 were made by complexing 8 μm sense and antisense oligonucleotides containing a site with perfect complementarity to the mature miRNA and random flanking sequence. PCR cycles were 2 times at 94°C 15 s, 67°C 30 s, 72°C 1 min and extension for 1 min, 72°C, then cloned downstream of Renilla luciferase.
Cell culture, transfections, and luciferase assays.
HEK 293 and NT2 [NTERA-2 c1.D1, lot number 4742175 from ATCC (CFL-1973)] cells were cultured in DMEM supplemented with 10% heat-inactivated fetal calf serum at 37°C and 5% CO2. Cells were transfected in triplicate on 24-well plates with Lipofectamine 2000 (Invitrogen) per the instructions of the manufacturer, with 4 ng of psiCHECK-3′ UTR plasmid and 4–400 ng of mU6 miRNA or pre-miR-9, 9* or negative controls (Ambion). Anti-miRNA rescue experiments were performed by cotransfecting psiCHECK-3′ UTR plasmids in conjunction with mU6 miR-9/miR-9* and anti-miRs 9–3, 9–3* or negative controls (Ambion). For NT2 differentiation, 0.01 mm all-trans retinoic acid (Sigma-Aldrich) was added to the media for 5 d.
Luciferase was measured 24 h later on a moon light luminometer (PharMingen). Renilla luciferase was normalized to firefly luciferase and is shown relative to controls.
“Target site protectors” (TSPs) were designed antisense to predicted 3′UTR MRE of REST and CoREST (supplemental methods, available at www.jneurosci.org as supplemental material). Optimized TSPs were designed with M. Behlke, Integrated DNA Technologies and used at 0.01–0.1 nm.
siRNAs.
HEK 293 and NT2 cells were transfected with 5 nm silencer select siRNAs to REST (s11933) (si-REST), or a scrambled negative control (si-NEG) (Ambion) as above. Total RNA was harvested 48 h later with TRIzol (Invitrogen), reverse transcribed with High Capacity cDNA Reverse Transcription Kit, then analyzed by QPCR using REST or 18 s rRNA TaqMan Gene Expression Assays (ABI).
Western blots.
Protein was harvested using RIPA buffer (Pierce) and 1× protease inhibitor using standard techniques and quantified using DC Protein Assay (Bio-Rad). Protein extracts were separated on a 7% acrylamide gel and transferred to Immobilon PDVF transfer membranes (Millipore). Primary antibodies to REST/NRSF (1:500; ab21635), CoREST (1:1000; ab56165) (both Abcam) and β-actin (1:10,000; Sigma) were used. Blots were developed using ECL Plus Western Blotting Detection System (GE Healthcare) and quantified by AlphaEase FC (Alpha Innotech, version 5.0.1).
Statistical analysis.
Data were analyzed by one-way ANOVA, followed by Dunnett's post hoc analysis (GraphPad InStat, GraphPad Software, version 3.06).
Results
miRNA expression changes with HD progression
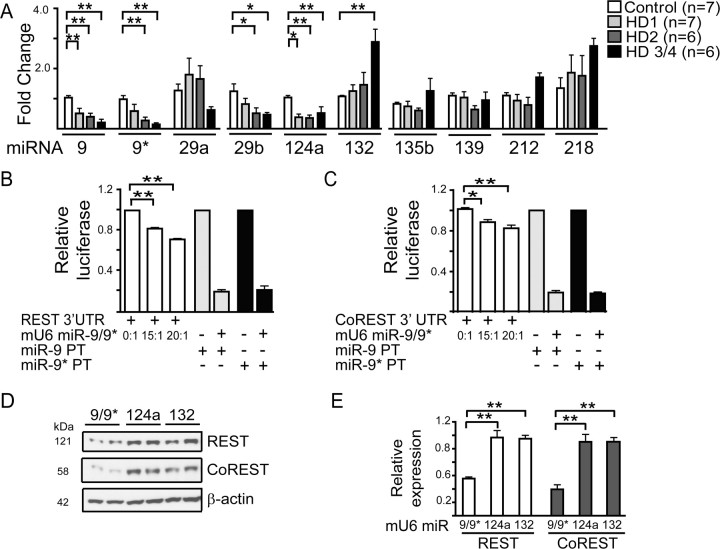
To determine whether miRNAs are affected with HD disease progression, we isolated total RNA from control and HD grade 1–4 brain samples. We specifically chose Brodmann's area 4 (BA4) cortex because prior work indicates widespread transcriptional changes in this region in HD patients (Hodges et al., 2006). We found five predicted REST-regulated miRs to be significantly different with increasing HD grade: miR-9 (F(3,22) = 9.876, p = 0.0003), miR-9* (F(3,22) = 6.799, p = 0.0021), miR-29b (F(3,22) = 3.658, p = 0.0281), miR-124a (F(3,16) = 7.833, p = 0.001), and miR-132 (F(3,22) = 7.611 p = 0.0011) (Fig. 1A). For miR-139, miR-135b, and miR-212 no significant differences were observed with increasing disease severity, while miR-218 demonstrated a trend toward increasing expression with disease grade (Fig. 1A). Previous work by Johnson and colleagues measured precursor miRNA levels in HD brain of unknown disease grade (Johnson et al., 2008). Interestingly, and in contrast to our findings, their work showed no change in miR-124a, elevated levels of miR-29a, and decreased levels of miR-132. We saw a trend toward increased miR-29a in tissue from early stage HD, followed by diminished levels. Also, we found no change in miR-132 at stages 1 and 2, but significant upregulation later. The differences in precursor (Johnson et al., 2008) versus mature miRNA levels (this study) is not surprising given recent reports describing posttranscriptional regulation of miRNAs (Obernosterer et al., 2006; Lee et al., 2008).
Figure 1.
Several REST-regulated miRNA levels change with HD progression, and miR-9/9* targets a silencing complex. A, Longitudinal expression profiles of REST-regulated miRNAs in HD and healthy control cortex (BA4) HD grade 1, 2, and 3/4 as measured by QPCR. Mature miRNA levels in HD brain relative to controls (*p < 0.05, **p < 0.01). B, C, miR-9/9* reduces expression of reporter proteins containing the 3′ UTR of REST or CoREST. B, HEK 293 cells were cotransfected with a vector expressing miR-9/9* (mU6 miR-9/9*) and REST 3′ UTR luciferase reporters. Relative luciferase activity was measured 24 h later. Increasing doses of mU6 miR-9/9* relative to target caused a dose-dependent decrease in relative luciferase. Control vectors containing a single site with perfect complementarity to mature miR-9 (miR-9-PT) or miR-9* (miR-9*-PT) were included as positive controls (n = 3). C, Experiments as in (B) with CoREST 3′ UTR luciferase reporters (n = 3). D, E, Transfection of HEK 293 cells with mU6 miR-9/9* reduces both REST and CoREST protein expression levels 24 h later. D, Representative Western blots and densitometry for (E) REST (n = 5) and CoREST (n = 5). Histograms show mean ±SEM after normalization to β-actin levels. *p < 0.05, **p < 0.01.
We next used a QPCR based miRNA array platform to evaluate the expression profiles of 365 mature miRNAs in BA4 cortex from control and early stage HD (grades 1 and 2). We found an additional 7 miRNAs that were decreased in HD1 and/or HD2 samples (supplemental Table 1, available at www.jneurosci.org as supplemental material). Also, miR-196a and miR-486 were significantly increased by nearly sixfold and threefold, respectively, in HD1 samples relative to controls. These results provide additional evidence for differential regulation of select miRNAs in HD.
REST and CoREST contain predicted miR-9 and 9* regulatory sites
MiR-9 and miR-9*, which decreased early in HD (Fig. 1A), are processed from the same primary transcript from 3 genomic loci (miR-9–1, 9–2 and 9–3); miR-9–1 and miR-9–3 both have upstream RE1 sequences that can be occupied by REST (Conaco et al., 2006; Johnson et al., 2008). We amplified the human miR-9–3 locus from HEK 293 cells and cloned it downstream of the mouse U6 promoter (mU6 miR-9/9*) for heterologous expression of miR-9/miR-9*. The cloned mU6 miR-9/9* plasmid resulted in efficient expression and processing of both miR-9 and miR-9* as determined by knockdown of perfect target controls (PT) (supplemental Fig. S1, available at www.jneurosci.org as supplemental material).
Among the targets predicted for miR-9 and 9* are REST and CoREST [TargetScan 4.1 (Lewis et al., 2003)]. This is interesting given the putative role of the REST repressor complex in misregulation of genes in HD (Zuccato et al., 2003). The 3′ UTRs of REST (supplemental Fig. S2, available at www.jneurosci.org as supplemental material) and CoREST (supplemental Fig. S3, available at www.jneurosci.org as supplemental material) contain miRNA recognition elements (MRE) for miR-9, miR-9*, miR-132 and miR-124a (CoREST only) (Wu and Xie, 2006). As a first test of miR-9/9* and REST or CoREST interaction, we cloned the 3′ UTR of REST and CoREST downstream from the gene encoding Renilla luciferase. Cotransfection of mU6 miR-9/9* with REST or CoREST-3′ UTR reporter constructs significantly decreased Renilla luciferase activity compared with controls (F(2,6) = 333.08, p = 0.0001 and F(2,6) = 16.134, p = 0.0034 for REST and CoREST, respectively) (Fig. 1B,C). Consistent with these reporter-based assays, over expression of mU6 miR-9/9*, but not mU6 miR-124a or 132, in HEK 293 cells significantly reduced REST (F(2,12) = 26.006, p = 0.0001) and CoREST (F(2,12) = 24.017, p = 0.0001) expression (Fig. 1D,E). Therefore, the predicted MREs in CoREST and REST for miR-9/9*, but not miR-124a and miR-132, are functional (Fig. 1D,E).
miR-9 targets REST
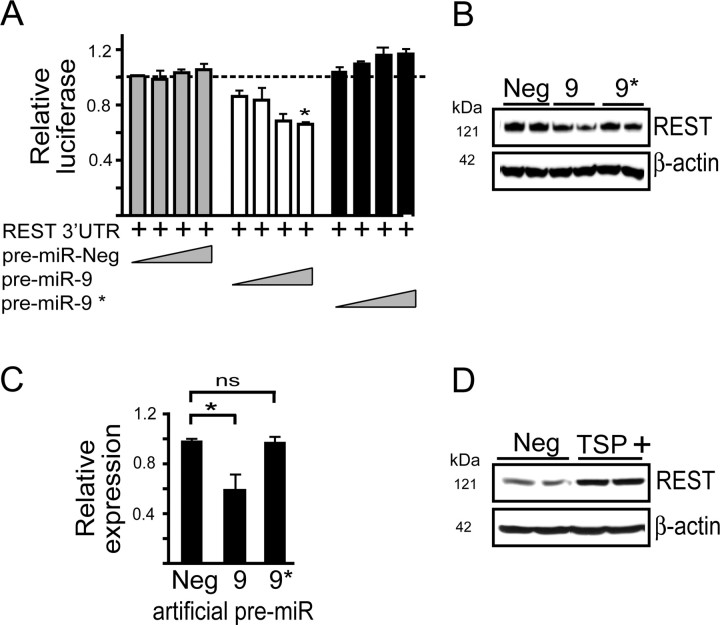
The full-length 3′ UTR of REST (supplemental Fig. S2, available at www.jneurosci.org as supplemental material) and CoREST (supplemental Fig S3, available at www.jneurosci.org as supplemental material) contain several predicted conserved MRE sites for miR-9 and miR-9*. Because mU6 miR-9/9* produces mature miR-9 and miR-9* upon processing (supplemental Fig. S1, available at www.jneurosci.org as supplemental material), we used artificial precursor miRNAs (pre-miR) that release a single mature miRNA (in this case miR-9 or miR-9*) and anti-miRNAs (which are antisense to each mature miRNA) to determine whether there is preferential activity for miR-9, miR-9*, or both on REST and/or CoREST. Cotransfection of 15 nm pre-miR-9 with luciferase reporters containing the 3′ UTR of REST significantly reduced reporter gene expression (F(2,9) = 35.524, p < 0.0001) in contrast to pre-miR-9* or a negative control pre-miR (Fig. 2A). [Specificity of the pre-miRs was confirmed using reporter constructs containing perfect miRNA target sites (supplemental Fig. S4, available at www.jneurosci.org as supplemental material)]. Also, anti-miR-9, but not anti-miR-9* rescued the inhibition of Renilla luciferase activity seen upon cotransfecting mU6 miR-9/9* with the REST 3′ UTR reporter plasmid (F(2,6) = 19.539, p = 0.0024) (supplemental Fig. S5, available at www.jneurosci.org as supplemental material). To extend the reporter studies to endogenous REST, we transfected HEK 293 cells with pre-miR-9. This resulted in significant knockdown of endogenous REST protein expression (F(2,9) = 7.552, p = 0.0119) compared with pre-miR-9* or the pre-miR negative control (Fig. 2B,C). Together, these results suggest that the miR-9 predicted MREs in the 3′ UTR of REST are functional, in contrast to the miR-9* predicted MREs.
Figure 2.
miR-9 targets the 3′ UTR of REST. A, Cotransfection of HEK 293 cells with luciferase reporters containing the 3′ UTR of REST in the presence of increasing concentrations (0.1, 1.0, 5.0, or 15.0 nm) of artificial pre-miR-9 reduces relative luciferase activity (n = 4). B, C, Knockdown of REST following transfection of HEK 293 cells with pre-miR-9 (30 nm). A representative Western blot (B) and the densitometry from (n = 4) independent experiments (C) after normalization to β-actin. Data are mean ±SEM; *p < 0.05, **p < 0.01. D, A representative Western blot showing that target site protectors (TSP) designed to block the predicted miR-9 binding site of REST (TSP +) rescues mU6 miR-9/9*-mediated knockdown of REST. HEK 293 cells were cotransfected with mU6 miR-9/9* and TSP+ or TSP negative control (Neg) and REST levels measured in cell lysates 24 h later. Densitometry from n = 4 independent experiments indicate increased REST in TSP+ (0.97 ± 0.065)-treated versus TSP Neg (0.54 ± 0.212)-treated cells.
We next designed a LNA-modified oligonucleotide as a TSP, with sequence complementarity to the predicted miR-9 binding site in the REST 3′ UTR (supplemental Fig. S6, available at www.jneurosci.org as supplemental material). TSPs contain sequences antisense to the MRE along with flanking sequences and serve to block specific miRNA-target site interactions within a 3′ UTR. These are distinct from anti-miRs, which impair all miRNA-target interactions. Cotransfection of HEK 293 cells with the REST 3′ UTR reporter plasmid, the mU6 miR-9/9* plasmid and TSP+ (targeting the terminal MRE in REST) (supplemental Fig. S2, available at www.jneurosci.org as supplemental material) showed protection from mRNA-mediated silencing compared with mU6 miR-9/9*- treated cells (F(2,9) = 38.395, p < 0.0001) (supplemental Fig. S6, available at www.jneurosci.org as supplemental material). Consistent with these observations, REST levels in HEK 293 cells, which decreased upon mU6 miR-9/9* transfection (Fig. 1D,E) was rescued by TSP+ (Student's t test, p < 0.05), but not a control TSP (TSP Neg) (Fig. 2D).
miR-9* targets CoREST
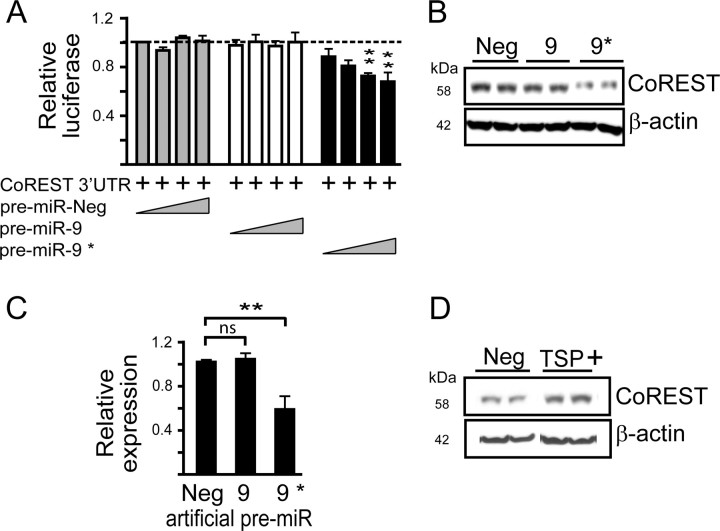
We used similar approaches to ascertain whether miR-9, miR-9* or both were functional in regulating CoREST. Cotransfection of the CoREST 3′ UTR reporter plasmid and 5 or 15 nm pre-miR-9* reduced reporter expression compared with controls (F(2,11) = 41.612, p = 0.0001 and F(2,11) = 7.423, p = 0.0091, respectively), unlike miR-9 (Fig. 3A). Also, only pre-miR-9* significantly reduced endogenous CoREST expression in HEK 293 cells (F(2,9) = 11.173, p = 0.0036) (Fig. 3B,C). Also, reduced CoREST expression after transfection of HEK 293 cells with mU6 miR-9/9* (Fig. 1D,E) was relieved upon anti-miR-9* cotransfection (supplemental Fig. S7, available at www.jneurosci.org as supplemental material).
Figure 3.
miR-9* targets the 3′ UTR of CoREST. A, Cotransfection of HEK 293 cells with luciferase reporters containing the 3′ UTR of CoREST in the presence of increasing concentrations (0.1, 1.0, 5.0, or 15.0 nm) of artificial pre-miR-9* reduces relative luciferase activity (n = 5). B, C, CoREST levels are reduced following transfection of HEK 293 cells with pre-miR-9* (30 nm). A representative Western blot (B) and densitometry (n = 4) (C) after normalized β-actin levels. Data are mean ± SEM; *p < 0.05, **p < 0.01. D, TSP designed the predicted miR-9* binding site of CoREST (TSP +) rescues pre-miR-9* mediated knockdown (B). HEK 293 cells were cotransfected with pre-miR-9* (15 nm) and TSP+ or TSP Neg and CoREST levels measured in cell lysates by Western blot 24 h later. Densitometry from n = 4 independent experiments indicate increased CoREST in TSP+ (1.02 ± 0.05)-treated versus TSP Neg (0.565 ± 0.08)-treated cells.
We next used TSPs against the conserved predicted miR-9* binding site in CoREST to test its functionality (supplemental Fig. S8, available at www.jneurosci.org as supplemental material). Cotransfection of mU6 miR-9/9* in the presence of TSP+ significantly rescued reporter gene expression compared with mU6 miR-9/9*-treated cells (F(2,6) = 32.882, p = 0.0006) (supplemental Fig. S8, available at www.jneurosci.org as supplemental material). Endogenous CoREST protein expression, which was reduced after pre-miR-9* transfection, increased upon cotransfection of TSP+ (Student's t test, p < 0.05) but not TSP Neg (Fig. 3D). These data provide support for a miR-9* MRE in regulating CoREST.
Finally, we tested how reducing REST expression impacts endogenous miR-9 levels in HEK 293 and NT2 cells. In both cell lines, short-term reduction in REST expression resulted in enhanced miR-9 expression (p < 0.05) and processing as determined by measuring level of mature miR (supplemental Fig. S9, available at www.jneurosci.org as supplemental material). These findings provide support to previous studies which showed REST occupancy on miR-9–1 and miR-9–3 (Conaco et al., 2006) and provide functional evidence for a negative feed back look between components of the REST complex and REST-regulated miRNAs.
Discussion
The transcriptional repressor REST is mislocalized in brains of patients with HD, with increased nuclear localization and occupancy at RE1 sites compared with healthy controls (Zuccato et al., 2003, 2007). REST interacts with CoREST and other transcriptional repressors to regulate neuronal gene expression and lineage commitment through its direct control of both mRNA and, as shown more recently, miRNA transcripts (Vo et al., 2005; Conaco et al., 2006; Mortazavi et al., 2006; Visvanathan et al., 2007). Indeed, canonical and noncanonical REST binding motifs have recently been mapped in close proximity to 22 miRNA sites in the human genome, including several of which are located within close proximity to neuron enriched miRNAs (Bruce et al., 2004; Jothi et al., 2008).
In this study we found several miRNAs to be significantly misregulated among RNA harvested from HD brains compared with those of healthy controls. Of these, the bifunctional miR-9/miR-9* targets two components of the REST repressor complex: miR-9 targets REST and miR-9* targets CoREST. MiR-132 has been shown to target MeCP2 (Klein et al., 2007), which can interact with REST and CoREST to suppress transcription (Lunyak et al., 2002). Double negative feedback networks have been reported in other systems where critical balancing of multiple factors is essential (Tsang et al., 2007).
Recent studies on the brain enriched miR-9 have revealed its importance in development and lineage commitment (Sempere et al., 2004; Krichevsky et al., 2006). In zebrafish, miR-9 is important for midbrain-hindbrain boundary definition (Leucht et al., 2008). In mouse cortical development, miR-9 appears necessary for appropriate differentiation of Cajal-Retzius cells (Shibata et al., 2008). In addition to miR-9/miR-9*, mature miR-124a is significantly reduced in HD cortices. miR-124 promotes neuronal differentiation, in part through targeting of a splicing factor (Makeyev et al., 2007). Also, over-expression of miR-124 in non-neuronal cells induces gene expression profiles to a neuronal phenotype (Lim et al., 2005). Interestingly, genes related to neurogenesis are highly overrepresented among the genes changed in expression in the cortex (BA4) of HD patients (Hodges et al., 2006). This may reflect a dedifferentiation mediated by aberrant REST localization, because REST mRNA levels do not change significantly in HD (Hodges et al., 2006), as well as global changes in miR-9/9* and miR-124a target levels. Whether diminished levels of miR-9/9*, miR-124a and other miRNAs in HD cortex result from, or contribute to, the dedifferentiation profile is currently under study.
In summary, we show that several REST-regulated miRNAs decrease with disease progression in HD brain. We demonstrate that among these, miR-9 and miR-9* target REST and CoREST, respectively. Our findings, together with recent work showing that REST can be regulated by ubiquitin-mediated proteolysis (Westbrook et al., 2008) underscore the importance of maintaining appropriate levels and intracellular localization (Zuccato et al., 2003) of this repressor complex for appropriate gene transcription and lineage commitment.
Footnotes
This work was supported by the Hereditary Disease Foundation and National Institutes of Health Grants HD 44093 and NS 050210. A.N.P. designed and performed experiments and wrote this manuscript; Y.X. performed experiments and edited this manuscript; S.Q.H. provided constructs and initiated REST-regulated miRNA studies; L.J. provided samples and edited this manuscript; B.L.D. initiated the studies, designed research, and wrote this manuscript. We thank G. Liu, S. Fineberg and B. Gilmore for technical expertise and Dr. Kathryn Chaloner, Professor and Head Department of Biostatistics, for assistance with statistical analysis.
The authors declare no competing financial interests.
References
- AndrésME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G (1999) CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci U S A 96:9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BallasN, Grunseich C, Lu DD, Speh JC, Mandel G (2005) REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121:645–657. [DOI] [PubMed] [Google Scholar]
- BartelDP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. [DOI] [PubMed] [Google Scholar]
- BruceAW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Göttgens B, Buckley NJ (2004) Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A 101:10458–10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ConacoC, Otto S, Han JJ, Mandel G (2006) Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A 103:2422–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HodgesA, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, Elliston LA, Hartog C, Goldstein DR, Thu D, Hollingsworth ZR, Collin F, Synek B, Holmans PA, Young AB, Wexler NS, Delorenzi M, Kooperberg C, Augood SJ, Faull RL, Olson JM, Jones L, Luthi-Carter R (2006) Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet 15:965–977. [DOI] [PubMed] [Google Scholar]
- JohnsonR, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ (2008) A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol Dis 29:438–445. [DOI] [PubMed] [Google Scholar]
- JothiR, Cuddapah S, Barski A, Cui K, Zhao K (2008) Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res 36:5221–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KimJ, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A (2007) A MicroRNA feedback circuit in midbrain dopamine neurons. Science 317:1220–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KleinME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH (2007) Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci 10:1513–1514. [DOI] [PubMed] [Google Scholar]
- KrichevskyAM, Sonntag KC, Isacson O, Kosik KS (2006) Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24:857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeeEJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD (2008) Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 14:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeuchtC, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L (2008) MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci 11:641–648. [DOI] [PubMed] [Google Scholar]
- LewisBP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115:787–798. [DOI] [PubMed] [Google Scholar]
- LimLP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773. [DOI] [PubMed] [Google Scholar]
- LunyakVV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG (2002) Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science 298:1747–1752. [DOI] [PubMed] [Google Scholar]
- MakeyevEV, Zhang J, Carrasco MA, Maniatis T (2007) The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27:435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MortazaviA, Leeper Thompson EC, Garcia ST, Myers RM, Wold B (2006) Comparative genomics modeling of the NRSF/REST repressor network: from single conserved sites to genome-wide repertoire. Genome Res 16:1208–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ObernostererG, Leuschner PJ, Alenius M, Martinez J (2006) Post-transcriptional regulation of microRNA expression. RNA 12:1161–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SempereLF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ShibataM, Kurokawa D, Nakao H, Ohmura T, Aizawa S (2008) MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J Neurosci 28:10415–10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72:971–983. [DOI] [PubMed] [Google Scholar]
- TsangJ, Zhu J, van Oudenaarden A (2007) MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 26:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VisvanathanJ, Lee S, Lee B, Lee JW, Lee SK (2007) The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev 21:744–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VoN, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S (2005) A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A 102:16426–16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WestbrookTF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW, Elledge SJ (2008) SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WuJ, Xie X (2006) Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol 7:R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZuccatoC, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E (2003) Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet 35:76–83. [DOI] [PubMed] [Google Scholar]
- ZuccatoC, Belyaev N, Conforti P, Ooi L, Tartari M, Papadimou E, MacDonald M, Fossale E, Zeitlin S, Buckley N, Cattaneo E (2007) Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington's disease. J Neurosci 27:6972–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]