Abstract
A recent crystal structure of HIV-1 protease (HIVp) was the first to experimentally observe a ligand targeting an open-flap conformation. Researchers studying a symmetric pyrrolidine inhibitor found that two ligands co-crystallized with the protease, forcing an unusual configuration and unique crystallographic contacts. One molecule is centered in the traditional binding site (α pose), and the other binds between the flaps (β pose). The ligands stack against each other in a region termed the “eye” site. Ligands bound to the eye site should prevent flap closure, but it is unclear if the pyrrolidine inhibitors or the crystal packing are causing the open state. Molecular dynamics simulations were used to examine the solution-state behavior of three possible binding modes: the ternary complex of HIVp+αβ and the binary complexes, HIVp+α and HIVp+β. We show that HIVp+α is the most stable of the three states. During conformational sampling, α takes an asymmetric binding pose, with one naphthyl ring occupying the eye site and the other reoriented down to occupy positions seen with traditional inhibitors. This finding supports previous studies that reveal a requirement for asymmetric binding at the eye site. In fact, if the α pose is modified to splay both naphthyl rings across the binding site like traditional inhibitors, one ring consistently flips to occupy the eye site. Our simulations reveal that interactions to the eye site encourage a conformationally-restrained state, and understanding those contacts may aid the design of ligands to specifically target alternate conformations of the protease.
Keywords: Molecular dynamics, protein flexibility, drug design, AIDS, ligand binding
Introduction
HIV-1 protease (HIVp) remains an important pharmaceutical target. Despite the existence of ten protease inhibitors (PIs) in clinical use1, there is little variety in their mechanism of action. Typical PIs are pseudosymmetric and compete with the substrate for binding at the base of the active site. Unfortunately, even the most recent protease inhibitors suffer from taxing sideeffects, poor pharmacokinetic properties, and development of drug resistance.2,3 There remains a need to discover novel therapeutics, especially compounds that bind in non-traditional modes and have the potential to target both wild-type and multi-drug-resistant (MDR) mutants and overcome resistance mechanisms.
The protease is a C2 symmetric dimer with the characteristic catalytic acids at the base of its active site. The active site is covered by two antiparallel β-hairpin turns, called the flaps (residues 43–58/43′–58′). The conformation of these flaps has been well-studied, and researchers have linked flap motion to activity of the protease.4 The flaps exist in three major conformations: open, semi-open, and closed. Structural studies have shown the semi-open conformation to be most populated in the native, apo state.5–8 The flaps shift 5–7 Å in position upon ligand binding and assist in its proper placement within the active site. Studies have shown that there is little difference in free energy between the three flap conformations.9 Researchers have shown that differences in flap mobility are a potential contribution to the mechanism of multi-drug resistance for certain HIVp mutants.10 Therefore, the possibility exists that protease activity can be easily affected by controlling flap conformation. By targeting HIVp through allosteric inhibition, it may be possible to avoid some of the difficulties that plague traditional PIs. Therefore, it is essential to investigate how compounds can alter flap behavior.
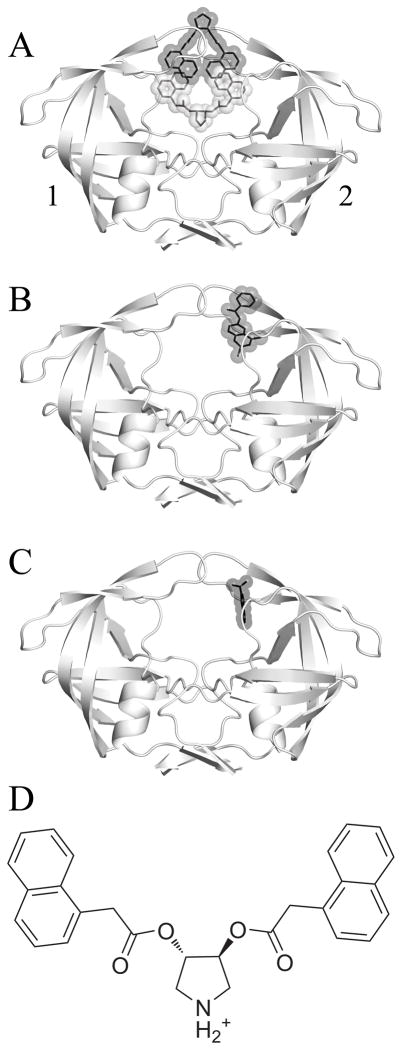
Böttcher et al. recently reported an interesting crystal structure demonstrating the simultaneous binding of two symmetric pyrrolidine diester inhibitors to the open-flap conformation of HIVp (PDB ID 3BC4).11 One is bound bridging the traditional active site and the “eye” site12, while the other is bound between the flaps (Figure 1a). The bridged binding pose places two napthyl rings into each eye site (Figure 1b), lending support to the possibility of targeting this for inhibitor design. Although the authors did not propose that this was necessarily a 2:1 complex in solution phase, the crystal structure merits investigation.
Figure 1.
A) The crystallized HIV-1 protease uniquely bound by two identical inhibitors, with pose α colored in grey and pose β in black. B) The crystal structure 3BC4 with Damm compound 112 (black) bound at the eye site. C) The 5-Nitroindole fragment (black) crystallized in the eye site by Perryman et al.28 D) A 2-dimensional representation of the pyrrolidine inhibitor that was co-crystallized with 3BC4. The affinity (Ki) of the compound for HIVp was measured by Klebe and coworkers as 20μM (WT), 41μM (I50V), and 4.5μM (I84V).11 For the following figures, we have used a convention of orienting the complex so that a naphthyl occupies the eye position on the right (ie. Monomer 2). We are labeling the monomers as “1” and “2” instead of “A” and “B” to avoid confusion with the α and β notation for the ligands.
We originally proposed the eye site as a possible new mode of HIVp inhibition.12 Our interest in designing compounds to target the eye site motivated us to study the conformational states occupied by this receptor-ligand complex in solution. Our goal is to explore the effect of these inhibitors on the conformation of the flaps. It is important to determine whether the conformation seen in the crystal structure is also seen in simulation when symmetry-related contacts of the crystal structure are absent. Retaining the notation used by Böttcher et al., we examined the two inhibitors bound to HIVp from the crystal structure (αβ), as well as a single inhibitor bound bridging the active site (α) or in the alternate position against the flaps (β). We hypothesize that the β pose is not stable, nor is the ternary crystal complex, given the poor contacts available to β without the influence of crystal contacts.
Methods
Our molecular dynamics protocol was based on work by Meagher et al.13 Our simulations were based on the protein structure crystallized by Böttcher et al. (PDB ID 3BC4). PyMOL14 was used to propagate the asymmetric unit cell. A combination of PyMOL and MolProbity15 was used to check/flip protonation states while MOE16 was used to modify the number of bound ligands. The catalytic aspartic acids were both deprotonated, as is appropriate in the presence of the positively charged ligand.
For each inhibitor-binding state (αβ, α-only, or β-only), eight independent, explicit-solvent simulations were performed. Parameters for the inhibitor were generated in antechamber with the Gaff force field17 and AM1-BCC charges18. Hydrogens were built in the tleap module of AMBER19. TIP3P waters20 were added as an orthogonal box with a 12 Å buffer to solvate the system. APBS-1.0.021 via the plugin for PyMOL22 was used to calculate an electrostatic surface 10 Å from the vdw surface of the protein, and chloride ions were placed at the most electropositive regions to neutralize the +4e charge of the protein and the +1e charge of each ligand. Molecular dynamics were performed in the sander module of AMBER using the ff99sb force field23 and a timestep of 2 fs. A non-bonded cutoff was applied at 10 Å. Particle Mesh Ewald24 was implemented, and bonds to hydrogen were constrained with SHAKE. Water was equilibrated prior to complete system equilibration to prevent protein collapse.13 Following minimization of hydrogens then side chains then the full system, equilibration was performed with a gradual removal of backbone restraints to achieve a stable trajectory. Over 500ps, the protein-ligand-solvent system was gradually heated from 10K to 310K, and backbone restraints were gradually softened from 2.0 kcal/mol*Å to 0.1 kcal/mol*Å in a total of five steps. After a two nanosecond (ns) equilibration of the protein with no restraints, the production phase lasted 25ns. This resulted in a total of 8 individual simulations of 25ns for each of the three sets of bound systems. Therefore, 200ns of total production time was collected for the HIVp+ α complex, HIVp+β complex, and ternary HIVp+ αβ complex.
Trajectories were analyzed using the AMBERTOOLS package. Ptraj allows for clustering simulations to determine the most prevalent conformations sampled within a specified time period.25 The trajectories were centered and aligned to the core of the protease (residues 1–45,55–99,1′–45′,55′–99′). The last 5 ns from each simulation for each complex were then sieved and clustered together with reference to the initial structure. Several clustering protocols were performed to determine the optimal algorithm and family size based on measures of the Davies-Bouldin index (DBI)26, pseudo F-statistic (pSF)27, and percentage of variance (SSR/SST). Clustering the simulations into ten families based on the average-linkage algorithm was judged to give the best performance. Ptraj was also used to evaluate the degree of flap opening, flap curling, ligand placement, and protein stability (see supporting information).
Results and Discussion
The recent crystal structure from Klebe and coworkers provided the first experimental support of the eye site as a target.11 The naphthyl rings of the two inhibitors crystallized into each eye site, confining the flaps to the semi-open conformation. Due to the implications of crystal packing effects, we performed MD simulations to determine the conformational behavior of both 1:1 complexes and the 2:1 complex in solution. Of interest in our study was the stability of the different potential complexes, the impact of the inhibitor(s) on flap conformation, and the potential for selective binding at the eye site region.
The impact of bound inhibitors at positions α, β, and αβ was examined over a series of eight unique simulations for each ligand pose. Simulations of α-only and β-only were examined as representatives of possible 1:1 complexes as compared to the 2:1 HIVp+αβ complex. The impact of inhibitor binding on backbone stability over the course of each simulation was measured by determining the Cα RMSD of the core residues from the initial crystal structure (see SI Figure S1). For the three different systems, the RMSD of the protease core is 1.65±0.28 Å (HIVp+αβ), 2.25±0.35 Å (HIVp+β), and 1.81±0.25 Å (HIVp+α), signifying core stability. The flaps were mobile as expected (see SI Figures S2–S4).
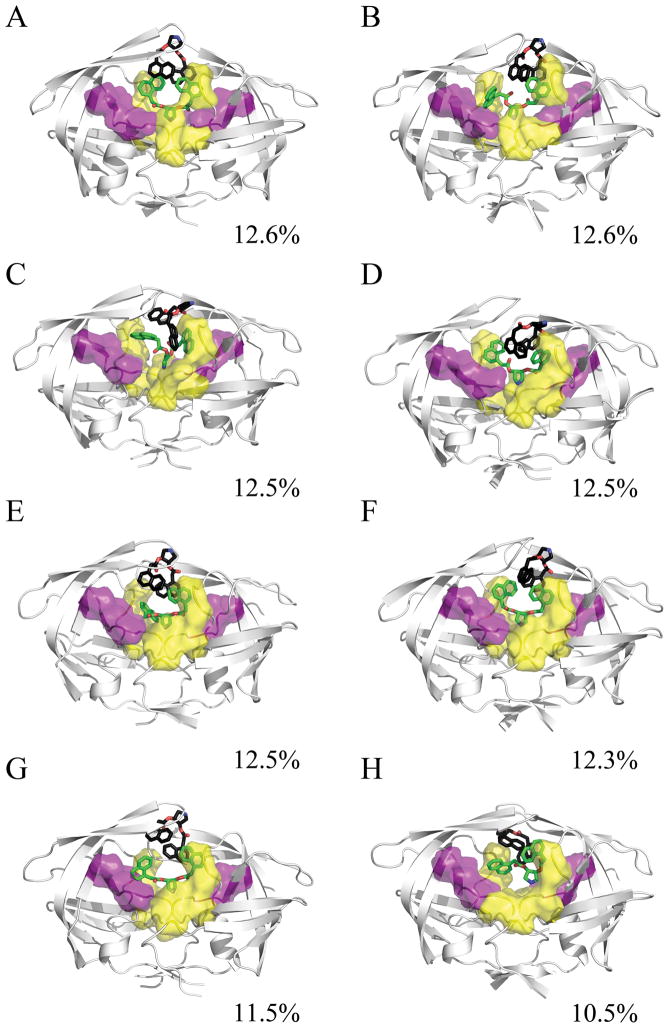
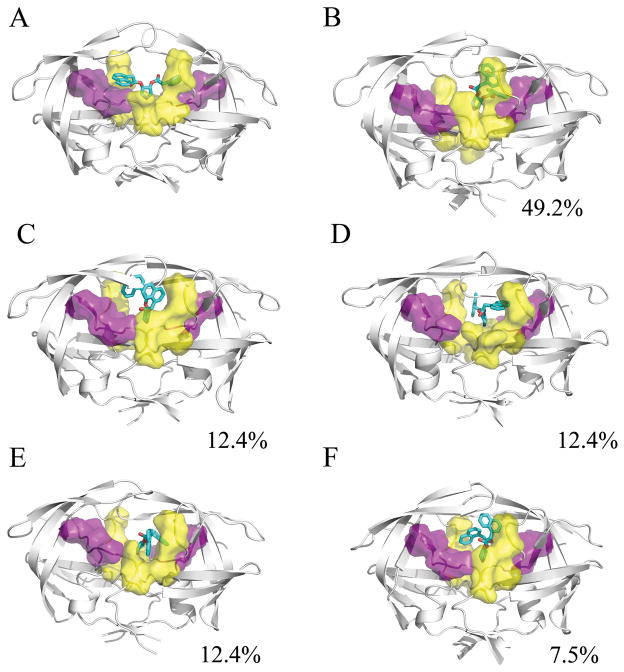
The impact of the ligands on the conformational ensemble was examined through clustering of the conformations sampled over the simulation trajectory. The atomic fluctuations of the protein for the complete simulation were calculated with ptraj to define the stable core residues. The trajectory was imaged and rms-fit to the protease backbone, and then ptraj was used to cluster the heavy atoms of the stable protease core and the ligand over the last 5ns of each 25ns trajectory. Clustering the final 10ns of the trajectory together required too much system memory; however, conformations observed from the last 10ns of individual runs were in agreement with clustering over all runs. A total of 10 families were generated using the average-linkage algorithm. This was accomplished for all simulations of the HIVp+αβ complex, the HIVp+α complex, and the HIVp+β complex. The representative structures were then examined to determine the similarity between binding modes among the families. In addition, calculations were performed to assess the stability of the bound ligands over time (see SI Figures S5–S6). The RMSD of each ligand was calculated against the average ligand position as well as the crystallographic pose. All values were calculated for the final 5ns of each trajectory. HIVp+αβ. The 2:1 complex of HIVp+αβ is unstable. The representative conformations illustrate a wide range of motion sampled by the β ligand, and a moderate amount of sampling by the α ligand. Compared to the average ligand position, the RMSD of the α ligand over the last 5ns ranged from 1.04 to 15.41 Å and β ligand ranged from 1.07 to 28.28 Å. The large fluctuation in position of the ligands illustrates the instability of the HIVp+αβ complex and hints at the instability of the binary HIVp+β complex as well. Analysis of the conformations present in representative families show that in our HIVp+αβ simulations, the α ligand has one naphthyl that occupies the eye site and the other naphthyl occupies the S1 or S2 pocket in approximately 62% of the trajectory sampled (Figure 2). The β ligand in these simulations is quite varied in position; contacts are typically maintained between the pyrrolidine amino group and flap tips or solution.
Figure 2.
Representative structures from the MD simulation of the HIVp+αβ complex, taken from the last 5ns of each 25ns trajectory. The α ligand is shown in green, the β ligand is shown in black, the S1/S1′ site is shown in yellow, and the S2/S2′ site is shown in purple. The conformational families demonstrate the instability of the 2:1 bound complex. The pyrrolidine ligands find a wide variety of ways to interact with the protease flaps, S1/S1′, S2/S2′, and/or the eye site.
Since our interest lay in understanding the impact of these inhibitors on flap conformation, we quantified flap motion over time. A common standard for evaluating flap conformation is the distance between the flap tips (Ile50/50′) and the catalytic aspartic acids (Asp25/25′).10 A typical distance for the closed flap form, based on the crystal structure 1PRO, is 14.1 Å. A typical distance for the semi-open form, based on the crystal structure 1HHP, is 17.8 Å. Over the last 5ns, the HIVp+αβ simulations sampled a median distance of 20.81 Å and 17.18 Å for monomers 1 and 2 respectively, implying an asymmetric, semi-open flap conformation for the duration of production time. The flap RMSD for the 2:1 complex demonstrates a considerable range of motion, with a mean of 3.71±0.98 Å relative to the crystal structure.
As expected, the ternary complex of HIVp+αβ was not stable. Klebe and co-authors did calculate their affinity data using a 1:1 ratio for kinetics in their experimental work. In a 1:1 ratio, the bound state is expected to be more similar to other holo crystal structures than the solved 3BC4 crystal structure. Klebe and co-authors also concluded their paper with a discussion of the possible influence of crystal packing on the crystallographic conformation and its potential instability in solution. In the crystal structure itself, it appears that the majority of symmetrypacking contacts are formed through ligand-ligand stacking. It may be that these contacts stabilize the observed bound handedness while requiring a semi-open flap elevation.
HIVp+β. Simulations of the β-only complex reveal a wide variation in the population ensemble. None of the representative structures occupy the same conformation as the crystallographic ligand position (see SI Figure S7). Over the last 5ns, the range in ligand deviation from the average pose is 1.73 to 7.56 Å, which shows that the β ligand alone is unstable, and this instability is reflected in the diversity of populated states from these simulations. While the β ligand samples widely, it always has a naphthyl ring in the eye site. The other moieties on the ligand are primarily involved in forming hydrophobic interactions with residues in the flap region. The presence of the β ligand skews the flaps asymmetrically, which is consistent with our previous simulation and proposed behavior of the eye site.11 Our simulations show that when bound alone, the pyrrolidine ligand is not likely to bind in the crystallographic β pose. The system is not stably bound, and the ligand moves to more favorable conformations. This can be seen from both the representative structures of the HIVp+β complex as well as the high RMSD of the ligand from the average conformation.
During the β simulations, the flaps display semi-open to open behavior with averages over the last 5ns for flap tip to catalytic aspartic acid distance of 20.39 Å and 15.43 Å for monomers 1 and 2. The conformation of the more open flap is explained by the presence of the β ligand its flap recognition pocket, which prevents traditional flap dynamics. The ligand interactions at the eye site skew the flaps of the protease into an asymmetric conformation. Over the last 5ns, the average RMSD of the flap residues to the crystal position for the β-only simulation is 4.73±1.06 Å; illustrating similar motion to that sampled by the HIVp+αβ complex. The β-only complex is far less favorable than α-only; in one of the simulations the ligand flips to occupy the α pose (SI Figure S7b) and in other populated states the ligand samples poses approaching α-only (SI Figure S7e).
HIVp+α. The α simulations most commonly sample a distance between the flap tip to the active site of 15–16 Å. This signifies that over the course of our simulations the flaps sample conformations in between closed and semi-open. They cannot close completely due to the presence of the ligand in an eye site. The flaps themselves have an average RMSD to the crystal structure of 4.44±0.47 Å, over the last 5ns. The low standard deviation signifies the greater stability of flap conformation throughout these simulations of the α-only complex, relative to the αβ and β-only simulations.
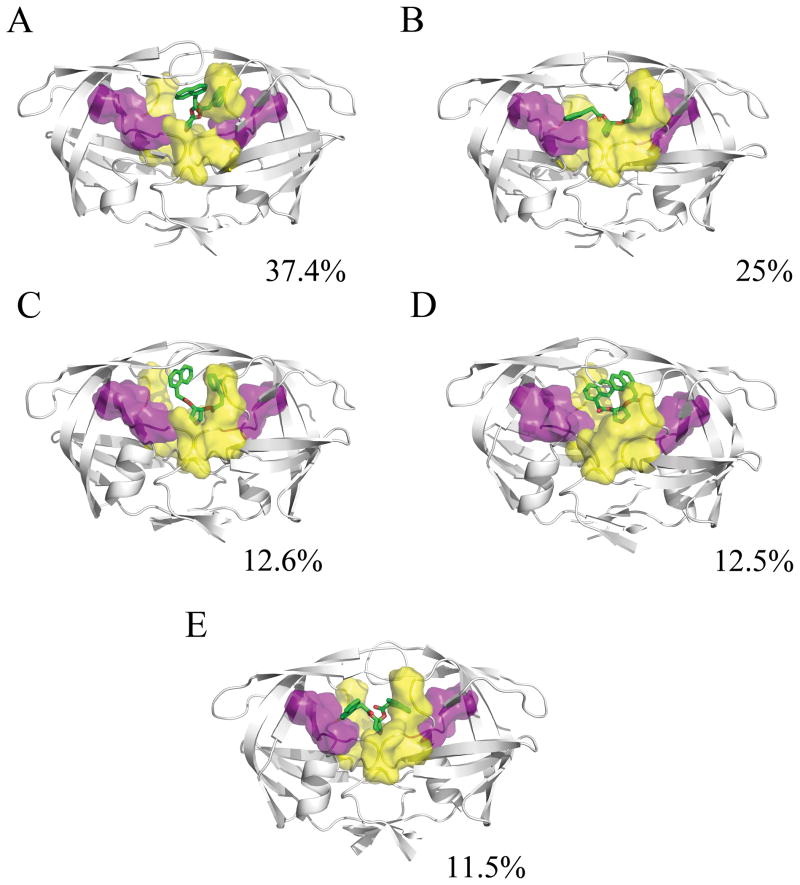
It is interesting that the most frequently sampled conformations in the α-only case illustrate a preference for asymmetric binding at the eye region (Figure 3). We find that one of the naphthyl rings of the ligand often reorients in a manner similar to positions of aromatic rings in known protease inhibitors. One naphthyl ring of the Klebe inhibitor remains stably bound at the eye site, while the second naphthyl ring dissociates from an eye site to occupy either the S1 or S2 site in approximately 75% of the simulation time sampled (Figures 3a and 3b). In addition to demonstrating a possible need to satisfy interactions at either the S1 or S2 binding pocket, this also agrees with two recent studies that illustrate a requirement for asymmetric binding to the eye site.12,28
Figure 3.
Representative structures from the MD simulation of the HIVp+α complex, taken from the last 5ns of each 25ns trajectory. The α ligand is shown in green, the S1/S1′ site is shown in yellow, and the S2/S2′ site is shown in purple. The conformational families for the α ligand illustrate its strong preference for forming one interaction between the naphthyl ring and the eye site, while the other naphthyl ring flips to interact at the S1/S1′ or S2/S2′ site, and the pyrrole maintains a hydrogen-bonding interaction with the catalytic aspartic acids.
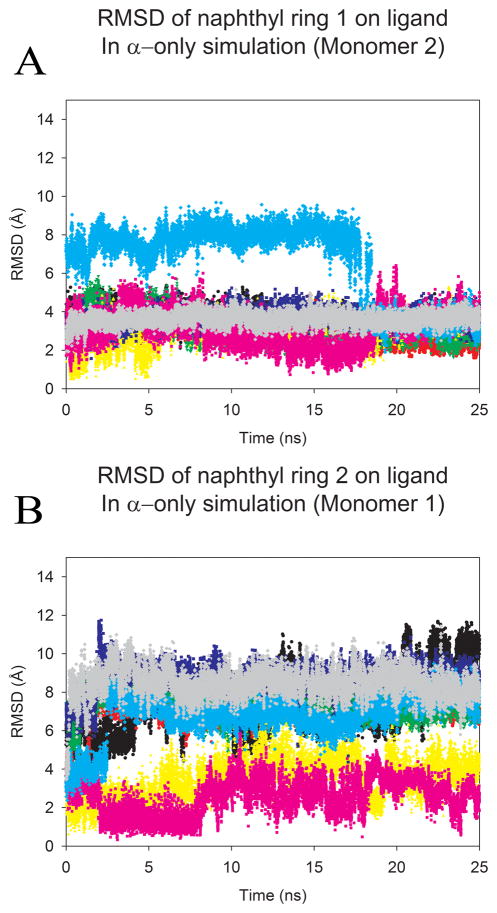
There is moderate deviation from the average ligand position in simulations of HIVp+α over the last 5ns, with a range of 1.78 to 5.86 Å. The loss of one naphthyl ring from an eye site results in the difference from the crystallographic pose. A low standard deviation of 0.73 Å validates the greater stability of this binding pose. RMSD values for the two naphthyl rings were calculated to better demonstrate alterations in ligand binding over time for the α-only case. RMSD traces were created to detail the ligand’s naphthyl ring position over time. The α ligand was fairly stable over time (see SI Figure S4), and the RMSD of the naphthyl rings clearly shows the continued binding of one ring in the eye site (Figure 4a) and the absence of binding at the other eye site (Figure 4b). In the least populated conformational family, both sides of the ligand do flip down into the traditional binding pocket (Figure 3e). This indicates that sufficient sampling has occurred and that this pose is less preferred than asymmetric binding to the eye and S1 or S2 site.
Figure 4.
The overall RMSD from the crystal pose calculated for each naphthyl ring of the ligand in HIVp+α over the length of the production run. Trajectories were first fit to the Cα core of the 3BC4 crystal structure. Each color represents a single production run; and denotes the same run for each plot. A) highlights the RMSD of the first naphtyhl ring over time, (B) highlights the RMSD of the second naphtyhl ring over time. As noted in figure 1, we have used the convention of labeling monomers 1 and 2 based on the behavoir of the ligand, where better agreement with the initial position in the eye is oriented to the right in the figures and labeled as monomer 2 in the graphs. An RMSD of 6.2–7.9 Å indicates occupation of the S2/S2′ site, while an RMSD of 7.8–10.1 Å indicates occupation of the S1/S1′ site.
Of course, it is possible that we observed incomplete sampling, and the inhibitor could actually prefer to be “extended” with both naphthyl rings occupying S1 and/or S2 sites. To determine this, we conducted a fourth series of MD. This system, HIVp+ α′, was obtained by modifying the HIVp+α crystal pose to flip both naphthyls into the S2/S2′ pockets. HIVp+ α′ was subjected to hydrogen minimization in the gas phase with AMBER to ensure that the MD simulations commenced from an unstrained system (Figure 5a). After this, the set-up and simulation of the HIVp+α′ complex followed the previously described protocol for complexes HIVp+ α, HIVp+β, and HIVp+ αβ. Again, eight independent simulations were conducted, and the last 5ns of each simulation was examined.
Figure 5.
A) The initial minimized conformation of the α′ ligand. B–F) Representative structures from the 200ns MD simulation of the HIVp-α′ complex. The α′ ligand is shown in cyan, the S1/S1′ site is shown in yellow, and the S2/S2′ site is shown in purple. Although the simulations were initiated with the naphthyl rings occupying traditional subsites of the active site, one naphthyl ring moves to form interactions at the eye site over the course of all eight independent simulations. The second ring remains in contact with the S1 or S2 site.
These additional simulations, beginning with both naphthyl rings interacting at the S2/S2′ pockets, resulted in at least one ring altering its position during simulation to interact with the eye region (Figure 5b–f). The most populated family type is extremely similar to the most populated families from HIVp+α complex simulations; wherein one ring interacts at the eye region while the other interacts at the S2 pocket. It is possible that the naphthyl rings are positioned one up, one down in solution. NMR data might show whether or not there is symmetry of the two rings’ environment in solution. We found that in the ternary complex, even
It is interesting to note that all of the protease conformations in the clustered families display flaps with the same handedness as the closed state. This signifies that the protease is occupying similar conformational space of the bound form, even though the flaps are typically in the semiopen position (also seen in the close-handed, wide-open structure 1TW7)29. However, this does not mean the flaps cannot flip handedness during the simulations, only that we do not observe it in the majority conformations viewed over the complete trajectory. The flaps do display the type of curling commonly observed during flap transitions. It may require more simulation time for the flaps to flip handedness, simply because of the presence of an inhibitor molecule bound in the flap region.
Despite the relative instability of the crystal conformation during MD simulations, the placement of moieties in the eye site intrigued us due to our previous work. We find that αβ and β probably do not exist, due to the instability of β and poor contacts available to β. Considerable flexibility in the flap region is observed in simulations with the alternate ligand (αβ and β) as compared to the simulations with α-only. Although these molecules have a unique crystallographic conformation, the structure in solution most likely resembles a conformer similar to the HIVp+α complex. The α pose is far more stable, and it most likely contacts one eye site as well as the traditional active site. Our results provide strong support for further exploration of the eye site as a new mode of inhibition for HIVp.
Conclusion
Although the original crystal structure of the pyrrolidine inhibitors is unlikely to exist in solution, we were interested in exploring the potential shown by this mode of binding because of its relationship to the eye site. Naphthyl groups are not ideal because of solubility and metabolic issues, but these inhibitors show that we can take advantage of the eye site in inhibitor design. The binding assays performed by Klebe and co-authors1 show the potential of these compounds for targeting HIVp. Investigating all of the potential bound states of this complex – HIVp+αβ, HIVp+β, HIVp+α, and HIVp+α′ – allows for an accurate study of the impact these ligands may have on flap conformation, and therefore, protease activity.
Our study utilized 200ns of simulation time per system to examine the conformational stability of several HIVp-ligand complexes based on a symmetric inhibitor from Klebe and coauthors10.
Our present results support previous findings that indicate the existence of an alternate binding site for HIVp: the eye site11. Furthermore, our data supports a preference of asymmetric binding at the eye site, as previously suggested.11,26 The representative structures of the HIVp+α and HIVp+α′ complexes illustrate that only one eye site tends to be occupied, while the other naphthyl ring prefers binding at the S1 or S2 site. This implies that traditional inhibitors could be modified to take advantage of this interaction and/or targeting the eye site may be improved by including some traditional S1 or S2 contacts. Inhibitors with improved contacts would be an important step towards demonstrating the viability of the eye site as a target for protease inhibition.
Supplementary Material
Acknowledgments
We thank Charles L. Brooks III of University of Michigan Ann Arbor for generously sharing his computer resources. We also thank Charles David Stout of the Scripps Research Institute for providing the coordinates of their fragment-based crystallography study that was used to create Figure 1C. This work has been supported by the National Institutes of Health (GM65372). KWL thanks Rackham Graduate School, the Pharmacological Sciences Training Program (GM07767), and the American Foundation for Pharmaceutical Education for funding. Molecular graphics images were produced using PyMOL.
Footnotes
Performed at the University of Michigan
Supporting Information Available: Supplementary analysis includes data for the conformational families of HIVp+β and additional measures of protein stability and ligand motion.
References
- 1.Eder J, Hommel U, Cumin F, Martoglio B, Gerhartz B. Aspartic proteases in drug discovery. Curr Pharm Des. 2007;13(3):271–285. doi: 10.2174/138161207779313560. [DOI] [PubMed] [Google Scholar]
- 2.McKeage K, Perry CM, Keam SJ. Darunavir: a review of its use in the management of HIV infection in adults. Drugs. 2009;69(4):477–503. doi: 10.2165/00003495-200969040-00007. [DOI] [PubMed] [Google Scholar]
- 3.Mehellou Y, De Clercq E. Twenty-six years of anti-HIV drug discovery: where do we stand and where do we go? J Med Chem. 53(2):521–538. doi: 10.1021/jm900492g. [DOI] [PubMed] [Google Scholar]
- 4.Hornak V, Simmerling C. Targeting structural flexibility in HIV-1 protease inhibitor binding. Drug Discov Today. 2007;12(3–4):132–138. doi: 10.1016/j.drudis.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedberg DI, Ishima R, Jacob J, Wang YX, Kustanovich I, Louis JM, Torchia DA. Rapid structural fluctuations of the free HIV protease flaps in solution: relationship to crystal structures and comparison with predictions of dynamics calculations. Protein Sci. 2002;11(2):221–232. doi: 10.1110/ps.33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishima R, Freedberg DI, Wang YX, Louis JM, Torchia DA. Flap opening and dimerinterface flexibility in the free and inhibitor-bound HIV protease, and their implications for function. Structure. 1999;7(9):1047–1055. doi: 10.1016/s0969-2126(99)80172-5. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson LK, Yamazaki T, Torchia DA, Grzesiek S, Bax A, Stahl SJ, Kaufman JD, Wingfield PT, Lam PY, Jadhav PK, et al. Flexibility and function in HIV-1 protease. Nat Struct Biol. 1995;2(4):274–280. doi: 10.1038/nsb0495-274. [DOI] [PubMed] [Google Scholar]
- 8.Sadiq AK, Wan S, Coveney PV. Insights into a mutation-assisted lateral drug escape mechanism from the HIV-1 protease active site. Biochemistry. 2007;46(51):14865–14877. doi: 10.1021/bi700864p. [DOI] [PubMed] [Google Scholar]
- 9.Rick SW, Erickson JW, Burt SK. Reaction path and free energy calculations of the transition between alternate conformations of HIV-1 protease. Proteins. 1998;32(1):7–16. doi: 10.1002/(sici)1097-0134(19980701)32:1<7::aid-prot3>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 10.Perryman AL, Lin JH, McCammon JA. HIV-1 protease molecular dynamics of a wildtype and of the V82F/I84V mutant: possible contributions to drug resistance and a potential new target site for drugs. Protein Sci. 2004;13(4):1108–1123. doi: 10.1110/ps.03468904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottcher J, Blum A, Dorr S, Heine A, Diederich WE, Klebe G. Targeting the open-flap conformation of HIV-1 protease with pyrrolidine-based inhibitors. ChemMedChem. 2008;3(9):1337–1344. doi: 10.1002/cmdc.200800113. [DOI] [PubMed] [Google Scholar]
- 12.Damm KL, Ung PM, Quintero JJ, Gestwicki JE, Carlson HA. A poke in the eye: inhibiting HIV-1 protease through its flap-recognition pocket. Biopolymers. 2008;89(8):643–652. doi: 10.1002/bip.20993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meagher KL, Carlson HA. Solvation influences flap collapse in HIV-1 protease. Proteins:Struct Funct Genet. 2005;58(1):119–125. doi: 10.1002/prot.20274. [DOI] [PubMed] [Google Scholar]
- 14.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2008. [Google Scholar]
- 15.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server issue):W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MOE. Montreal, Canada: Chemical Computing Group, Inc; 2008. [Google Scholar]
- 17.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 18.Jakalian A, Bush BL, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic Charges. AM1-BCC model: I. Method. Journal of Computational Chemistry. 2000;21(2):132–146. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 19.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell J, Ross W, Kollman P. AMBER 8. version 8. San Francisco, CA: 2004. [Google Scholar]
- 20.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics. 1983;79(2):926–935. [Google Scholar]
- 21.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerner M. APBS plugin. Ann Arbor, MI: 2004. [Google Scholar]
- 23.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins:Struct Funct Genet. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N. Log(N) Method for Ewald Sums in Large Systems. Journal of Chemical Physics. 1993;98(12):10089–10092. [Google Scholar]
- 25.Shao J, Tanner SW, Thompson N, Cheatham TE. Clustering molecular dynamics trajectories: I. Characterizing the performance of different clustering algorithms. J Chem Theory Comp. 2007;3:2312–2334. doi: 10.1021/ct700119m. [DOI] [PubMed] [Google Scholar]
- 26.Davies DL, Bouldin DW. A Cluster Separation Measure. Pattern Analysis and Machine Intelligence, IEEE Transactions on 1979. PAMI-1(2):224–227. [PubMed] [Google Scholar]
- 27.Calinski T, Harabasz J. A dendrite method for cluster analysis. Communications in Statistics. 1974;3(1):1–27. [Google Scholar]
- 28.Perryman AL, Zhang Q, Soutter HH, Rosenfield R, McRee DE, Olson AJ, Elder JE, Stout CD. Fragment-Based Screen against HIV Protease. Chem Biol Drug Des. 2010;75(3):257–268. doi: 10.1111/j.1747-0285.2009.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin P, Vickrey JF, Proteasa G, Jimenez YL, Wawrzak Z, Winters MA, Merigan TC, Kovari LC. “Wide-open” 1.3 A structure of a multidrug-resistant HIV-1 protease as a drug target. Structure. 2005;13(12):1887–1895. doi: 10.1016/j.str.2005.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.