Abstract
Human ABH2 repairs DNA lesions by using an Fe(II)- and αKG-dependent oxidative demethylation mechanism. The structure of the active site features the facial triad of protein ligands consisting of the side chains of two histidine and one aspartate residues that is common to many nonheme Fe(II) oxygenases. X-ray absorption spectroscopy (XAS) of metallated (Fe and Ni) samples of ABH2 were used to investigate the mechanism of ABH2 and its inhibition by Ni(II) ions. The data are consistent with a sequential mechanism that features a five coordinate metal center in both the presence and absence of the α-ketoglutarate cofactor. This aspect is not altered in the Ni(II)-substituted enzyme, and both metals are shown to bind the cofactor. When substrate is bound to the native Fe(II) complex with α-ketoglutarate bound, a five-coordinate Fe(II) center is retained that features an open coordination position for O2 binding. However, in the case of the Ni(II)-substituted enzyme, the complex that forms in the presence of cofactor and substrate is six-coordinate, and therefore features no open coordination site for oxygen activation at the metal.
Gene silencing via DNA methylation is part of the normal epigenetic regulation of gene expression (1 – 5). Cellular DNA can also sustain damage from alkylation by chemicals that modify DNA bases (6), and this damage can be inherited (7). The effects of alkylation damage can be mutagenetic, cytotoxic, or both (8 – 10). Cells have evolved systems to repair DNA alkylation damage (11). E. coli has an adaptive response pathway to protect DNA against high levels of alkylation damage (12). In humans, the enzyme ABH2 plays the role of repairing endogenously formed 1-methyladenine and 3-methylcytosine lesions in duplex DNA (13).
ABH2 utilizes Fe(II) and αKG to catalyze the oxidative demethylation of methylated DNA bases (Figure 1) (14). Thus, ABH2 belongs to the large class of αKG-dependent non-heme Fe(II) oxygenases that activate O2 in order to catalyze the oxidation of a broad range of biological substrates (15 – 20). The active site metal is typically bound by a facial triad of protein ligands consisting of the side chains of two histidines and one glutamate or aspartate residues and water molecules (15 – 18, 20, 21). A crystal structure of the ABH2-αKG complex with the non-native metal ion Mn(II) reveals a six-coordinate structure where the protein ligands are provided by His171, His236 and Asp173, the αKG cofactor binds in a bidentate fashion, with the sixth position occupied by an aqua ligand (22). Prior XAS studies have shown that the native metal ion, Fe(II), is five-coordinate in the resting enzyme (23).
Figure 1.
Demethylation of 3- methylcytosine showing the step catalyzed by ABH2.
Substitution of iron by other metals in ABH2 would be expected to yield an enzyme with altered function because of changes in the ability of the metal site to bind and decarboxylate αKG, and/or bind and activate O2. It has been reported that Ni(II) inhibits ABH2 by replacing the active site metal (23), although the nature of the mechanistic change is not known. Substitution of the active site Fe(II) ion by Ni(II) has also been proposed to be involved in nickel carcinogenesis via the silencing of tumor suppressor genes (24, 25). Herein, we use XAS as a structural probe of the active site metal center in WT-recombinant ABH2 (Fe-ABH2) and Ni(II)-substituted WT-recombinant ABH2 (Ni-ABH2) in order to examine the reaction mechanism and ascertain the intermediate(s) affected by nickel substitution.
Experimental
Expression and purification of ABH2
The pET28a-His6-ABH2 vector was obtained from Dr. Timothy R. O’Connor (13). E. coli BL21(DE3)pLysS competent cells (Novagen™) were transformed with pET28a-His6-ABH2 and were plated and grown overnight at 37°C on LB media containing 30 µg/mL kanamycin and 34 µg/mL chloramphenicol. Single colonies were grown overnight in 150 mL cultures (LB kanamycin/ chloramphenicol) and diluted 1:100 in 2 L of fresh media. The cells were grown at 37 °C to an optical density of 0.5 at 600 nm and then induced with isopropyl β-D-1-thiogalactopyranoside (final conc. 1 mM). The cells were grown for an additional 3 h at 25 °C, and were harvested by centrifugation and resuspended in lysis buffer (50 mM HEPES, 300 mM NaCl, 2 mM 2-mercaptoethanol, 2 mM EDTA and 5% glycerol at a pH of 7.8) and stored at −80 °C. Upon thawing, the cells were lysed in the presence of PMSF and DNAse and then centrifuged to collect the lysate. The lysate was then dialyzed against lysis buffer without EDTA. After dialysis the soluble proteins were loaded on to a Ni-NTA column. The column was washed sequentially with buffers (50 mM HEPES, 300 mM NaCl, 2 mM 2-mercaptoethanol and 5% glycerol at a pH of 7.8) containing 10 mM, 40 mM and 60 mM imidazole. Pure protein was eluted using the same wash buffer, but with 250 mM imidazole added. The His6-tag was cleaved from purified protein using a Thrombin CleanCleave Kit (Sigma) and removed by dialysis. Purity was confirmed by a single band with MW = 33 kD on an SDS-PAGE gel. Apo-protein was prepared by adding a 5-fold excess of EDTA to the protein solution and incubating at 4 °C overnight. The EDTA was then removed by dialysis against a buffer solution containing 50 mM HEPES, 100 mM NaCl and 2 mM ascorbic acid at pH = 7.8. Protein concentration was determined by the Bradford assay (26) using BSA as a standard. Metallated samples were prepared in a Coy™ chamber and frozen under N2 atmosphere. Fe-ABH2 was prepared by adding a solution of (NH4)2FeSO4 (10 mM, 1.25 equiv.) in the buffer defined above to 18 µM apo-protein and incubating for 6 h. Unbound and non-specifically bound iron was then removed by adding Chelex 100 beads to the sample, incubating 20 min., and then decanting the solution via pipette. The demethylase activity of the samples was confirmed using an assay described previously (23) (see supporting information).
The same procedure, only substituting Ni(OAc)2 for (NH4)2FeSO4 was used to prepare Ni-ABH2.
XAS Sample preparation
From the Fe-ABH2 preparation described above, a total of three samples were prepared for XAS experiments. The first sample contained only the metallated protein – hereafter referred to as Fe-ABH2. The second sample contained Fe-ABH2 plus 1.0 equiv. of αKG, hereafter referred to as Fe-ABH2-αKG. Finally, the third sample contained Fe-ABH2, 1.0 equiv. of αKG plus 5 equiv. of a substrate, 5'-AAAGCACmeCGGTCGAAAAAGCGAAA-3' (MeC = 3-methylcytosine; Midland Certified Regent Company; Km ~ 4 – 5 µM (27)), and is hereafter referred to as Fe-ABH2-αKG + substrate. The samples had an iron content of ~ 0.3 mM, as determined by ICP-OES. Three analogous samples were prepared from Ni-ABH2 (vide supra). The samples had a nickel content of ~ 0.3 mM Ni. Based on a Bradford assay of protein concentration, the samples had metal:protein ratios of 0.89:1 (Fe-ABH2) and 0.93:1 (Ni-ABH2). The samples were syringed into polycarbonate sample holders with kapton windows and frozen in liquid N2 for XAS data collection.
X-ray Absorption Spectroscopy
XAS data collection and analysis were performed as reported previously (28). Iron and nickel K-edge data were collected at 10 K using a liquid He cryostat (Oxford Instruments) on beam line 9-3 at the Stanford Synchrotron Radiation Laboratory. The ring conditions were 3 GeV and 80–100 mA. Beam line optics consisted of a Si(220) double crystal monochromator and two Rh-coated mirrors for focusing and harmonic rejection. X-ray fluorescence was collected using a 30-element Ge detector (Canberra), with the exception of Fe-ABH2-αKG + substrate, where a 100 element detector was employed. The data obtained using the latter detector has significantly higher signal:noise than other data collected at the Fe K-edge, and allowed for a higher resolution analysis. Scattering was minimized by placing a set of Soller slits with a Z-1 element (Mn or Co) filter in between the sample chamber and the detector. X-ray absorption near edge spectroscopy (XANES) data was collected from −200 eV to +200 eV with respect to the edge energy of the absorbing metal. The energy of each metal K-edge was calibrated to the first inflection point of the relevant metal foil (7112.5 eV for Fe and 8331.6 eV for Ni). Extended X-ray absorption fine structure (EXAFS) data were collected to k = 16 Ǻ−1 for Ni-ABH2, and k = 14 Å−1 above the edge energy for all other samples.
XAS data analysis was performed using EXAFS123 (29) for XANES analysis and SixPack (30) for EXAFS analysis. Scattering parameters for SixPack fitting were generated using the FEFF 8 software package (31). For the data shown, 18 scans were averaged for the Fe-ABH2 and 12 scans were averaged for Fe-ABH2-αKG and for Fe-ABH2-αKG + substrate; 5 scans were averaged for Ni-ABH2 and Ni- ABH2-αKG, and 9 scans were averaged for Ni-ABH2-αKG + substrate. The averaged spectra were background corrected and normalized in Sixpack, which uses the AUTOBK routine of IFEFFIT (30, 32, 33). For XANES analysis, the edge energy reported was taken to be the maximum of the first-derivative of the XANES spectrum. For pre-edge XANES analysis, a 75% Gaussian and 25% Lorenzian function was used to fit the rise in the fluorescence at the edge. Gaussian peaks were added to the pre-edge fit to account for any peaks in the pre-edge region, and the areas of these peaks were taken as a measure of their intensities (see supporting information). The intensity of the peak associated with a 1s → 3d electronic transition was then used to indicate the coordination number/geometry of metal sites (34 – 36). For Ni XANES analysis, the presence/absence of a feature associated with a 1s → 4pz transition is also useful in identifying complexes with five-coordinate pyramidal or four-coordinate planar geometries (34).
For EXAFS analysis, the averaged spectra were first converted to k-space. For comparison purposes, all of the data for each metal K-edge was refined over the same limits, which were determined by the sample with the poorest signal:noise. For Ni K-edge data, EXAFS data was fit over a k-range of 2 - 12.5 Å−1. For data collected at the Fe K-edge, a limit of 2 – 12 Å−1 was used. The primary effect of truncating the data is to lower the resolution of similar shells in the fits. For data that would support a higher resolution analysis, fits employing all the usable data (maximum resolution) were also performed (see supporting information). Structural models of the metal sites involving coordination numbers from 2 – 7 were systematically evaluated for all possible combinations of N/O- and S-donors by holding the number of scattering atoms in each shell to integer values (see supporting information). The number of histidine imidazole ligands involved in the coordination sphere was estimated by multiple-scattering analysis as previously described (37 – 39). Amplitudes and phase shifts for multiple scattering paths for Fe-Im ligands were generated using FEFF (v. 8.0) with the coordinates of an Fe-Im ligand obtained from the crystal structure of human PHF8 with Fe(II) and αKG (3K3O). For Ni-Im ligands, the crystal structure of JMJD2A with Ni(II) and substrate bound (2Q8C) was used. Scattering paths of similar length were combined in one shell, as described by Tierney et al. (38, 39). Therefore, no single label can be meaningfully applied to these combined paths. During the fitting process, coordination numbers were constrained to be integer values and a scale factor of 0.9 was used. Bond lengths, σ2 and a single value of ΔE0 were allowed to vary in each fit. For αKG and ascorbate ligands, fits including imidazole ligands were initially screened for the presence of additional second coordination sphere C atoms not accounted for by His ligands using individual single scattering C atoms at ~2.5 Å and ~3.1 Å, respectively (see supporting information). The distances for the αKG C atoms were obtained from the crystal structures of human PHF8 with Fe(II) and αKG (3K3O) and from JMJD2A with Ni(II), substrate, and αKG bound (2Q8C). The crystal structure of the iron complex of trimethyl reductic acid (an ascorbic acid analogue) (40) was used for the ascorbate model. The best models using single-scattering C atoms were further examined using multiple-scattering analysis derived from rigid O-C-CO five-membered chelate rings with parameters obtained using FEFF (v. 8.0) and the above referenced structures (Table 1). In this analysis, distances in the chelate ring were constrained to vary with a single value of Δr and σ2. In all cases where single scattering indicated the presence of second-coordination sphere C atoms, the fits were improved by the substitution of two N/O donors in the first coordination sphere by the chelate ring, which adds two C atoms to the second coordination sphere.
Table 1.
XAS Analysis of ABH2 Samples
| Sample | XANES Analysis | EXAFS Analysis | Ref. | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Edge E(eV) |
1s→3d peak area (×10−2 eV) |
Coord. No. |
Shell | r (Å)a | σ2b (×10−3 Å2) |
ΔE0 (eV) |
%Rc | Red. χ2 |
||
| Fe-ABH2 (k = 2–13 Å−1) | 7125.9(2) | 9(1) | 5 | 3 N/O | 1.95(2) | 0(2) | −8(3) | 10.45 | 97.44 | 23 |
| 2 N/O (2 Im) | 2.08(2) | 2(2) | ||||||||
| 3 N/O (2 Im) | 2.09(3) | 5(2) | −4(3) | 10.14 | 56.8 | This work | ||||
| 1 O | [1.88(2)]d | 2(1) | ||||||||
| 1 O | [1.99(2)] | |||||||||
| 1 C | [2.66(2)] | |||||||||
| 1 C | [2.75(2)] | |||||||||
| Fe-ABH2- αKG (k = 2–14 Å−1) | 7122.5(2) | 14.5(7) | 5 | 2 N/O | 1.96(3) | 3(2) | −15 | 2.96 | 53.86 | This work |
| 3 N/O (2 Im) | 2.08(2) | 4(3) | ||||||||
| 3 N/O (2 Im) | 2.06(2) | 1(1) | −5(2) | 2.07 | 36.33 | This work | ||||
| 1 O | [1.94(2)] | 4(1) | ||||||||
| 1 O | [2.15(2)] | |||||||||
| 1 C | [2.66(2)] | |||||||||
| 1 C | [2.76(2)] | |||||||||
| Fe-ABH2- αKG + Substrate (k = 2–12 Å−1) | 7121.5(2) | 10.5(6) | 5 | 2 N/O | 1.85(3) | 3(2) | −30(5) | 17.15 | 82.04 | This work |
| 3 N/O (2 Im) | 2.04(3) | 4(2) | ||||||||
| 3 N/O (2 Im) | 1.93(3) | 4(2) | −14(5) | 5.8 | 42.36 | This work | ||||
| 1 O | [1.85(2)] | 11(3) | ||||||||
| 1 O | [2.06(2)] | |||||||||
| 1 C | [2.57(2)] | |||||||||
| 1 C | [2.67(2)] | |||||||||
| Ni-ABH2 (k = 2–12.5 Å−1) | 8344.7(2) | 5(1) | 5 | 5 N/O (2 Im) | 2.08(2) | 5(1) | −4(1) | 2.3 | 8.03 | 23 |
| 3 N/O (2 Im) | 2.05(2) | 2(1) | 1(3) | 4.1 | 16.45 | This work | ||||
| 1 O | [1.86(4)] | 5(4) | ||||||||
| 1 O | [1.91(4)] | |||||||||
| 1 C | [2.53(4)] | |||||||||
| 1 C | [2.55(4)] | |||||||||
| Ni-ABH2- αKG (k = 2–14 Å−1) | 8345.3(2) | 5.4(8) | 5 | 5 N/O (2 Im) | 2.10(2) | 6(1) | −6(1) | 3.15 | 48.82 | 23 |
| 3 N/O (2 Im) | 2.08(2) | 2(1) | 1(2) | 1.35 | 22.47 | This work | ||||
| 1 O | [1.89(2)] | 9(2) | ||||||||
| 1 O | [1.94(2)] | |||||||||
| 1 C | [2.56(2)] | |||||||||
| 1 C | [2.58(2)] | |||||||||
| Ni-ABH2- αKG + Substrate(k = 2–14 Å−1) | 8343.5(2) | 3.6(7) | 6 | 2 N/O | 2.08(2) | 3(1) | −4(2) | 3.82 | 468.9 | This work |
| 4 N/O (2 Im) | 2.02(8) | 10(7) | ||||||||
| 4 N/O (2 Im) | 2.04(2) | 2(1) | 6(1) | 2.34 | 60.22 | This work | ||||
| 1 O | [2.07(2)] | 3(2) | ||||||||
| 1 O | [2.12(2)] | |||||||||
| 1 C | [2.74(2)] | |||||||||
| 1 C | [2.76(2)] | |||||||||
r (Å) is the radial distance between metal and ligand.
σ2 is the root mean square disorder in the metal-ligand distance.
R is the goodness of fit. Numbers in parentheses represent standard deviation for least square fits.
Distances in [ ] correspond to atoms in a O-CC-O chelate ring and were constrained to vary with a single value of Δr for the chelate ring.
To compare different fits to the same data set, ifeffit uses three goodness of fit parameters, χ2 (Equation 1), reduced χ2, and R
| (1) |
(Equation 2), where Nidp = number of independent data points, Nε2= number of uncertainties to
| (2) |
minimize; Re(fi) =real part of the EXAFS fitting function; Im(fi) = imaginary part of the EXAFS fitting function. Reduced χ2 = χ2/(Nind Nvarys) (where Nvarys = the number of refining parameters) and represents the degrees of freedom in the fit. Ifeffit also calculates R for the fit, which is given by Equation 2 and is scaled to the magnitude of the data, making it proportional to χ2. To compare different models (fits) the R-factor and reduced χ2 parameters can be assessed, in which case both parameters should be minimized. Although R will always improve with an increasing number of shells (adjustable parameters), reduced χ2 will go through a minimum and then increase, indicating that the model is over fitting the data. Best fits were judged by using two goodness of fit parameters, reduced χ2 and R, and the deviation of σ2 from typical values.
Results
XANES Analysis
XAS experiments were performed to investigate the structures of Fe(II) and Ni(II) bound to ABH2 and upon addition of cofactor or cofactor and substrate. The analysis of XANES data provides information about the coordination number and geometry of a metal site (34 – 36). Both Fe(II) and Ni(II) have vacancies in the 3d manifold that give rise to peaks associated with 1s → 3d electronic transitions that are observed in the pre-edge XANES region of the K-edge spectra in all the ABH2 samples (Figure 2, supporting information). The peak areas (intensities) of the 1s → 3d transitions depend on the coordination number and geometry of the metal sites (34 – 36). By comparing the 1s → 3d transition peak areas of the ABH2 samples with typical values for known coordination numbers/geometries we were able to determine the coordination numbers of all the six ABH2 samples from the XANES data. EXAFS analysis also provides a measure of the number of ligands present in the primary coordination sphere (N), and in all of the cases presented the value determined for N are in agreement with the XANES analysis.
Figure 2.
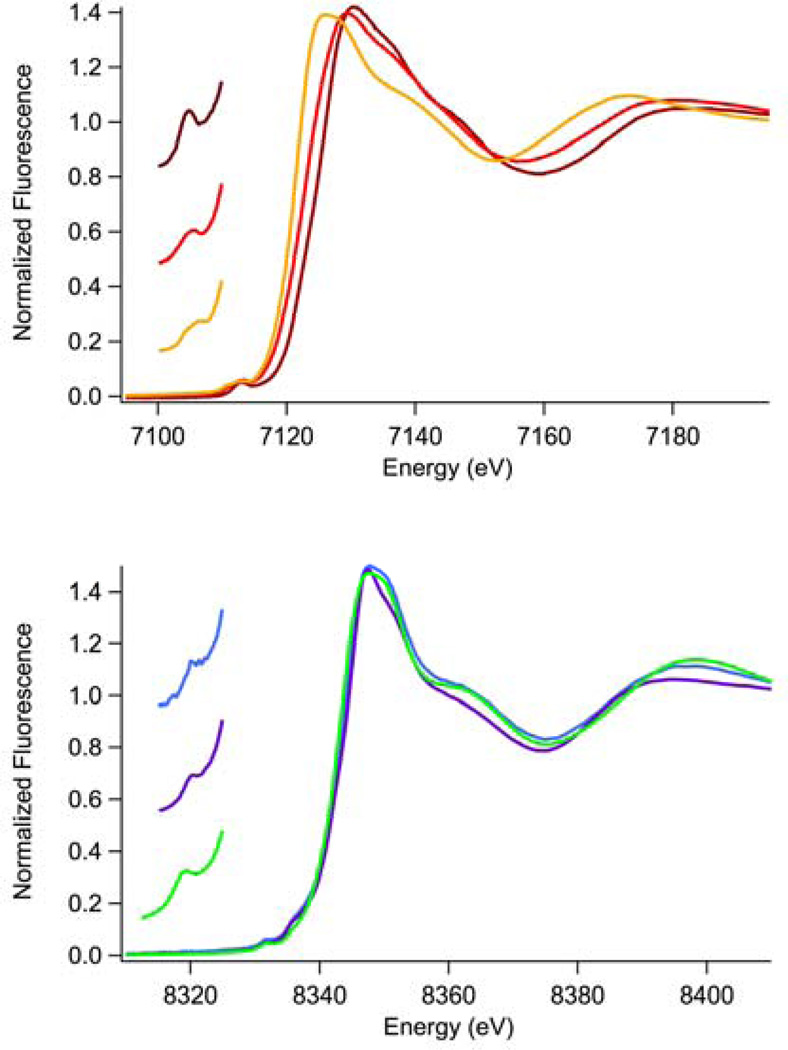
Top: Fe K-Edge XANES spectra; Fe(ABH2) (brown), Fe(ABH2)-αKG (red) and Fe(ABH2)- αKG + substrate (orange). Bottom: Ni K-Edge XANES spectra; Ni(ABH2) (blue), Ni(ABH2)-αKG (green) and Ni(ABH2)-αKG + substrate (purple). Inserts: Expansions of the pre-edge XANES region showing peaks associated with 1s → 3d electronic transitions.
Comparisons of XANES data (Table 1 and Figure 2) for the three iron-containing samples show that each spectrum is distinct. Thus, the structure of the iron site is sensitive to the presence of both αKG and the substrate. There is a systematic shift to lower K-edge energy from Fe-ABH2 → Fe-ABH2-αKG → Fe-ABH2-αKG + substrate that indicates an increase in electron density at the iron site in each of these samples. The edge energy observed for Fe(ABH2) (7125.9(2) eV) is very high for Fe(II); more typical of Fe(III) complexes (e.g., 7124 eV for Fe(III) protocatechuate 3, 4-dioxygenase (PCD) (41), 7126.6 eV for a hexaquo Fe(III) model complex (42), 7125.7 eV for a non-heme alkylperoxo)Fe(III) intermediate (43)) and higher than for other non-heme Fe(II) enzymes (e.g. 7120.5 eV for Fe(II)PCD (44)). In addition, the magnitude of the edge energy shift for Fe(ABH2) → Fe(ABH2)-αKG is quite large (3.4(4) eV) and could be interpreted as a reduction of Fe(III) → Fe(II). However, the samples were prepared anaerobically and under reducing conditions (2 mM ascorbate) where no Fe(III) should be present. Thus, formation of Fe(III) in the enzyme would require some redox center intrinsic to the protein. Further, X-band EPR spectra obtained on one of the samples of Fe(ABH2) used for XAS did not exhibit features at either g – 4.3 or g ~ 2 (see supporting information), suggesting that little, if any, Fe(III) is present. Another possible explanation might be that the ligand envionment, which is composed of 3 O-donors (including ascorbate) and 2 N-donors (vide infra) leads to an unusually ionic environment that has a high + charge on the Fe center. In either event, the observed energy shifts to lower energy would be expected for a site that will eventually bind and reductively activate O2. No changes in the coordination number of the Fe(II) center are apparent upon binding the αKG cofactor or upon binding substrate to Fe-ABH2-αKG. In all the three samples a peak associated with the 1s → 3d transition is observed near 7113 eV. In the resting sample, Fe-ABH2, the 1s → 3d peak area (~ 9 × 10−2 eV) is indicative of a five-coordinate complex (typical values are ~8 – 13 × 10−2 eV) (35, 36). The complex remains five-coordinate with a 1s → 3d peak area of ~14 × 10−2 eV upon the addition of αKG. Addition of substrate decreases the 1s → 3d peak area to ~10 × 10−2 eV, a value that is still consistent with a five-coordinate site.
Unlike Fe, which remains five-coordinate in all of the ABH2 samples, Ni-ABH2 shows a change in coordination number upon binding substrate. Again, the spectrum of each of the nickel-containing sample is distinct. However, the Ni K-edge shows no systematic shift to lower energy upon binding αKG or substrate (Table 1), indicating that little change in electron density of the Ni(II) site occurs in response to binding αKG or substrate. Like the analogous iron complexes, XANES analysis shows that Ni-ABH2 has a five-coordinate Ni(II) site (1s → 3d peak area (near 8332 eV) ~ 5 × 10−2 eV, typical range = 4 – 10 × 10−2 eV (34)) that remains five-coordinate upon binding αKG (1s → 3d peak area ~ 5 × 10−2 eV). However, upon binding substrate, the Ni(II) site becomes six-coordinate, with a decrease observed in the 1s → 3d peak area to 3 – 4 × 10−2 eV; the typical range for six-coordinate Ni complexes is 0.6 – 4 × 10−2 eV (34)). In cases of samples with five-coordinate Ni(II) sites, additional small shoulders near 8338 eV, may be observed that are associated with 1s → 4pz transitions. Shoulders corresponding to these transitions are observed for Ni-ABH2 and Ni-ABH2-αKG, but not for Ni-ABH2- αKG + substrate, consistent with the change in coordination number from five for Ni-ABH2 and Ni-ABH2-αKG to six for Ni-ABH2- αKG + substrate (Figure 2, supporting information) (34). In addition, observation of these features indicates a geometry that is closer to a square pyramidal than to trigonal bipyramidal (34) for the five-coordinate complexes.
EXAFS Analysis
The analysis of EXAFS provides information about the number and types of ligands bound to a metal, and metric details of the metal site structure. This information is obtained from the best fits of the data and is summarized in Figure 3 and Table 1. The best fit for Fe-ABH2 EXAFS data consists of five N/O donor ligands of which two are imidazoles from multiple-scattering analysis (Table 1). This result is consistent with the crystal structure of Mn-ABH2 (22) and with the active site structures of other resting nonheme Fe-dioxygenase enzymes (45), which show ligation by the 2 His + 1 carboxylate (Asp or Glu) facial triad plus water molecules. Fits obtained using single scattering C atoms in the second coordination sphere indicated the presence of two - three second sphere carbon atoms for Fe-ABH2 in addition to those associated with two imidazole ligands. Given the prior crystal structure of Mn-ABH2 showing the position of the carboxylate carbon of Asp 173 (Mn--C ~ 2.97 Å (22)), only one of these C atoms could be assigned to the carboxylate C atom. The other carbon atoms can be attributed to the carbon atoms from a coordinated ascorbate ligand (46, 47). Multiple-scattering analysis derived from rigid O-C-C-O five-membered chelate ring was performed to further test the fit indicating the presence of ascorbate, which is based on the presence of second coordination sphere C atoms in the models with single-scattering C atoms (see supporting information). These fits use a single distance parameter to position all the atoms in the chelate, and a single value of s2 for the ligand, and thus do not contain more adjustable parameters than the corresponding fit without the C atoms. The results show that the quality of the fits (both R and χ2) are improved by the replacement of two first coordination shell N/O donors by one O-C-C-O unit (Table 1). The crystal structure of iron bound to tetramethyl reductic acid shows that carbon atoms in the chelate ring are at a distance of ~ 3.0 Å from the iron centers (40). Using the chelate model, C atoms are found at ~2.7 Å. (Table 1. Note that the model obtained from the crystal structure features two M-C distances, and this was used without modification. The difference in the M-C distance is below the resolution of the EXAFS data, and coupled with the errors indicate that difference in the distances to the C atoms is not resolved.) Thus, in the resting iron enzyme prepared in the presence of ascorbate, the aqua ligands are replaced by a bidentate ascorbate ligand (Figure 4). The addition of a third C atom to model the C atom from the aspartate ligand does lead to a significant improvement in the fit (see supporting information).
Figure 3.
EXAFS analysis. Left: Unfiltered, k3-weighted EXAFS spectra (colored lines) and fits (black lines, from Table 1). Right: Fourier-transformed EXAFS data and fits.
Figure 4.
Comparison of the complexes of ascorbate and α-ketoglutarate with the Fe(II) site in ABH2.
When ABH2 was reconstituted with Ni(II), the same buffer containing ascorbate was used, but the Ni K-edge EXAFS lacks the features associated with the extra C atoms in the FT-EXAFS spectrum (Figure 3). The fit obtained using an O-C-C-O chelate ring is worse that that obtained using only the ligand donor-atoms (Table I). Thus, there is no indication that ascorbate binds to Ni-ABH2, and the Ni(II) site is best modeled by 2 His imidazoles, 1 carboxylate and 2 aqua ligands.
Addition of αKG to metallated ABH2 (Fe or Ni) results in changes in the EXAFS spectra arising from scattering atoms in both the first-and second-coordination spheres in the Fourier-transformed spectra (Figure 3). These changes are consistent with the binding of αKG in a bidentate fashion, where changes in the primary coordination sphere reflect the ligand exchange, and the observation of C atoms in the second coordination sphere arise from the ordering of all the atoms in the five-membered chelate ring. Although the improvement in the fit is not as dramatic as for the addition of ascorbate to Fe-ABH2, both R and χ2 are improved in every case (Table 1). The distance observed for the two C atoms in the model incorporating a five-membered chelate ring is (~ 2.66 – 2.76 Å, Figure 4), and is similar to that observed in the crystal structure of human PHF8 complexed with αKG (2.47; 2.57 Å) (48) and TauD complexed with Fe(II) and substrate (2.57, 2.85 Å) (49). Again, the addition of a third C atom to model the aspartate β-C atom does not lead to significant improvements in the fit (see supporting information).
Addition of substrate to the Fe- or Ni-ABH2-αKG complexes results in additional structural perturbations, particularly for the Fe sample (Figure 3). The best fits of the EXAFS data suggest the presence of five N/O donor ligands for Fe(II)-ABH2-αKG + substrate, but six N/O-donors for the Ni(II)-ABH2- αKG + substrate, in agreement with the XANES analysis (Table 1).
The O-C-C-O five-membered chelate rings seen for the αKG complexes are retained when substrate is added. The best fit for Fe-ABH2-αKG +substrate shows the presence of two second sphere carbon atoms at ~2.5–2.6 Ǻ. Similarly, the best fit for Ni-ABH2-αKG + substrate also shows the presence of two second coordination sphere carbon atoms at a distance of ~2.6–2.7 Ǻ. Again, the addition of a C scatterer to model the aspartate β-C does not significantly improve the fits (see supporting information). Curiously, the best fit for Fe-ABH2-αKG + substrate also shows one very short N/O-Fe distances at ~1.85 Ǻ, a shortening of ~ 0.09 Å from the distance in Fe-ABH2-αKG. While shorter than found in the Fe-ABH2- αKG fit, similar distances has been observed in crystal structures of non-heme iron halogenase (1.87, 1.73 Å) (50), in clavaminate synthase (1.80 Å) Å (51) and in TauD (1.73 Å) (49). The shortening of these bonds may indicate increased covalency between Fe(II) and αKG, and may account in part for the observed decrease in K-edge energy observed upon substrate binding.
Discussion
ABH2 is a member of a group of Fe(II)- and αKGdependent dioxygenases that use the 2-His-1-carboxylate motif to bind the metal cofactor. Previously we reported that ABH2 and JMJD2A are sensitive to Ni(II) ion inhibition (23). This inhibition may lead to defective DNA demethylation and mutations in the case of ABH2 inhibition, or gene silencing by JMJD2A inhibition; the latter is consistent with the known epigenetic mechanism of nickel carcinogenesis (24 – 25). XAS was used to examine the structures of Fe(II)- and Ni(II)-ABH2 complexes for further mechanistic insights. Characterization of a number non-heme Fe(II) oxygenases have revealed that they operate by a sequential reaction mechanism that is designed to prevent reductive oxygen activation until the ternary complex (composed of enzyme, αKG cofactor and substrate) has formed (Scheme 1) (16, 52 – 54). In the first step of the consensus mechanism, αKG binds in a bidentate fashion to the 6-coordinate metal site composed of the 2-His-1-carboxylate facial triad and three aqua ligands (Scheme 1, A), displacing two aqua ligands (Scheme 1, B). This step is followed by binding of the substrate to a site close to the active site metal. The presence of both substrate and cofactor induces the loss of the remaining aqua ligand, which opens a coordination site that serves to bind O2 (Scheme 1, C). Thus, the saturation of the coordination sphere of the Fe(II) center is used to prevent O2 binding. Subsequently, oxidative decarboxylation of αKG takes place, leading to the formation of an Fe(IV)=O species (15). This high-valent iron species is the oxidant involved in catalysis. In the case of ABH2, the Fe(IV)=O species will hydroxylate the methyl group on the N-atom of the DNA base. The resulting alcohol then undergoes a further spontaneous reaction to yield formaldehyde and the dealkylated base (Figure 1).
Scheme 1.
Consensus mechanism of nonheme Fe(II) oxygenases.
Fe-ABH2 appears to operate by a variation of the consensus mechanism (Scheme 2). XANES and EXAFS analysis are consistent with a five-coordinate site composed of N/O-donor ligands for both Fe(II) and Ni(II) in ABH2 in the absence of cofactor or substrate (Scheme 2, A). Analysis of features from scatterers in the second and third coordination shells indicate that two of these ligands are His imidazoles, consistent with the available crystallographic data, and the remaining ligands are presumably derived from Asp and water. The data from the complexes formed with αKG are consistent with the cofactor binding to both metals in a bidentate fashion, while retaining the five-coordinate geometries of the complexes (Scheme 2, B). Thus, αKG displaces two ligands, presumably aqua ligands, from the resting complex. Similar results have been reported by Scott and workers with TfdA, where no change in coordination number was observed for Cu(II)- or Fe(II)-TfdA upon addition of αKG (37). Therefore, binding of αKG does not change the coordination number of metal center but perturbs the electronic environment of the metal center by substituting for other ligands (e.g., aqua or ascorbate). Upon binding substrate, the Fe(II) site remains five-coordinate. (Scheme 2, C). However, the K-edge energy in the presence of substrate is lower than that for the other two samples (Fe-ABH2 with and without αKG). This indicates a change in the electronic environment of the Fe(II) center in presence of the substrate. Differences are also observed in the maximum intensity of the rising edge (white line), and also at energies above the edge (e.g., at ~7160 eV). These differences also reflect changes in the structure of the Fe(II) site upon addition of substrate (55). The five-coordinate Fe(II) site is in contrast to the known structure of Mn-substituted ABH2, which reveals a six-coordinate Mn(II) site in the ternary complex formed with αKG and substrate (22).
Scheme 2.
Variation of consensus mechanism for ABH2.
The reaction sequence in Scheme 2 is distinct from the consensus mechanism (Scheme 1) in that a 5-coordinate complex is present in the resting enzyme and is maintained in the catalytic cycle through the formation of the enzyme-substrate complex (vide supra). This implies that Fe-ABH2 uses a different mechanism than found in other non-heme Fe(II) oxygenases to prevent reductive activation of O2 prior to formation of the ternary complex.
One possible alternative mechanism would be to use protein conformation to block O2 access to the Fe site, and remove the impediment when substrate binds via a protein conformational change. The presence of a five-coordinate Fe(II) site in the resting enzyme (Scheme 2, A) and its maintenance through ternary complex formation is not unprecedented in DNA or histone demethylases. The crystal structure of JMJD2A with αKG (no substrate) also reveals a five-coordinate Fe(II) site (56). An allosteric mechanism also has a precedent in bacterial DNA demethylases. E. coli AlkB is a bacterial enzyme that catalyzes demethylation of methylated DNA bases. Like human ABH2, AlkB is a mononuclear non-heme Fe(II) enzyme that utilizes αKG and O2 to carry out the oxidative demethylation of DNA. Both of these enzymes also utilize the facial 2-His-1-carboxylate motif to bind Fe(II). There are several crystal structures of AlkB available in literature where Fe(II) has been replaced by Mn(II), Co(II) or Ni(II) in presence of αKG and also in presence of both αKG and substrate (22, 57 – 59). All of the metal-substituted AlkB enzymes are catalytically inactive. Comparison of crystal structures of AlkB containing Fe(II), Co(II) or Mn(II) in presence of both αKG and substrate shows that the positioning of Trp178 and Leu184 is different in case of Mn(II) or Co(II) than that in case of Fe(II) (59, 60). In the case of Mn(II) and Co(II) Trp178 blocks the O2 binding site and Leu184 blocks the O2 diffusion tunnel.
For Ni(II), the mechanism of ABH2 inhibition is different in three important ways: First, Ni(II) in a ligand environment consisting of the 2-His-1-caboxylate protein binding site and an αKG ligand would not be expected to possess a redox potential capable of reducing O2 (60 – 62). Second, the electron density build-up that occurs on the Fe(II) center upon binding αKG and then substrate is not observed for Ni(II). Third, the ternary complex formed between Ni(II), the protein and αKG contains a six-coordinate nickel site (Figure 5) that has no open coordination site for binding O2. The six-coordinate complex formed with Ni(II) is indistinguishable from that the structure of Mn(II) center in the Mn-substituted enzyme. Thus, inhibition by metal ions other than Fe(II) appears to involve blocking access to the metal via coordination of a sixth ligand and, in the case of Ni(II), both electronic and steric components may be involved.
Figure 5.
Proposed structures of the Fe and Ni sites of ABH2. M-ABH2 (left), in presence of αKG (middle), and in presence of both αKG and substrate (right). Iron is brown, Nickel cyan, carbon gray, oxygen red and nitrogen blue.
Supplementary Material
Acknowledgments
This work was supported by NIH grant ES005512-,P30ES000260 (MC). The Stanford Synchrotron Radiation Laboratory, is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural and Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program.
List of Abbreviations
- ABH2
Alpha ketoglutarate dependent dioxygenase AlkB human homologue 2
- αKG
2-Oxopentanedioic acid
- BSA
Bovine Serum Albumin
- ICP-OES
Inductively Coupled Plasma – Optical Emission Spectroscopy
- PCD
Protocatechuate 3, 4-dioxygenase
- EXAFS
Extended X-Ray Absorption Fine Structure
- XANES
X-Ray Absorption Near Edge Structure
- XAS
X-Ray Absorption Spectroscopy
Footnotes
Supporting Information Available: Supporting information contains tables of EXAFS fits to Fe and Ni K-edge data used to select the best-fit models, Figures of XANES analyses, Figure of enzyme activity assay of ABH2. This information is available free of charge via the Internet at: http://pubs.acs.org.
REFERENCES
- 1.Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293:1070–1074. doi: 10.1126/science.293.5532.1070. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Gene Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Tariq M, Paszkowski J. DNA and histone methylation in plants. Trends Genet. 2004;20:244–251. doi: 10.1016/j.tig.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Bender J. DNA methylation and epigenetics. Annu. Rev. Plant Biol. 2004;55:41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- 5.Chan SWL, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in arabidopsis thaliana. Nat. Rev. Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 6.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 7.Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, Krokan HE. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 8.Rydberg B, Lindahl T. Non-enzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L- methionine is a potentially mutagenic reaction. Embo J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sedgwick B. Repairing DNA-methylation damage. Nat. Rev. Mol. Cell Biol. 2004;5:148–157. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]
- 10.Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE. Alkylation damage in DNA and RNA - repair mechanisms and medical significance. DNA Repair. 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu. Rev. Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- 12.Lee DH, Jin SG, Cai S, Chen Y, Pfeifer GP, O'Connor TR. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J. Biol.Chem. 2005;280:39448–39459. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- 13.Ringvoll J, Nordstrand LM, Vagbo CB, Talstad V, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjork A, Doughty RW, Falnes PO, Krokan HE, Klungland A. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. Embo J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollinger JM, Price JC, Hoffart LM, Barr EW, Krebs C. Mechanism of taurine: alpha-ketoglutarate dioxygenase (TauD) from E. coli. Eur. J. Inorg. Chem. 2005:4245–4254. doi: 10.1021/bi050227c. [DOI] [PubMed] [Google Scholar]
- 16.Hausinger RP. Fe(II)/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 17.Roach PL, Clifton IJ, Hensgens CMH, Shibata N, Schofield CJ, Hajdu J, Baldwin JE. Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature. 1997;387:827–830. doi: 10.1038/42990. [DOI] [PubMed] [Google Scholar]
- 18.Koehntop KD, Emerson JP, Que L. The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. J. Biol. Inorg. Chem. 2005;10:87–93. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- 19.Neidig ML, Brown CD, Kavana M, Choroba OW, Spencer JB, Moran GR, Solomon EI. Spectroscopic and electronic structure studies of the role of active site interactions in the decarboxylation reaction of alpha-keto acid-dependent dioxygenases. J. Inorg. Biochem. 2006;100:2108–2116. doi: 10.1016/j.jinorgbio.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 20.Neidig ML, Solomon EI. Structure-function correlations in oxygen activating non-heme iron enzymes. Chem. Commun. 2005:5843–5863. doi: 10.1039/b510233m. [DOI] [PubMed] [Google Scholar]
- 21.Neidig ML, Brown CD, Light KM, Fujimori DG, Nolan EM, Price JC, Barr EW, Bollinger JM, Krebs C, Walsh CT, Solomon EI. CD and MCD of CytC3 and taurine dioxygenase: Role of the facial triad in alpha-KG-dependent oxygenases. J. Am. Chem. Soc. 2007;129:14224–14231. doi: 10.1021/ja074557r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang CG, Yi CQ, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–964. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen HB, Giri NC, Zhang RH, Yamane K, Zhang Y, Maroney M, Costa M. Nickel ions inhibit histone demethylase JMJD1A and DNA repair enzyme ABH2 by replacing the ferrous iron in the catalytic centers. J. Biol. Chem. 285:7374–7383. doi: 10.1074/jbc.M109.058503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YW, Broday L, Costa M. Effects of nickel on DNA methyltransferase activity and genomic DNA methylation levels. Mutat. Res-Gen. Tox. En. 1998;415:213–218. doi: 10.1016/s1383-5718(98)00078-3. [DOI] [PubMed] [Google Scholar]
- 25.Zingg JM, Jones PA. Genetic and epigenetic aspects of DNA methylation on genome expression, evolution, mutation and carcinogenesis. Carcinogenesis. 1997;18:869–882. doi: 10.1093/carcin/18.5.869. [DOI] [PubMed] [Google Scholar]
- 26.Smith PK, et al. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 27.Roy TW, Bhagwat AS. Kinetic studies of E. coli AlkB using a new fluorescence-based assay for DNA demethylation. Nucleic Acids Res. 2007;35:e147. doi: 10.1093/nar/gkm1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leitch S, Bradley MJ, Rowe JL, Chivers PT, Maroney MJ. Nickel-specific response in the transcriptional regulator, E. coli NikR. J. Am. Chem. Soc. 2007;129:5085–5095. doi: 10.1021/ja068505y. [DOI] [PubMed] [Google Scholar]
- 29.Padden KM, Krebs JF, MacBeth CE, Scarrow RC, Borovik AS. Immobilized metal complexes in porous organic hosts: Development of a material for the selective and reversible binding of nitric oxide. J. Am. Chem. Soc. 2001;123:1072–1079. doi: 10.1021/ja003282b. [DOI] [PubMed] [Google Scholar]
- 30.Webb SM. SIXpack: a graphical user interface for XAS analysis using IFEFFIT. Phys. Scr. 2005;T115:1011–1014. [Google Scholar]
- 31.Ankudinov AL, Ravel B, Rehr JJ, Conradson SD. Real-space multiple-scattering calculation and interpretation of x-ray-absorption near-edge structure. Phys. Rev. 1998;B 58:7565–7576. [Google Scholar]
- 32.Newville M. EXAFS analysis using FEFF and FEFFIT. J. Synchrotron Radiat. 2001;8:96–100. doi: 10.1107/s0909049500016290. [DOI] [PubMed] [Google Scholar]
- 33.Zabinsky SI, Rehr JJ, Ankudinov A, Albers RC, Eller MJ. Multiple-Scattering Calculations of X-Ray-Absorption Spectra. Phys. Rev. 1995;B 52:2995–3009. doi: 10.1103/physrevb.52.2995. [DOI] [PubMed] [Google Scholar]
- 34.Colpas GJ, Maroney MJ, Bagyinka C, Kumar M, Willis WS, Suib SL, Baidya N, Mascharak PK. X-ray spectroscopic studies of nickel-complexes, with application to the structure of nickel sites in hydrogenases. Inorg. Chem. 1991;30:920–928. [Google Scholar]
- 35.Randall CR, Shu LJ, Chiou YM, Hagen KS, Ito M, Kitajima N, Lachicotte RJ, Zang Y, Que L. X-ray-absorption pre-edge studies of high-spin iron(II) complexes. Inorg. Chem. 1995;34:1036–1039. [Google Scholar]
- 36.Westre TE, Kennepohl P, DeWitt JG, Hedman B, Hodgson KO, Solomon EI. A multiplet analysis of Fe K-edge 1s->3d pre-edge features of iron complexes. J. Am. Chem. Soc. 1997;119:6297–6314. [Google Scholar]
- 37.Cosper NJ, Stalhandske CMV, Saari RE, Hausinger RP, Scott RA. X-ray absorption spectroscopic analysis of Fe(II) and Cu(II) forms of a herbicide-degrading alpha-ketoglutarate dioxygenase. J. Biol. Inorg. Chem. 1999;4:122–129. doi: 10.1007/s007750050295. [DOI] [PubMed] [Google Scholar]
- 38.Costello A, Periyannan G, Yang KW, Crowder MW, Tierney DL. Site-selective binding of Zn(II) to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia. J. Biol. Inorg. Chem. 2006;11:351–358. doi: 10.1007/s00775-006-0083-z. [DOI] [PubMed] [Google Scholar]
- 39.Costello AL, Sharma NP, Yang KW, Crowder MW, Tierney DL. X-ray absorption spectroscopy of the zinc-binding sites in the class B2 metallo-beta-lactamase ImiS from Aeromonas veronii bv. sobria. Biochemistry. 2006;45:13650–13658. doi: 10.1021/bi061547e. [DOI] [PubMed] [Google Scholar]
- 40.Kim YJ, Feng XD, Lippard SJ. Synthesis, structure, and properties of a mixed-valent triiron complex of tetramethyl reductic acid, an ascorbic acid analogue, and its relationship to a functional non-heme iron oxidation catalyst system. Inorg. Chem. 2007;46:6099–6107. doi: 10.1021/ic700622a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wasinger EC, Davis MI, Pau MYM, Orville AM, Zaleski JM, Hedman B, Lipscomb JD, Hodgson KO, Solomon EI. Spectroscopic studies of the effect of ligand donor strength on the Fe-NO bond in intradiol dioxygenases. Inorg. Chem. 2003;42:365–376. doi: 10.1021/ic025906f. [DOI] [PubMed] [Google Scholar]
- 42.Westre TE, Kennepohl P, DeWitt JG, Hedman B, Hodgson KO, Solomon EI. A multiplet analysis of Fe K-edge 1s->3d pre-edge features of iron complexes. J. Am. Chem. Soc. 1997;119:6297–6314. [Google Scholar]
- 43.Shan X, Rohde JU, Koehntop KD, Zhou Y, Bukowski MR, Costas M, Fujisawa K, Que L. X-ray absorption spectroscopic studies of high-spin Nonheme (Alkylperoxo)iron(III) intermediates. Inorg. Chem. 2007;46:8410–8417. doi: 10.1021/ic700649w. [DOI] [PubMed] [Google Scholar]
- 44.Davis MI, Wasinger EC, Westre TE, Zaleski JM, Orville AM, Lipscomb JD, Hedman B, Hodgson KO, Solomon EI. Spectroscopic investigation of reduced protocatechuate 3,4-dioxygenase: Charge-induced alterations in the active site iron coordination environment. Inorg. Chem. 1999;38:3676–3683. doi: 10.1021/ic981464p. [DOI] [PubMed] [Google Scholar]
- 45.Que L, Ho RYN. Dioxygen activation by enzymes with mononuclear non-heme iron active sites. Chem. Rev. 1996;96:2607–2624. doi: 10.1021/cr960039f. [DOI] [PubMed] [Google Scholar]
- 46.Khan MMT, Martell AE. Metal ion and metal chelate catalyzed oxidation of ascorbic acid by molecular oxygen .I. cupric and ferric ion catalyzed oxidation. J. Am. Chem. Soc. 1967;89:4176–4185. doi: 10.1021/ja00992a036. [DOI] [PubMed] [Google Scholar]
- 47.Khan MMT, Martell AE. Metal ion and metal chelate catalyzed oxidation of ascorbic acid by molecular oxygen .II. cupric and ferric chelate catalyzed oxidation. J. Am. Chem. Soc. 1967;89:7104–7111. doi: 10.1021/ja01002a046. [DOI] [PubMed] [Google Scholar]
- 48.Yu L, Wang Y, Huang S, Wang JJ, Deng ZQ, Zhang Q, Wu W, Zhang XL, Liu Z, Gong WM, Chen ZZ. Structural insights into a novel histone demethylase PHF8. Cell Res. 20:166–173. doi: 10.1038/cr.2010.8. [DOI] [PubMed] [Google Scholar]
- 49.O'Brien JR, Schuller DJ, Yang VS, Dillard BD, Lanzilotta WN. Substrate-induced conformational changes in Escherichia coli taurine/alpha-ketoglutarate dioxygenase and insight into the oligomeric structure. Biochemistry. 2003;42:5547–5554. doi: 10.1021/bi0341096. [DOI] [PubMed] [Google Scholar]
- 50.Wong C, Fujimori DG, Walsh CT, Drennan CL. Structural Analysis of an Open Active Site Conformation of Nonheme Iron Halogenase CytC3. J. Am. Chem. Soc. 2009;131:4872–4879. doi: 10.1021/ja8097355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang ZH, Ren JS, Stammers DK, Baldwin JE, Harlos K, Schofield CJ. Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase. Nat. Struct. Biol. 2000;7:127–133. doi: 10.1038/72398. [DOI] [PubMed] [Google Scholar]
- 52.Costas M, Mehn MP, Jensen MP, Que L. Dioxygen activation at mononuclear nonheme iron active sites: Enzymes, models, and intermediates. Chem. Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 53.Hanauskeabel HM, Gunzler V. A stereochemical concept for the catalytic mechanism of prolylhydroxylase - applicability to classification and design of inhibitors. J. Theor. Biol. 1982;94:421–455. doi: 10.1016/0022-5193(82)90320-4. [DOI] [PubMed] [Google Scholar]
- 54.Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee SK, Lehnert N, Neese F, Skulan AJ, Yang YS, Zhou J. Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem. Rev. 2000;100:235–349. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 55.Chow MS, Eser BE, Wilson SA, Hodgson KO, Hedman B, Fitzpatrick PF, Solomon EI. Spectroscopy and kinetics of wild-type and mutant tyrosine hydroxylase: Mechanistic insight into O-2 activation. J. Am. Chem. Soc. 2009;131:7685–7698. doi: 10.1021/ja810080c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen ZZ, Zang JY, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao QL, Pan CH, Dai SD, Hagman J, Hansen K, Shi Y, Zhang GY. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 57.Yu B, Edstrom WC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature. 2006;439:879–884. doi: 10.1038/nature04561. [DOI] [PubMed] [Google Scholar]
- 58.Yu B, Hunt JF. Enzymological and structural studies of the mechanism of promiscuous substrate recognition by the oxidative DNA repair enzyme AlkB. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14315–14320. doi: 10.1073/pnas.0812938106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holland PJ, Hollis T. Structural and mutational analysis of E. coli AlkB provides insight into substrate specificity and DNA damage searching. PLoS One. 2010;5:e8680. doi: 10.1371/journal.pone.0008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mirza SA, Day RO, Maroney MJ. Oxidation of a dimeric nickel thiolate complex with O2. Inorg. Chem. 1996;35:1992–1995. [Google Scholar]
- 61.Grapperhaus CA, Darensbourg MY. Oxygen capture by sulfur in nickel thiolates. Acc. Chem. Res. 1998;31:451–459. [Google Scholar]
- 62.Goldcamp MJ, Robison SE, Bauer JAK, Baldwin MJ. Oxygen reactivity of a nickel(II)-polyoximate complex. Inorg. Chem. 2002;41:2307–2309. doi: 10.1021/ic015590w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.