Abstract
Summary and comment on a recent Cell paper entitled ‘PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease’ (Shin et al., 2011).
Parkinson’s disease (PD) is the second most common neurodegenerative disorder, second only to Alzheimer’s disease. The clinical triad of resting tremor, bradykinesia and rigidity characterizes PD. These symptoms are considered to be a direct consequence of neurodegeneration and dopaminergic cell loss. Another pathognomonic lesion of PD is the presence in the cytoplasm of abnormal protein aggregates called Lewy bodies, which are composed of the presynaptic protein α-synuclein (Lees et al., 2009; Shulman et al., 2011).
Over 90% of PD cases are classified as sporadic PD, in which aging and the environment, combined with minor contributions from multiple relatively low-risk genetic factors, interact to cause the disease (Hardy, 2010). Recent genome-wide association studies have started to determine the identity of these low-risk pathogenic loci. The first of these to be associated with sporadic PD was the SNCA gene, which encodes α-synuclein, demonstrating that genetic variations at this site not only contribute to early-onset PD, but also to late-onset sporadic PD (Hardy, 2010; Nalls et al., 2011).
By contrast, a small proportion of PD cases are familial. Mutations in SNCA and leucine-rich repeat kinase 2 (LRRK2; also known as dardarin) cause an autosomal-dominant form of PD. Mutations in other PD-associated genes contribute to pathology by affecting mitochondrial homeostasis. These genes include those encoding PINK1, parkin and DJ-1, and are known to cause autosomal-recessive PD (AR-PD) (Park et al., 2009; Hardy, 2010). In fact, mutations in parkin cause the most common form of AR-PD. Carriers of parkin mutations usually develop early-onset disease, even before the age of 40, with a slow and benign disease course and good response to the common PD therapeutic, L-DOPA. Although there are some reports that Lewy bodies are present in the cells of these patients, the main pathological characteristic of mutant parkin carriers is the degeneration of substantia nigra dopaminergic neurons (Lees et al., 2009; Hardy, 2010).
In recent years, there has been an explosion in cellular and model-organism-based research investigating the role of PINK1 and parkin in PD (Park et al., 2009). Initially, two elegant studies carried out in the fruit fly Drosophila melanogaster determined that Pink1 and Parkin interact in the same molecular pathway, and that Parkin acts downstream of Pink1 (Park et al., 2006; Clark et al., 2006). Since then, other work has shown that Pink1 is expressed in both the cytoplasm and mitochondria. Although it seems to have relevant functions outside mitochondria, its mitochondrial localization is central for the Pink1-Parkin interaction (Whitworth et al., 2008; Deas et al., 2011). When mitochondrial membrane potential is compromised, Pink1 recruits Parkin to damaged mitochondria to initiate mitophagy (Fig. 1A), the autodegradative process by which mitochondria are cleared (for reviews, see Deas et al., 2011; Youle and Narendra, 2011). Defects in the function of either of these proteins might disrupt this process.
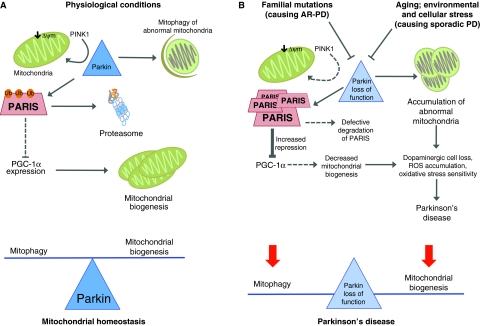
Fig. 1.
Parkin regulates the proteasomal degradation of PARIS and PGC-1α-dependent mitochondrial biogenesis. (A) Normal physiological conditions that maintain mitochondrial homeostasis: PINK1 recruits parkin to the mitochondria, where these two proteins interact to eliminate abnormal mitochondria through mitophagy. Alterations in mitochondrial membrane potential (ΔΨm; a key indicator of mitochondrial physiology and cell viability) initiate the PINK1-parkin cascade of events that lead to mitophagy. Furthermore, parkin ubiquitylates and thereby promotes proteasomal degradation of PARIS. Because PARIS represses the expression of PGC-1α, degradation of PARIS by parkin allows PGC-1α-dependent gene expression and enables mitochondrial biogenesis. Parkin seems to be an integral regulator of mitochondrial homeostasis, controlling both degradation and biogenesis. (B) Loss of parkin function as a result of familial mutations (in the case of AR-PD) or aging, environmental or cellular stress (in the case of sporadic PD) leads to the accumulation of abnormal mitochondria, owing to faulty mitophagy. In addition, PARIS accumulates and represses PGC-1α, preventing mitochondrial biogenesis. Loss of parkin function does not tip the balance between mitochondrial biogenesis and degradation to either side, but leads to a general breakdown of mitochondrial homeostasis that can ultimately lead to PD.
Evidence has accumulated to support mitochondrial involvement in the pathogenesis of PD. Specifically, the hypothesis has emerged that an age-dependent accumulation of mitochondrial mutations (equating to mitochondrial damage) in substantia nigra neurons, coupled with defective mitophagy (perhaps caused by a defective PINK1-parkin pathway and possibly other non-PINK1-parkin-dependent autophagic mechanisms), are sufficient to cause PD (Youle and Narendra, 2011). However, recent data from Ted and Valina Dawson’s group have found that there is more to the role of parkin than just mediating mitophagy when it comes to controlling mitochondrial homeostasis (Shin et al., 2011).
Using a yeast two-hybrid screen, the authors of this study found that parkin interacts with the protein PARIS (also known as ZNF746). PARIS is a 644 amino acid protein that has not previously been associated with PD and that has a Kruppel-associated box (KRAB) domain at its N-terminus. It has a widespread distribution throughout human and mouse tissues. Moreover, it has a differential pattern of expression in the brain and is also expressed in the pars compacta nigral neurons. The authors used co-immunoprecipitation analyses to demonstrate a direct interaction between parkin and PARIS in SH-SY5Y cells (a human neuroblastoma cell line), in mouse brain and in human striatum. This interaction required the RING1 or RING2 domains of parkin (Shin et al., 2011).
Once it was established that PARIS binds to parkin, it was important to determine the physiological relevance of this interaction. Parkin is an E3 ubiquitin ligase that participates in the proteasomal degradation system and for which several substrates have been identified [see Dawson and Dawson (Dawson and Dawson, 2010) and references therein]. Shin et al. showed that parkin ubiquitylates PARIS, and that parkin mutants that are associated with PD in humans have a substantially reduced ubiquitylation activity against PARIS (Shin et al., 2011). Furthermore, proteasomal inhibition increased the levels of PARIS expression by up to threefold, demonstrating that parkin ubiquitylates PARIS to target it for proteasomal degradation.
The authors then characterized the interaction in further detail using co-expression experiments in which they showed that the expression levels of parkin and PARIS are inversely proportional. Increasing parkin expression led to decreased levels of PARIS. The opposite experiments – i.e. reducing parkin expression using short hairpin RNA (shRNA) in SH-SY5Y cells, or downregulating parkin expression in vivo in adult conditional parkin-knockout (KO) mice – showed that reduction of parkin expression was accompanied by PARIS upregulation. Notably, they also found that PARIS expression levels were increased by twofold in the cingulate cortex of AR-PD patients lacking functional parkin. Similarly, they reported that PARIS levels are increased by more than twofold in the striatum and nigra neurons of patients with sporadic cases of PD. Because they did not find alterations in PARIS at the mRNA level, this suggests that the increase is due to defective proteasomal degradation caused by parkin loss of function.
Having established that PARIS is a novel parkin-interacting protein, and that its levels increase in the absence of functional parkin, the next step was to determine the relevance of PARIS upregulation in PD. Proteins with KRAB domains are known to function as transcriptional repressors. In line with this, Shin et al. found that PARIS binds to the promoter of peroxisome proliferator-γ (PPARγ) coactivator-1α (PGC-1α) in SH-SY5Y cells, as well as in mouse and human brain, repressing its activity (Shin et al., 2011). Notably, they also showed by chromatin immunoprecipitation that the increased levels of PARIS found in striatal cells of PD patients were correlated with increased endogenous (PGC-1α) promoter occupancy, compared with those in control cells. This is in line with a recent report from the Global PD Gene Expression Consortium showing that PGC-1α responsive genes are underexpressed in PD patients and in patients with incipient Lewy body disease (Zheng et al., 2010). The authors further showed that the repressive effect of PARIS on PGC-1α is highly specific and, furthermore, that one of the main genes co-regulated by PARIS and PGC-1α in PD is nuclear respiratory factor-1 (NRF1), which has roles in mitochondrial function and oxidant metabolism. Both PGC-1α and NRF1 are important mediators of mitochondrial biogenesis. In addition, parkin has previously been shown to be involved in mitochondrial biogenesis by regulating mitochondrial DNA transcription and regulation (Kuroda et al., 2006).
Finally, Shin et al. developed a mouse model overexpressing PARIS in the substantia nigra by using adenovirus-mediated gene delivery (Shin et al., 2011). When PARIS was overexpressed on its own, a dramatic reduction in dopaminergic neurons was observed. This loss could be rescued if parkin or PGC-1α were co-delivered. This demonstrates that parkin is sufficient to eliminate PARIS repression of PGC-1α expression, and that increasing the amount of PGC-1α overcomes the repressive effects of PARIS on PGC-1α expression.
In summary, these results indicate that, under normal (healthy) conditions, parkin ubiquitylates PARIS, leading to its proteasomal degradation, which in turn releases the repressive effect of PARIS on PGC-1α expression, enabling mitochondrial biogenesis (Fig. 1A). It seems likely that parkin plays a key role in overall mitochondrial dynamics in the cell, because it can both promote mitophagy of damaged mitochondria (via interactions with PINK1) and promote mitochondrial biogenesis through its effects on PARIS and PGC-1α. Although the processes of mitophagy and mitochondrial biogenesis might seem to be on opposite ends of a balance (Fig. 1A), when parkin is lost, the balance is not tipped either way – both processes are affected (Fig. 1B). This disruption leads to an accumulation of mitochondrial mutations and increased levels of reactive oxygen species (ROS) (owing to mitochondrial malfunction) and cell death, and ultimately leads to PD (Fig. 1B).
The specific repression of PGC-1α is very interesting given that it has been documented that lack of PGC-1α results in basal ganglia degeneration, particularly in the striatum (Lin et al., 2004; St-Pierre et al., 2006). PGC-1α regulates energy, metabolism and mitochondrial biogenesis in a diverse range of tissues, including brain, heart and muscle, as well as tissues that are involved in metabolism, such as liver and adipose tissue (Lin et al., 2005). PGC-1α-knockout mice have a reduced capacity to sustain running exercise and a low fatigue resistance index, which are features that correlate with morphological and functional mitochondrial abnormalities in skeletal muscle (Leone et al., 2005). Moreover, the expression of genes that are known to be important in metabolizing ROS – such as cytoplasmic superoxide dismutase 1 (SOD1), mitochondrial SOD2 and peroxisomal catalase – are reduced in the brain of PGC-1α-knockout mice. This suggests a role for PGC-1α in ROS defense, implying that cells lacking the ability to induce PGC-1α expression would have increased sensitivity to oxidative stress (St-Pierre et al., 2006). Notably, other sources of cellular stress have been found to regulate PGC-1α expression, such as nutrition, temperature, physical exercise and specific oxidative stressors used in chemically induced models of PD such as rotenone and the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) (Lin et al., 2005; Zheng et al., 2010). All of the above emphasizes the importance of PGC-1α regulation, particularly in the brain and basal ganglia.
Although the findings of Shin et al. (Shin et al., 2011) and Zheng et al. (Zheng et al., 2010) highlight the importance of PGC-1α in PD pathogenesis, they also raise more questions. Given that the PINK1-parkin interaction is important in the context of PD pathology, one avenue that would be worth exploring is whether PINK1 also plays a role in parkin-dependent degradation of PARIS. Furthermore, although an interaction between parkin and α-synuclein still awaits confirmation (Dawson and Dawson, 2010), it is possible that overexpression of parkin might be able to rescue α-synuclein-mediated neurodegeneration through mediating PARIS degradation and, in turn, upregulating PGC-1α expression. Notably, Zheng et al. showed that adenoviral co-transduction of midbrain primary neuronal cultures with PGC-1α could rescue cell loss caused by a PD-associated SNCA mutation (A53T) (Zheng et al., 2010). Because α-synuclein is emerging as a major contributor to PD [see Hardy (Hardy, 2010) and references therein], it is highly relevant to evaluate whether there is a connection between these disease-associated factors. Finally, because mitochondrial abnormalities are not exclusive to PD (Lin and Beal, 2006), it would be of great value to explore the connection between PARIS and PGC-1α in other neurodegenerative diseases.
Acknowledgments
I thank Kerri J. Kinghorn and Ivana Bjedov for critical reading of this paper and for the endless discussions in the lab.
REFERENCES
- Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. (2006). Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 [DOI] [PubMed] [Google Scholar]
- Dawson T. M., Dawson V. L. (2010). The role of parkin in familial and sporadic Parkinson’s disease. Mov. Disord. 1, S32–S39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas E., Wood N. W., Plun-Favreau H. (2011). Mitophagy and Parkinson’s disease: the PINK1-parkin link. Biochim. Biophys. Acta 1813, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J. (2010). Genetic analysis of pathways to Parkinson disease. Neuron 68, 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda Y., Mitsui T., Kunishige M., Shono M., Akaike M., Azuma H., Matsumoto T. (2006). Parkin enhances mitochondrial biogenesis in proliferating cells. Hum. Mol. Genet. 15, 883–895 [DOI] [PubMed] [Google Scholar]
- Lees A. J., Hardy J., Revesz T. (2009). Parkinson’s disease. Lancet 373, 2055–2066 [DOI] [PubMed] [Google Scholar]
- Leone T. C., Lehman J. J., Finck B. N., Schaeffer P. J., Wende A. R., Boudina S., Courtois M., Wozniak D. F., Sambandam N., Bernal-Mizrachi C., et al. (2005). PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 3, e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Wu P. H., Tarr P. T., Lindenberg K. S., St-Pierre J., Zhang C. Y., Mootha V. K., Jäger S., Vianna C. R., Reznick R. M., et al. (2004). Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell 119, 121–135 [DOI] [PubMed] [Google Scholar]
- Lin J., Handschin C., Spiegelman B. M. (2005). Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1, 361–370 [DOI] [PubMed] [Google Scholar]
- Lin M. T., Beal M. F. (2006). Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795 [DOI] [PubMed] [Google Scholar]
- Nalls M. A., Plagnol V., Hernandez D. G., Sharma M., Sheerin U. M., Saad M., Simón-Sánchez J., Schulte C., Lesage S., Sveinbjörnsdóttir S., et al. (2011). Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet 377, 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., et al. (2006). Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 [DOI] [PubMed] [Google Scholar]
- Park J., Kim Y., Chung J. (2009). Mitochondrial dysfunction and Parkinson’s disease genes: insights from Drosophila. Dis. Model. Mech. 2, 336–340 [DOI] [PubMed] [Google Scholar]
- Shin J. H., Ko H. S., Kang H., Lee Y., Lee Y. I., Pletinkova O., Troconso J. C., Dawson V. L., Dawson T. M. (2011). PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell 144, 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman J. M., De Jager P. L., Feany M. B. (2011). Parkinson’s disease: genetics and pathogenesis. Annu. Rev. Pathol. 6, 193–222 [DOI] [PubMed] [Google Scholar]
- St-Pierre J., Drori S., Uldry M., Silvaggi J. M., Rhee J., Jäger S., Handschin C., Zheng K., Lin J., Yang W., et al. (2006). Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127, 397–408 [DOI] [PubMed] [Google Scholar]
- Whitworth A. J., Lee J. R., Ho V. M., Flick R., Chowdhury R., McQuibban G. A. (2008). Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson’s disease factors Pink1 and Parkin. Dis. Model. Mech. 1, 168–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R. J., Narendra D. P. (2011). Mechanisms of mitophagy. Nat. Rev. Mol. Cell. Biol. 12, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Liao Z., Locascio J. J., Lesniak K. A., Roderick S. S., Watt M. L., Eklund A. C., Zhang-James Y., Kim P. D., Hauser M. A., et al. (2010). PGC-1α, a potential therapeutic target for early intervention in Parkinson’s disease. Sci. Transl. Med. 2, 52ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]