Abstract
Zebrafish studies in the past two decades have made major contributions to our understanding of hematopoiesis and its associated disorders. The zebrafish has proven to be a powerful organism for studies in this area owing to its amenability to large-scale genetic and chemical screening. In addition, the externally fertilized and transparent embryos allow convenient genetic manipulation and in vivo imaging of normal and aberrant hematopoiesis. This review discusses available methods for studying hematopoiesis in zebrafish, summarizes key recent advances in this area, and highlights the current and potential contributions of zebrafish to the discovery and development of drugs to treat human blood disorders.
Introduction
Hematopoiesis is defined as the formation of all types of blood cells from hematopoietic stem cells (HSCs) (Orkin and Zon, 2008). This complex process involves the coordination of intrinsic master blood transcriptional regulators and signaling molecules from the surrounding tissues (Huang and Zon, 2008). Understanding the molecular and cellular mechanisms that drive normal hematopoiesis and hematopoietic malignancies is crucial for developing new treatments for blood diseases. Despite the identification of many essential genes involved in hematopoiesis, the underlying mechanisms still remain poorly understood.
Zebrafish are being increasingly used as a model organism to study vertebrate hematopoiesis. This genetically tractable model has many appealing features, such as the ease of manipulation of transparent embryos and the capacity to carry out large-scale genetic and chemical screens (Lieschke and Currie, 2007). Similar to other vertebrates, zebrafish have sequential waves of hematopoiesis (Fig. 1A) (Davidson and Zon, 2004; de Jong and Zon, 2005); however, hematopoiesis in zebrafish occurs in sites that are distinct from other vertebrates. For example, zebrafish HSCs reside in the kidney marrow, which is functionally equivalent to the bone marrow niche of mammals. Nonetheless, the developmental processes and genetic programs of hematopoiesis are highly conserved in zebrafish (Fig. 1B) (Chen and Zon, 2009; Huang and Zon, 2008; Paik and Zon, 2010). In the past two decades, a wealth of studies has been published that has supplied many novel insights and discoveries into the field of hematology. Additionally, the parallels between the hematopoietic system in zebrafish and humans prompt the increasing use of zebrafish to model hematological disorders. Excellent recent reviews cover the different aspects of these studies in great detail (Bertrand and Traver, 2009; Carradice and Lieschke, 2008; Chen and Zon, 2009; Ellett and Lieschke, 2010; Huang and Zon, 2008; Paik and Zon, 2010; Payne and Look, 2009). In this Primer article, we survey various methods for research in zebrafish, illustrate the advantages of using zebrafish to study normal and pathogenic hematopoiesis, highlighting key recent advances, and discuss the contribution and potential of zebrafish to identify new disease treatments.
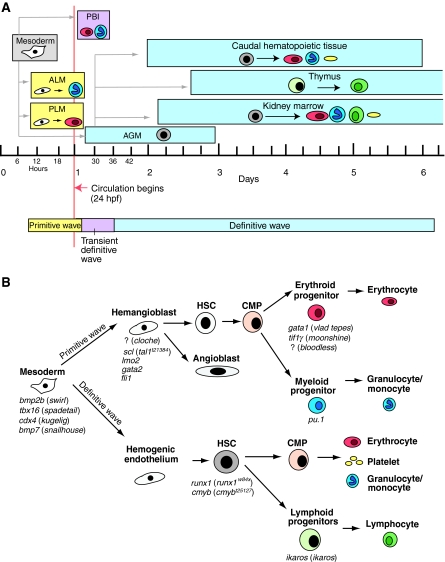
Fig. 1.
The ontogeny of hematopoiesis in zebrafish. (A) Hematopoiesis in zebrafish occurs in consecutive waves. Embryonic primitive hematopoiesis (yellow) starts at around 11 hours post fertilization (hpf) when hemangioblasts (which have the potential to become either endothelial vascular cells or hematopoietic cells, shown in white) appear in the anterior lateral mesoderm (ALM) and posterior lateral mesoderm (PLM), which collectively are analogous to the blood islands in the mammalian yolk sac. Hemangioblasts in the PLM later converge medially to form the intermediate cell mass (ICM; not shown), where primitive erythrocytes (red) predominantly arise. The ALM, which later becomes the rostral blood island (RBI; not shown), is the major site for primitive myeloid cells (blue). At around 24 hpf, embryonic erythrocytes enter circulation. As circulation begins, hematopoiesis within the ICM gradually diminishes. A transient definitive wave (pink) initiates shortly after multi-lineage erythromyeloid progenitors appear in the posterior blood island (PBI). Starting from 26 hpf, definitive HSCs (gray) emerge from hemogenic endothelial cells of the dorsal aorta in the aorta-gonad-mesonephros (AGM) region. Shortly thereafter, HSCs migrate to and seed the caudal hematopoietic tissue at 48 hpf, which is an expansion of the PBI and acts as a transient hematopoietic site that gives rise to erythroid, myeloid and thromboid (yellow) cells. The caudal hematopoietic tissue is equivalent to mouse fetal liver or placenta. HSCs from the AGM region colonize kidney around 48 hpf. Kidney marrow, which is functionally similar to mammalian bone marrow, gives rise to all blood lineages, including erythroid, myeloid, thromboid and lymphoid (green) cells for the larval and adult zebrafish. At around 54 hpf, common lymphoid progenitor (CLP) cells from the AGM region seed the thymus, which is the site for maturation of lymphoid T cells. (B) Crucial genes during different stages of hematopoietic development in zebrafish. The names of corresponding available zebrafish mutants are shown in parenthesis; the mutated genes are unknown for the mutants preceded by ‘?’. Adapted from Chen and Zon (Chen and Zon, 2009) and Paik and Zon (Paik and Zon, 2010), with permission.
Zebrafish forward genetic screens: powerful approaches to identify new players in hematopoiesis
The zebrafish is amenable to forward genetic screens on a scale that is infeasible in any other vertebrate models that have the same low cost and space requirements (Patton and Zon, 2001). The embryos can survive without red blood cells by passive diffusion of oxygen for the first week, allowing the identification of mutations that would be embryonic lethal in mice. Screens using the chemical mutagen ENU (N-ethyl-N-nitrosourea) or retroviral mutagenesis have successfully yielded a plethora of blood mutants (Amsterdam et al., 2004; Ransom et al., 1996; Weinstein et al., 1996). These mutants have helped elucidate the genetic pathways that regulate HSCs as well as different hematopoietic lineages, and many of them are also excellent models of human blood diseases (Carradice and Lieschke, 2008; North and Zon, 2003; Shafizadeh and Paw, 2004). In some instances, zebrafish mutants have revealed genes that have later been shown to be involved in human disease. For example, cloning of the hypochromic anemic mutant weissherbst (weh) identified a previously unknown iron transporter, ferroportin1 (Donovan et al., 2000). ferroportin1 is required for iron transport from the yolk store to the circulation. Subsequent to the zebrafish findings, mutations in this gene were found in patients with type IV hemochromatosis (iron overload) (Gordeuk et al., 2003; Montosi et al., 2001).
Genetic modifier screens that are routinely used in invertebrates such as flies and worms are effective for identifying genes that suppress or enhance the function of the gene of interest, and thus have the potential to reveal new therapeutic targets. The first zebrafish suppressor screen was recently carried out in moonshine mutants, which are deficient for transcriptional intermediary factor 1-γ (tif1γ; also known as trim33) (Bai et al., 2010). These embryos have severely impaired erythropoiesis because they fail to appropriately express Gata1, a crucial factor for erythropoiesis (Ransom et al., 2004). Tif1γ also promotes erythropoiesis in mammals (He et al., 2006), but its molecular mechanism remained elusive. Bai et al. created a viable homozygous tif1γ−/− line by rescuing zebrafish by using a green fluorescent protein (GFP)-labeled wild-type tif1γ transgene, and subsequently performed a haploid genetic suppressor screen (see Fig. 2). The screen identified that mutation of the cdc73 gene [which encodes a component of the PAF (Pol-II-associated factor) complex], restores erythropoiesis in tif1γ−/− embryos. Knocking down the expression of other PAF subunits or components of the 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) sensitivity-inducing factor (DSIF) complex restored red blood cells to wild-type levels in tif1γ−/− embryos. Mechanistically, PAF and DSIF stall RNA polymerase II (Pol II) at target gene promoters. tif1γ recruits transcription elongation factors such as positive transcription elongation factor b (p-TEFb) and facilitates chromatin transcription (FACT) complex to erythroid genes to release the paused Pol II, thereby promoting transcription. Importantly, tif1γ-regulated elongation is conserved in human cells. The suppressor screen thus uncovered an unexpected role of tif1γ in transcriptional elongation during erythroid cell differentiation.
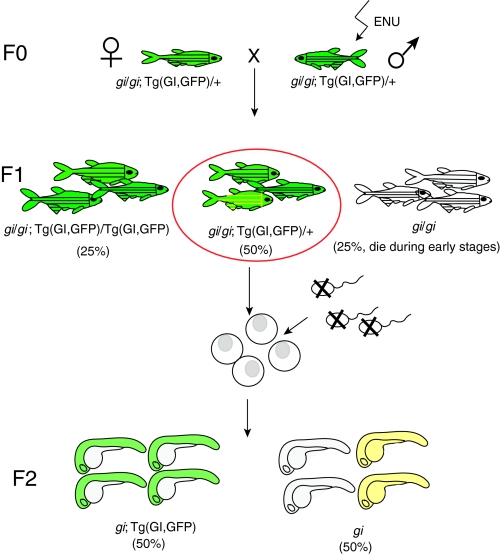
Fig. 2.
Scheme of a genetic suppressor screen. Lethal mutations in a gene of interest (gi) can be rescued by co-expressing a transgene in which a wild-type copy of the gene (GI) marked with GFP is driven by an appropriate promoter [note that a bacterial artificial chromosome (BAC) transgene that contains the entire locus of the GI gene could be used in the absence of suitable promoter, as in the case for moonshine (Bai et al., 2010)], enabling survival to adulthood (F0). Then, male F0 fish are mutagenized with the chemical ENU and are crossed to untreated female fish to generate F1 progeny. The F1 female fish that carry one copy of the transgene are used for subsequent in vitro fertilization. Eggs are squeezed out from the mother and fertilized with UV-treated sperm (which still activates the eggs but does not contribute chromosomes). All of the eggs develop into haploid embryos (F2), which stay alive until 5 days post-fertilization. 50% of the F2 embryos are GFP-positive (i.e. they express the rescuing transgene) and develop normally. The other 50% of haploid embryos are GFP-negative (i.e. they do not express the rescuing transgene) and develop recessive mutant phenotypes. However, if the mother carries a suppressor mutation in its genome (contributed by the ENU-mutagenized F0 male), 50% of the GFP-negative haploid embryos will be rescued (yellow).
Zebrafish reverse genetics: transient and stable genetic manipulations
The function of candidate genes during hematopoietic development and the genetic interactions that affect disease phenotypes can be quickly examined in zebrafish embryos and larvae through microinjections of mRNA (overexpression) and morpholino antisense oligomers (knockdown) (Nasevicius and Ekker, 2000), without the need to make stable animal strains. Human blood diseases have been phenocopied and studied in zebrafish using morpholino injection. For instance, ribosomal proteins are frequently mutated in patients with Diamond-Blackfan anemia (DBA), but it is not clear how the ribosomal defects lead to the disease. Two reports have found that knockdown of ribosomal protein 19 (RPS19) in zebrafish results in dramatic loss of red blood cells as well as morphological abnormalities that resemble DBA (Danilova et al., 2008; Uechi et al., 2008). In addition, the maturation and proliferation of erythroid lineages are severely blocked, as found in humans with the disease. The dose effect of RPS19 on disease phenotype was demonstrated by the injection of different amounts of morpholinos. Loss of RPS19 induces broad activation of the p53 pathway and apoptosis in embryos, including of erythroid cells, which is consistent with observations from murine models. Suppression of the p53 pathway partially rescues RPS19-deficient phenotypes. In addition, mutations in other ribosomal proteins all led to similar activation of p53 family proteins. These studies pointed out that the p53 protein family could be a target for DBA therapies.
Although gene targeting by homologous recombination is widely used in mice, this method is still lacking in zebrafish. However, recent studies have developed the techniques TILLING (Targeting Induced Local Lesions In Genome) (Stemple, 2004) and zinc-finger nuclease (ZFN)-mediated mutation (Doyon et al., 2008; Meng et al., 2008; Sander et al., 2011) to engineer stable strains carrying defects at specific gene loci. Numerous mutants have been made using these approaches. Recently, the von Hippel-Lindau tumor suppressor (VHL) mutant was identified through TILLING (van Rooijen et al., 2009); this represents the first viable embryonic animal model carrying a VHL mutation. The mutants develop polycythemia, resembling the clinical manifestation of VHL-associated Chuvash polycythemia, thereby offering the first in vivo system to study the progression of this disease.
Furthermore, injection of mRNA and cDNA into zebrafish embryos can be used to quickly dissect the functions of specific mutations identified in patients. In the aforementioned human disorder hemochromatosis, different mutations in ferroportin1 (SLC40A1) lead to iron overload in either macrophages or hepatocytes. Determining the effect of specific mutations often requires monitoring the localization of the protein in cultured cells and the level of cellular iron accumulation. De Domenico and colleagues injected the mutant ferroportin1 cDNA into zebrafish embryos and found that only mutations affecting macrophages, not hepatocytes, severely limit erythropoiesis, demonstrating a rapid method by which to diagnose the effect of specific ferroportin1 mutations found in humans (De Domenico et al., 2007).
Recent insights into leukemogenesis from transgenic zebrafish
Zebrafish cancer models are predominantly generated through transgenic expression of oncogenes. Generating stable transgenic zebrafish is efficiently done with Tol2 transposons (Kawakami, 2007). Several approaches, such as the Cre-loxP and Gal4-UAS systems, have been developed to control the spatiotemporal expression of transgenes (Halpern et al., 2008; Jopling et al., 2010; Mosimann et al., 2011). The first zebrafish model of leukemia was of T-cell acute lymphoblastic leukemia (T-ALL), and involved transgenic zebrafish expressing the mouse Myc gene tagged with enhanced GFP (EGFP) under the control of the rag2 promoter (rag2:EGFP-Myc) (Langenau et al., 2003). The leukemia induced in this model mirrors many aspects of the human disease, with the added benefit that the EGFP-labeled leukemic cells can be directly visualized. The induction of leukemia in this model is so efficient that the majority of the injected animals die before reaching sexual maturity. A conditional Cre-loxP system was subsequently introduced: in double transgenics expressing rag2:loxP-dsred-loxP-GFP-myc and heat-shock-inducible Cre recombinase (hsp70:cre), T-cell lymphoblastic lymphoma (T-LBL) was efficiently induced after heat-shock induction, and T-LBL rapidly progressed to T-ALL (Feng et al., 2007). The authors of this study then used the transgenic zebrafish to dissect the molecular pathways involved in the disease progression by testing candidate genes (Feng et al., 2010) because, in humans, the molecular cause for the progression from the highly localized thymic T-LBL to disseminated T-ALL is not clear. The zebrafish experiments revealed that apoptosis regulator BCL2, sphingosine 1-phosphate receptor 1 (S1P1) and intercellular adhesion molecule 1 (ICAM1) are highly expressed in T-LBL; this inhibits apoptosis and prevents the tumor cells from invading the vasculature owing to increased cell-cell adhesion. Akt activation can overcome the block in intravasation and promote the dissemination of T-LBL cells. These studies indicate that treating T-LBL patients with a combination of BCL2 and Akt inhibitors could promote lymphoblast death and also prevent the dissemination of tumor cells. The identified markers can further be used to determine T-LBL- and T-ALL-specific therapies.
Myeloid leukemia has also been modeled in zebrafish. The AML1-ETO (AE) translocation (also known as RUNX1-RUNX1T1) is the most common chromosomal translocation in acute myeloid leukemia (AML). Ubiquitous overexpression of the oncogenic AE protein in zebrafish embryos leads to early lethality and thus requires inducible expression by the heat-shock promoter in transgenic fish (hsp70: AML1-ETO) (Yeh et al., 2008). The induced AE protein in embryos redirects erythroid progenitor cells to develop into granulocytic cells, strongly resembling the cell fate redirections observed in AE-associated AML patients. This study further found that downregulation of the hematopoietic master regulator gene scl is required for the cell fate switch.
Chemical screening in zebrafish: in vivo drug discovery
Zebrafish embryos are permeable to water-soluble chemicals and can be housed in 96-well plates, making them ideal for high-throughput chemical screening for novel bioactive compounds (Zon and Peterson, 2005). Compared with cell-based drug screening, a whole-animal model substantially reduces the likelihood of encountering common issues that arise during late stages of drug development, such as drug toxicity and off-target side effects (Bowman and Zon, 2008). Chemical screens based on zebrafish disease models have the potential to identify lead compounds for novel therapeutics. For example, in a chemical screen that was conducted to identify new regulators of HSCs, North et al. found that prostaglandin E2 (PGE2) increased HSC number (North et al., 2007). PGE2 interacts with the Wnt signaling pathway to regulate HSCs through cAMP-PKA-mediated β-catenin phosphorylation (Goessling et al., 2009). A stable derivative of PGE2 (dmPGE2) increases the frequency of long-term repopulating HSCs in irradiated murine bone marrow, strongly suggesting a therapeutic potential (North et al., 2007). dmPGE2 is currently in clinical trials for treating patients receiving cord blood stem cell transplants. As another example, Yeh and colleagues utilized the transgenic hsp70:AML1-ETO line discussed above and screened for compounds that suppressed the inducible erythropoiesis-to-granulopoiesis conversion (Yeh et al., 2009). They found that chemicals that inhibit PGE2 or β-catenin pathways block the capacity of AE to redirect hematopoiesis, a strategy that could potentially provide therapeutic benefits in AML patients. These two independent screens collectively demonstrate the crucial roles of PGE2 and β-catenin interaction in normal and leukemic hematopoiesis.
In addition to identifying therapeutic compounds, chemical screens can also reveal genetic pathways that are involved in hematopoiesis. In the same chemical screen that unveiled PGE2 as a regulator of HSCs, compounds that regulate blood flow were also found to affect HSC formation during embryogenesis. Further analysis demonstrated that the initiation of circulation seems to trigger the formation of HSCs and that circulation regulates HSCs via nitric oxide (NO) signaling (North et al., 2009).
Live imaging and hematopoietic cell transplantation
Zebrafish embryos, which are transparent, allow direct visualization of hematopoietic cells in live animals. Various fluorescent transgenic lines have been generated that facilitate the imaging of hematopoietic cells (Bertrand and Traver, 2009). Three studies in 2010 reported imaging of the birth of HSCs during development (Bertrand et al., 2010; Boisset et al., 2010; Kissa and Herbomel, 2010). The precise origin of HSCs – whether they arise from the aortic floor or the underlying mesenchyme within the aorta-gonad-mesonephros (AGM) region –has been a longstanding question in the field. Two studies directly imaged HSC creation in zebrafish embryos using fluorescent reporter transgenes to mark nascent HSCs (cmyb:eGFP or Cd41:GFP) and vascular precursors (flk1:memcherry, flk1:GFP or Lmo2:dsred). The authors found that HSCs directly emerge from aortic endothelial cells through unique cell movement in which single endothelial cells bend out of the aortic ventral wall to the sub-aortic space (Bertrand et al., 2010; Kissa and Herbomel, 2010). These results were corroborated in a third study, in which the mouse embryos were cut into transverse sections and the deeply located aorta was imaged in the cultured live sections (Boisset et al., 2010). These studies provide a better understanding of HSC emergence during development, which will aid efforts to eventually instruct HSC formation from embryonic stem cells or induced pluripotent stem cells (iPSCs) for clinical applications.
Several techniques have been developed in zebrafish to label single hematopoietic cells. Photo-activation of caged fluorescent compounds has been employed in zebrafish embryos to follow the migration and cell fate of individual hematopoietic precursor cells or HSCs (Bertrand et al., 2008; Jin et al., 2007; Kissa et al., 2008; Vogeli et al., 2006; Warga et al., 2009). In addition, an inducible Cre transgene (encoding Cre protein fused to estrogen receptor) and Lox-switch transgene (loxP-stop-loxP-GFP) have been used to permanently label hematopoietic cells (Bertrand and Traver, 2009). In principle, by carefully choosing Cre and loxP transgenes, a small set of hematopoietic cells can be labeled during a specific temporal window. Then, the migration, proliferation and fate potentials of these cells can be studied in their physiological environments.
Furthermore, kidney marrow transplantation into sub-lethally irradiated animals has been developed in zebrafish (Pugach et al., 2009; Traver et al., 2004), and each major adult blood lineage can be isolated by flow cytometry (Traver et al., 2003). These assays allow the functional studies of adult HSCs, and are analogous to bone marrow transplantation assays in mice. Furthermore, a transparent adult zebrafish casper line was recently created, which enables the visualization of transplanted HSCs during homing and engraftment process in adult animals (White et al., 2008).
Conclusions
Studies using zebrafish have greatly infused our understanding of hematopoiesis and the perturbation of hematopoietic programs in disease. Previous screens mainly identified genes that are crucial for embryonic erythropoiesis. Future chemical and genetic screens will continue to identify additional genes and new compounds that modulate the generation of HSCs and other downstream lineages. The speed and scale of the screens will be enhanced by incorporating fluorescent transgenic lines, particularly with the new automated high-throughput manipulation and imaging platforms (Pardo-Martin et al., 2010). Functional assays of long-term HSC self-renewal in zebrafish (i.e. competitive and serial kidney marrow transplantation) are under development, and screens for HSC function are highly anticipated.
The use of zebrafish models for studying blood diseases such as leukemia is frequently limited by the low penetrance and long latency of the disease phenotype. Therefore, it is necessary to develop stable transgenic lines using different approaches to express pathogenic genes at sufficiently high levels to enhance the use of zebrafish models. The low penetrating phenotype might also indicate that multiple genetic mutations are required to provide a predisposing background for disease induction. Zebrafish are particularly amenable to multigenetic analysis through a combination of convenient microinjections of morpholinos and mRNA or cDNA. However, a significant challenge in using zebrafish is the current lack of target-specific conditional mutagenesis technology, although the new ZFN method has the potential to enable the development of a knock-in approach (Woods and Schier, 2008). Furthermore, modeling a disease in embryonic zebrafish provides a convenient platform in which to carry out genetic or chemical screens for suppressors or enhancers of the disease phenotype. However, it is important to understand how the disease and zebrafish models differ at a mechanistic level in order to select the optimal assays for high-throughput screens. Finally, these recently developed genetic tools allow modeling of human chronic blood disease in zebrafish adults. However, a better understanding of anatomy and physiology of adult hematopoiesis will be required to make optimal use of them.
Advantages of using zebrafish to study hematopoiesis and hematological malignancy.
High fecundity, small size and fast embryonic development make zebrafish suitable for large-scale forward and modifier genetic screens with low cost and space requirements. Phenotype-based screens provide unbiased identification of candidate genetic pathways.
Transient genetic manipulations mediated by morpholino, mRNA or cDNA injections offer convenient methods for functional analysis of candidate genes without the need to make transgenic animals.
Human blood diseases have been modeled in zebrafish, and small-molecule screens in such models have the potential to reveal new drugs for human disease treatment.
The transparency of zebrafish embryos and of the adult zebrafish casper line allows live imaging of hematopoietic cells and of the pathologies of the diseases that affect them.
In addition to the lack of knock-in technology, several other major limitations need to be overcome in order to fully exploit the potential of zebrafish in hematological research. For example, few antibody reagents are available for biochemical and immunohistochemistry studies in zebrafish systems. Production of target-protein-specific antibodies, including those that recognize cell-surface markers, for zebrafish is ongoing. In addition, studies in zebrafish have been hindered by the lack of methods that are available to culture and differentiate hematopoietic cells in vitro. Two recent papers reported in vitro culture and clonal assays of zebrafish hematopoietic progenitor cells (Stachura et al., 2009; Stachura et al., 2011). The identification of additional growth factors and the optimization of in vitro techniques will eventually enable the culture, proliferation, differentiation and therefore more in-depth study of all zebrafish blood-cell lineages.
Acknowledgments
We thank Xiaoying Bai, Teresa Bowman, Scott Lacadie, Christian Mosimann and Owen Tamplin for critical reading of the manuscript. L.I.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
COMPETING INTERESTS
L.I.Z. is a founder and stock holder of Fate, Inc. and a scientific advisor for Stemgent.
REFERENCES
- Amsterdam A., Nissen R. M., Sun Z., Swindell E. C., Farrington S., Hopkins N. (2004). Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA. 101, 12792–12797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Kim J., Yang Z., Jurynec M. J., Akie T. E., Lee J., LeBlanc J., Sessa A., Jiang H., DiBiase A., et al. (2010). TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell 142, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J. Y., Traver D. (2009). Hematopoietic cell development in the zebrafish embryo. Curr. Opin. Hematol. 16, 243–248 [DOI] [PubMed] [Google Scholar]
- Bertrand J. Y., Kim A. D., Teng S., Traver D. (2008). CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development 135, 1853–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J. Y., Chi N. C., Santoso B., Teng S., Stainier D. Y., Traver D. (2010). Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset J. C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., Robin C. (2010). In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 [DOI] [PubMed] [Google Scholar]
- Bowman T. V., Zon L. I. (2008). Swimming into the future of drug discovery: in vivo chemical screens in zebrafish. ACS Chem. Biol. 5, 159–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carradice D., Lieschke G. J. (2008). Zebrafish in hematology: sushi or science? Blood 111, 3331–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. T., Zon L. I. (2009). Zebrafish blood stem cells. J. Cell. Biochem. 108, 35–42 [DOI] [PubMed] [Google Scholar]
- Danilova N., Sakamoto K. M., Lin S. (2008). Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood 112, 5228–5237 [DOI] [PubMed] [Google Scholar]
- Davidson A. J., Zon L. I. (2004). The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene 23, 7233–7246 [DOI] [PubMed] [Google Scholar]
- De Domenico I., Vaughn M. B., Yoon D., Kushner J. P., Ward D. M., Kaplan J. (2007). Zebrafish as a model for defining the functional impact of mammalian ferroportin mutations. Blood 110, 3780–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong J. L., Zon L. I. (2005). Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu. Rev. Genet. 39, 481–501 [DOI] [PubMed] [Google Scholar]
- Donovan A., Brownlie A., Zhou Y., Shepard J., Pratt S. J., Moynihan J., Paw B. H., Drejer A., Barut B., Zapata A., et al. (2000). Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 403, 776–781 [DOI] [PubMed] [Google Scholar]
- Doyon Y., McCammon J. M., Miller J. C., Faraji F., Ngo C., Katibah G. E., Amora R., Hocking T. D., Zhang L., Rebar E. J., et al. (2008). Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 26, 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellett F., Lieschke G. J. (2010). Zebrafish as a model for vertebrate hematopoiesis. Curr. Opin. Pharmacol. 10, 563–570 [DOI] [PubMed] [Google Scholar]
- Feng H., Langenau D. M., Madge J. A., Quinkertz A., Gutierrez A., Neuberg D. S., Kanki J. P., Look A. T. (2007). Heat-shock induction of T-cell lymphoma/leukaemia in conditional Cre/lox-regulated transgenic zebrafish. Br. J. Haematol. 138, 169–175 [DOI] [PubMed] [Google Scholar]
- Feng H., Stachura D. L., White R. M., Gutierrez A., Zhang L., Sanda T., Jette C. A., Testa J. R., Neuberg D. S., Langenau D. M., et al. (2010). T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell 18, 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessling W., North T. E., Loewer S., Lord A. M., Lee S., Stoick-Cooper C. L., Weidinger G., Puder M., Daley G. Q., Moon R. T., et al. (2009). Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell 136, 1136–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordeuk V. R., Caleffi A., Corradini E., Ferrara F., Jones R. A., Castro O., Onyekwere O., Kittles R., Pignatti E., Montosi G., et al. (2003). Iron overload in Africans and African-Americans and a common mutation in the SCL40A1 (ferroportin 1) gene. Blood Cells Mol. Dis. 31, 299–304 [DOI] [PubMed] [Google Scholar]
- Halpern M. E., Rhee J., Goll M. G., Akitake C. M., Parsons M., Leach S. D. (2008). Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 5, 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Dorn D. C., Erdjument-Bromage H., Tempst P., Moore M. A., Massague J. (2006). Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell 125, 929–941 [DOI] [PubMed] [Google Scholar]
- Huang H. T., Zon L. I. (2008). Regulation of stem cells in the zebra fish hematopoietic system. Cold Spring Harb. Symp. Quant. Biol.. 73, 111–118 [DOI] [PubMed] [Google Scholar]
- Jin H., Xu J., Wen Z. (2007). Migratory path of definitive hematopoietic stem/progenitor cells during zebrafish development. Blood 109, 5208–5214 [DOI] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Marti M., Raya A., Belmonte J. C. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. (2007). Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 8 Suppl. 1, S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K., Herbomel P. (2010). Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115 [DOI] [PubMed] [Google Scholar]
- Kissa K., Murayama E., Zapata A., Cortes A., Perret E., Machu C., Herbomel P. (2008). Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood 111, 1147–1156 [DOI] [PubMed] [Google Scholar]
- Langenau D. M., Traver D., Ferrando A. A., Kutok J. L., Aster J. C., Kanki J. P., Lin S., Prochownik E., Trede N. S., Zon L. I., et al. (2003). Myc-induced T cell leukemia in transgenic zebrafish. Science 299, 887–890 [DOI] [PubMed] [Google Scholar]
- Lieschke G. J., Currie P. D. (2007). Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 8, 353–367 [DOI] [PubMed] [Google Scholar]
- Meng X., Noyes M. B., Zhu L. J., Lawson N. D., Wolfe S. A. (2008). Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 26, 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montosi G., Donovan A., Totaro A., Garuti C., Pignatti E., Cassanelli S., Trenor C. C., Gasparini P., Andrews N. C., Pietrangelo A. (2001). Autosomal-dominant hemochromatosis is associated with a mutation in the ferroportin (SLC11A3) gene. J. Clin. Invest. 108, 619–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C., Kaufman C. K., Li P., Pugach E. K., Tamplin O. J., Zon L. I. (2011). Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A., Ekker S. C. (2000). Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26, 216–220 [DOI] [PubMed] [Google Scholar]
- North T. E., Zon L. I. (2003). Modeling human hematopoietic and cardiovascular diseases in zebrafish. Dev. Dyn. 228, 568–583 [DOI] [PubMed] [Google Scholar]
- North T. E., Goessling W., Walkley C. R., Lengerke C., Kopani K. R., Lord A. M., Weber G. J., Bowman T. V., Jang I. H., Grosser T., et al. (2007). Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North T. E., Goessling W., Peeters M., Li P., Ceol C., Lord A. M., Weber G. J., Harris J., Cutting C. C., Huang P., et al. (2009). Hematopoietic stem cell development is dependent on blood flow. Cell 137, 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S. H., Zon L. I. (2008). Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik E. J., Zon L. I. (2010). Hematopoietic development in the zebrafish. Int. J. Dev. Biol. 54, 1127–1137 [DOI] [PubMed] [Google Scholar]
- Pardo-Martin C., Chang T. Y., Koo B. K., Gilleland C. L., Wasserman S. C., Yanik M. F. (2010). High-throughput in vivo vertebrate screening. Nat. Methods 7, 634–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton E. E., Zon L. I. (2001). The art and design of genetic screens: zebrafish. Nat. Rev. Genet.. 2, 956–966 [DOI] [PubMed] [Google Scholar]
- Payne E., Look T. (2009). Zebrafish modelling of leukaemias. Br. J. Haematol. 146, 247–256 [DOI] [PubMed] [Google Scholar]
- Pugach E. K., Li P., White R., Zon L. (2009). Retro-orbital injection in adult zebrafish. J. Vis. Exp. 34, 10.3791/1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom D. G., Haffter P., Odenthal J., Brownlie A., Vogelsang E., Kelsh R. N., Brand M., van Eeden F. J., Furutani-Seiki M., Granato M., et al. (1996). Characterization of zebrafish mutants with defects in embryonic hematopoiesis. Development 123, 311–319 [DOI] [PubMed] [Google Scholar]
- Ransom D. G., Bahary N., Niss K., Traver D., Burns C., Trede N. S., Paffett-Lugassy N., Saganic W. J., Lim C. A., Hersey C., et al. (2004). The zebrafish moonshine gene encodes transcriptional intermediary factor 1gamma, an essential regulator of hematopoiesis. PLoS Biol. 2, E237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D., Dahlborg E. J., Goodwin M. J., Cade L., Zhang F., Cifuentes D., Curtin S. J., Blackburn J. S., Thibodeau-Beganny S., Qi Y., et al. (2011). Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat. Methods 8, 67–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafizadeh E., Paw B. H. (2004). Zebrafish as a model of human hematologic disorders. Curr Opin Hematol. 11, 255–261 [DOI] [PubMed] [Google Scholar]
- Stachura D. L., Reyes J. R., Bartunek P., Paw B. H., Zon L. I., Traver D. (2009). Zebrafish kidney stromal cell lines support multilineage hematopoiesis. Blood 114, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachura D. L., Svoboda O., Lau R. P., Balla K. M., Zon L. I., Bartunek P., Traver D. (2011). Clonal analysis of hematopoietic progenitor cells in the zebrafish. Blood (Epub ahead of print). [DOI] [PMC free article] [PubMed]
- Stemple D. L. (2004). TILLING-a high-throughput harvest for functional genomics. Nat Rev Genet. 5, 145–150 [DOI] [PubMed] [Google Scholar]
- Traver D., Paw B. H., Poss K. D., Penberthy W. T., Lin S., Zon L. I. (2003). Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol.. 4, 1238–1246 [DOI] [PubMed] [Google Scholar]
- Traver D., Winzeler A., Stern H. M., Mayhall E. A., Langenau D. M., Kutok J. L., Look A. T., Zon L. I. (2004). Effects of lethal irradiation in zebrafish and rescue by hematopoietic cell transplantation. Blood 104, 1298–1305 [DOI] [PubMed] [Google Scholar]
- Uechi T., Nakajima Y., Chakraborty A., Torihara H., Higa S., Kenmochi N. (2008). Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of Diamond-Blackfan anemia. Hum. Mol. Genet. 17, 3204–3211 [DOI] [PubMed] [Google Scholar]
- van Rooijen E., Voest E. E., Logister I., Korving J., Schwerte T., Schulte-Merker S., Giles R. H., van Eeden F. J. (2009). Zebrafish mutants in the von Hippel-Lindau tumor suppressor display a hypoxic response and recapitulate key aspects of Chuvash polycythemia. Blood 113, 6449–6460 [DOI] [PubMed] [Google Scholar]
- Vogeli K. M., Jin S. W., Martin G. R., Stainier D. Y. (2006). A common progenitor for haematopoietic and endothelial lineages in the zebrafish gastrula. Nature 443, 337–339 [DOI] [PubMed] [Google Scholar]
- Warga R. M., Kane D. A., Ho R. K. (2009). Fate mapping embryonic blood in zebrafish: multi- and unipotential lineages are segregated at gastrulation. Dev. Cell 16, 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein B. M., Schier A. F., Abdelilah S., Malicki J., Solnica-Krezel L., Stemple D. L., Stainier D. Y., Zwartkruis F., Driever W., Fishman M. C. (1996). Hematopoietic mutations in the zebrafish. Development 123, 303–309 [DOI] [PubMed] [Google Scholar]
- White R. M., Sessa A., Burke C., Bowman T., LeBlanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C. E., et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods I. G., Schier A. F. (2008). Targeted mutagenesis in zebrafish. Nat Biotechnol. 26, 650–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh J. R., Munson K. M., Chao Y. L., Peterson Q. P., Macrae C. A., Peterson R. T. (2008). AML1-ETO reprograms hematopoietic cell fate by downregulating scl expression. Development 135, 401–410 [DOI] [PubMed] [Google Scholar]
- Yeh J. R., Munson K. M., Elagib K. E., Goldfarb A. N., Sweetser D. A., Peterson R. T. (2009). Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat. Chem. Biol. 5, 236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon L. I., Peterson R. T. (2005). In vivo drug discovery in the zebrafish. Nat. Rev. Drug. Discov.. 4, 35–44 [DOI] [PubMed] [Google Scholar]