SUMMARY
Fluorescent proteins (FPs) have great utility in identifying specific cell populations and in studying cellular dynamics in the mouse. To quantify the factors that determine both the expression and relative brightness of FPs in mouse embryonic stem cells (mESCs) and in mice, we generated eight different FP-expressing ROSA26 alleles using recombinase-mediated cassette exchange (RMCE). These alleles enabled us to analyze the effects on FP expression of a translational enhancer and different 3′-intronic and/or polyadenylation sequences, as well as the relative brightness of five different FPs, without the confounding position and copy number effects that are typically associated with randomly inserted transgenes. We found that the expression of a given FP can vary threefold or more depending on the genetic features present in the allele. The optimal FP expression cassette contained both a translational enhancer sequence in the 5′-untranslated region (UTR) and an intron-containing rabbit β-globin sequence within the 3′-UTR. The relative expressed brightness of individual FPs varied up to tenfold. Of the five different monomeric FPs tested, Citrine (YFP) was the brightest, followed by Apple, eGFP, Cerulean (CFP) and Cherry. Generation of a line of Cherry-expressing mice showed that there was a 30-fold variation of Cherry expression among different tissues and that there was a punctate expression pattern within cells of all tissues examined. This study should help investigators make better-informed design choices when expressing FPs in mESCs and mice.
INTRODUCTION
Fluorescent proteins (FPs) are widely used to monitor gene expression and study cellular dynamics in cells and tissues (Tsien, 1989; Heim et al., 1994; Tsien and Miyawaki, 1998; Shaner et al., 2004). However, the successful use of FPs requires that their relative brightness in a cell exceeds the experimental detection threshold (Niswender et al., 1995). Many variables, both intrinsic and extrinsic to the FP, influence the brightness of FPs when expressed in cells or mice. Factors intrinsic to the FP include quantum efficiency, the necessity for some FP variants to dimerize or fold in a specific manner, and the nature of the protein to which they are attached. Factors extrinsic to the FP include the transcriptional activity of the promoter being used and both the half-life and translational efficiency of the mRNA generated.
Although some of the intrinsic factors that influence FP brightness can readily be determined, extrinsic factors have been more difficult to quantify. This is largely owing to the confounding effects of transgene copy number and the genetic insertion site, which can cause large variations in gene expression. Although there have been prior attempts to compare different FP reporter constructs in vitro (Falcone and Andrews, 1991; Yew et al., 1997), the results obtained have been neither quantitative nor conclusive. Similarly, although a large amount of data has been collected on the properties of bacterially-expressed FPs (Olenych et al., 2007), there have been fewer studies to compare the relative brightness of FPs in mammalian cells. As a result, it is not uncommon for suboptimal reporter design to result in wasted time and effort when generating new lines of FP-expressing mice.
The method of recombinase-mediated cassette exchange (RMCE) enables the efficient generation of an allelic series in embryonic stem cells (ESCs) and mice (Araki et al., 2002). RMCE is a two-step procedure. The first step is the generation, by gene targeting, of an acceptor allele containing the appropriate recombinase recognition sites and chemically selectable markers. The second step is to exchange a cassette containing DNA sequences of interest into the acceptor allele by RMCE. An advantage of this method is that it enables multiple allelic variants to be generated at a defined genetic location with greater ease than can be achieved by repetitive gene targeting.
To quantify the effects of intrinsic and extrinsic variables that affect both the expression and brightness of FPs, we generated a loxed cassette acceptor (LCA) allele for the ubiquitously expressed Gt(ROSA)26Sor (ROSA26) gene (Zambrowicz et al., 1997; Kohlhepp et al., 2001; Safran et al., 2003). Utilizing the ROSA26LCA allele, we generated eight different FP reporter alleles by RMCE, which were used to assess the effects of several regulatory elements on FP expression and to compare the brightness of five different FP variants. As part of this effort, we also made several plasmids that can be used to generate both targeting vectors and exchange cassettes by BAC recombineering. The plasmids, mouse ESCs (mESCs), expression cassettes and findings that we describe should be of general use in generating other LCA alleles, basal exchange cassettes, and for optimizing the expression of FPs and other reporter alleles in mESCs and mice.
RESULTS
Generation of a ROSA26LCA allele
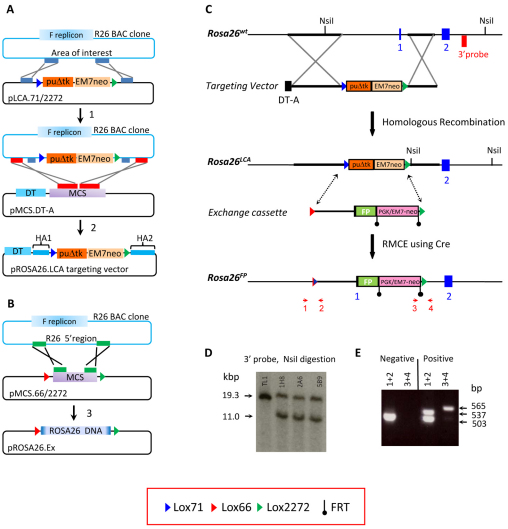
A targeting vector was made using a two-step BAC recombineering method, as shown in Fig. 1A. First, tandem lox71 and lox2272 sites, flanking both PGK-puΔtk and EM7-neoR selection cassettes, were inserted into a ROSA26 BAC clone (Fig. 1A) using kanamycin to select for recombinant clones. Second, the entire LCA selection cassette along with flanking upstream and downstream homology arms of 8.232 kb and 3.770 kb, respectively, was retrieved into pMCS.DT-A by recombineering-mediated gap repair. The targeting vector that was generated replaced a 5.165 kb region of the ROSA26 gene, containing both the promoter and first exon, with the dual PGK-puΔtk and EM7-neoR selection cassette. The heteromeric lox sites that were present in the targeting vector have been previously reported to allow efficient, unidirectional RMCE (Araki et al., 2002). After electroporation, 270 puromycin-resistant mESC clones were obtained. Southern blot analysis identified three that had undergone the desired homologous recombination event (Fig. 1C,D). Clones 5B9 and 2A6, which demonstrated germline transmission, were used subsequently for all RMCE experiments.
Fig. 1.
BAC recombineering, gene targeting and RMCE strategies. (A) Steps for making a ROSA26 gene targeting vector by BAC recombineering. Short inner homology arms were amplified from a BAC containing the ROSA26 gene and cloned into the pLCA.71/2272 vector. In the first recombination step (1), the cassette containing a puΔtk fusion gene surrounded by tandem lox71 and lox2272 sites was introduced into the BAC. After cloning short outer homology arms into pMCS.DT-A, a second recombination step (2) enabled retrieval of the targeting vector. DT, diptheria toxin A; MCS, multiple cloning site; HA1/2, short homology arm 1 or 2. (B) Generation of the ROSA26 exchange vector by BAC recombineering. pMCS.66/2272 was used to retrieve a 5.165-kb fragment of the ROSA26 gene, thereby generating a basal exchange vector. A variant of this plasmid was used to make all FP exchange vectors as shown in Fig. 2. (C) Gene targeting and RMCE strategy. A ROSA26LCA allele containing the puΔtk fusion gene flanked by tandem lox71 and lox2272 sites was made by gene targeting in mESCs. Puromycin-resistant mESC clones were screened by Southern blot analysis. DNA was digested with NsiI and hybridized with the 3′ probe indicated in the figure. Correctly targeted clones containing the ROSA26LCA allele were used to perform RMCE to derive eight different FP-expressing alleles. PCR using primers 1–4 (red arrows) were used for the screening of clones that were resistant to puromycin and sensitive to gancyclovir. (D) Southern blot analysis. The presence of an 11 kb band indicates a correctly targeted mESC clone with the ROSA26LCA allele. (E) DNA PCR analysis. Representative PCR amplification results are shown, indicating correct exchange of the exchange cassette into the ROSA26LCA allele. Prior to RMCE, cells with the ROSA26LCA allele exhibit a 503 bp band after PCR using primers 1 and 2. Correctly exchanged clones exhibit both a 537 bp and 503 bp product using primers 1 and 2, and a 565 bp product with primers 3 and 4.
Design of an allelic series of RMCE vectors and insertion into the ROSA26 gene locus
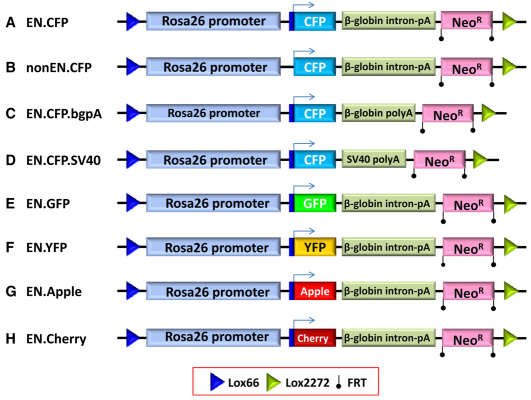
We next used pMCS.66/2272, a plasmid with tandem lox66 and lox2272 sites, to retrieve by BAC recombineering a 4.1 kb region of the ROSA26 gene that is absent in the ROSA26LCA mESCs (Fig. 1B). The resulting plasmid, pROSA26.Ex, was used as the basis for making eight different RMCE vectors that contained a dual promoter (PGK and EM7)-driven neoR cassette so as to enable G418 to be used as a positive selectable marker during RMCE (Fig. 2). The correct exchange of each vector into the ROSA26LCA allele after RMCE was confirmed by PCR analysis (Fig. 1C,E). To simplify assembly, only a 1.065 kb region of the ROSA26 gene, containing the sequence downstream of the promoter, was replaced, thereby allowing expression of the FPs to occur without the need to include splice donor and acceptor sites (Nyabi et al., 2009). Other aspects of the cassette were varied in order to assess the impact of different transcriptional and translational components. First, we examined the impact of a putative 5′-untranslated region (UTR) translational enhancer sequence from the Xenopus β-globin gene, hereafter termed EN (Falcone and Andrews, 1991). Second, we examined three different 3′-UTR configurations: a Simian virus 40 polyadenylation sequence (SV40), a rabbit β-globin polyadenylation sequence (bgpA) and an intron-containing rabbit β-globin polyadenylation sequence. Lastly, we examined the relative brightness of five different FP variants: Apple, Cerulean (CFP), Cherry, Citrine (YFP) and enhanced GFP (GFP).
Fig. 2.
FP-expressing ROSA26 alleles. This figure illustrates the design of the eight different FP-expressing alleles made and used in this study. In all alleles, a 4.081 kb region of the ROSA26 gene was inserted into the ROSA26LCA allele in order to reconstitute promoter activity. The blue arrow indicates the start site for FP translation. (A–D) Four alleles were generated whose design was varied slightly in order to assess the role of (1) a translational enhancer [small dark-blue box upstream (left) of arrow] and (2) the effect of three different 3′-UTR configurations. (A) The EN.CFP allele contains the translational enhancer sequence, CFP (Cerulean), and both a splice and polyadenylation site from the rabbit β-globin gene. (B) The nonEN.CFP allele lacks the translational enhancer. (C) EN.CFP.bgpA contains a β-globin 3′-UTR and polyadenylation signal sequence but lacks exon 2 and intron 2 that are present in EN.CFP. (D) EN.CFP.SV40 contains an SV40 polyadenylation site. (E–H) FP color variants tested. Each allele is of the same design as EN.CFP (A) but contains GFP (E), Citrine (F), Apple (G) or Cherry (H).
Effects of extrinsic factors on FP expression
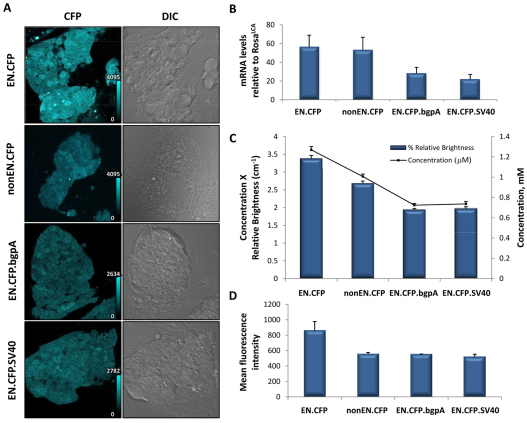
We first sought to determine the effects of the allele design by comparing the expression of the four alleles that express CFP (Cerulean). Cerulean has been shown to be a bright CFP variant with efficient folding at 37°C (Rizzo et al., 2004). Correctly exchanged clones were analyzed by fluorescence-activated cell sorting (FACS) for the absence of mosaicism (data not shown). Several that exhibited considerable non-fluorescent cell populations were not used in the subsequent studies. The mRNA expression of two different clones of each allele was determined by quantitative PCR and compared with that of the EN.CFP allele (which contains the Xenopus β-globin 5′-UTR translational enhancer sequence, CFP, and an intron-containing rabbit ′-globin polyadenylation sequence). As shown in Fig. 3B, the EN.CFP allele showed the highest level of expression, followed by nonEN.CFP (lacking the translational enhancer), EN.CFP.bgpA (containing a rabbit β-globin polyadenylation sequence) and finally EN.CFP.SV40 (containing a SV40 polyadenylation sequence). One-tailed Student’s t-tests showed no significant difference between EN.CFP and nonEN.CFP mRNA levels. Similarly, there was no significant difference in the expression of EN.CFP.bgpA and EN.CFP.SV40. However, the presence of the β-globin intron resulted in a twofold higher level of expression as compared with the allele that only contains a polyadenylation sequence. Similarly, the mRNA expression level of both the EN.CFP and nonEN.CFP alleles was approximately 2.5-fold higher than that of EN.CFP.SV40.
Fig. 3.
Effects of extrinsic factors on FP expression. (A) Confocal images of mESC clones containing the four Cerulean-expressing alleles shown in Fig. 2. EN.CFP was visibly brighter than the other three variants. Calibration bars indicate the fluorescence intensity level that corresponds to one color shade in the image; intensity is expressed in arbitrary units. (B) mRNA levels relative to the colorless ROSA26LCA cells as determined by real-time PCR. Each data point represents the average of two independent mESC clones. P<0.05 for EN.CFP versus either EN.CFP.bgpA or EN.CFP.SV40. Bars indicate s.e.m. (n=6 for each bar). (C) Quantitative confocal microscopy measurements of FP concentration (black line) and relative actual expressed brightness (concentration × extinction coefficient × quantum yield; blue bars). The mean intensities for two clones, with ten images acquired for each clone, were averaged together for each data point. P<0.01 for all comparisons except between EN.CFP.bgpA and EN.CFP.SV40. Bars indicate s.e.m. (n=20 each). (D) Quantification of mean fluorescence intensities by FACS. Expression results for two clones were averaged together for each data point. Bars indicate s.e.m. (n=2 each).
Next, we explored the effects of regulatory elements on protein expression by using quantitative confocal microscopy. Relative FP concentrations were calculated on the basis of fluorescence intensities as compared with the ROSA26LCA allele, which does not contain a FP and was used as a non-expressing control. Ten confocal images for each of the mESC clones were taken at 40× magnification with no saturation areas to eliminate non-linearity in intensity measurements (Fig. 3A). In parallel, we imaged serial dilutions of purified FP with known concentrations and measured the fluorescence intensity for each concentration (Patterson et al., 1997). A linear equation of fluorescence intensity against concentration was plotted and used to calculate average concentration of each sample, thereby allowing direct comparison between FPs. The percentage of photons emitted, which correlates to relative brightness, was calculated using the following equation: relative brightness = quantum yield × molar extinction coefficient × concentration. The concentration and relative brightness for each of the four different CFP-containing alleles are shown in Fig. 3C. The EN.CFP allele showed the highest level of protein expression, with 3.4% relative brightness, followed by nonEN.CFP, EN.CFP.bgpA and then EN.CFP.SV40 (Fig. 3C). The EN.CFP allele, which contains the translational enhancer sequence, had 1.26-fold higher protein expression levels than nonEN.CFP, which lacked the Xenopus translational enhancer sequence (P<3.8×10−8). Mirroring the real-time PCR data, the EN.CFP allele, which has an intron-containing rabbit β-globin polyadenylation sequence, exhibited 1.75-fold higher (P<4.0×10−19) protein expression than EN.CFP.bgpA, which only contained the polyadenylation signal sequence from the rabbit β-globin gene as the 3′-UTR. Both the EN.CFP and nonEN.CFP alleles had 1.7-fold (P<1.7×10−16) and 1.4-fold (P<5.62×10−10) higher protein concentrations than the EN.CFP.SV40 allele, respectively. The EN.CFP.bgpA and EN.CFP.SV40 alleles did not show significant differences in protein expression when compared with each other.
To further assess the effects of the different regulatory elements on FP expression, we used FACS. Two clones for each allele were profiled, and the mean fluorescence intensity for each cell line was calculated (Fig. 3D). The results paralleled the confocal measurements, with the EN.CFP allele having the highest level of expression, followed by the nonEN.CFP, EN.CFP.bgpA and EN.CFP.SV40 alleles.
Comparison of intrinsic brightness of five different FPs
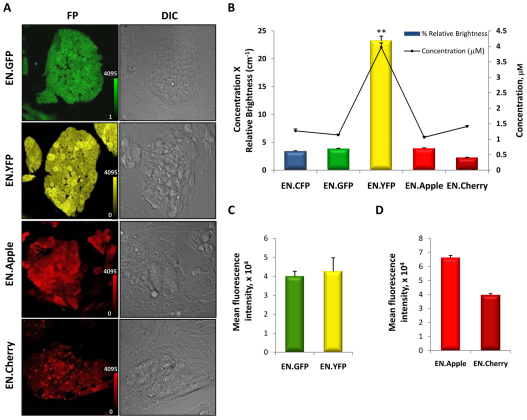
We next sought to examine the relative brightness of four other monomeric FPs (Cherry, Apple, Citrine and eGFP) with that of Cerulean. These FPs were chosen because they represent different parts of the visible light spectrum and have well-defined physical characteristics. For instance, eGFP is a widely used mutational variant of wild-type GFP owing to its superior brightness, photostability and relatively small sensitivity to pH (Cormack et al., 1996).
To limit variation in extrinsic variables that influence FP brightness, each allele was identical in design to the EN.CFP allele, with the only modification being the FP itself (Fig. 2A). As is well known, a number of factors influence apparent brightness of an FP beyond protein expression, including the efficiency and rate of maturation, the molar extinction coefficient values within the excitation wavelength range, and the quantum yield (Olenych et al., 2007; Rizzo et al., 2009). The known physical properties of the five FPs studied are summarized in Table 1. To directly compare the efficiency of different color variants of FPs (Fig. 2A,E-H), as represented by relative brightness, we employed the same quantitative confocal microscopy method described in the previous section. eGFP, Apple, Citrine and Cerulean (Fig. 3A and Fig. 4A) all exhibited uniform fluorescence within the mESC colonies. However, Cherry (Fig. 4A) showed a distinctly punctate pattern that seemed to be confined to cell bodies or vesicles. To compare FP intensities, a maximum threshold was set for the intensity measurements of Cherry because the extreme aggregation of proteins within cells rendered saturation unavoidable. Calculations of average protein concentrations, on the basis of calibration intensities of purified corresponding FPs, allowed direct comparison between different color variants. Fig. 4B shows the average concentrations and relative brightness of each FP variant. Citrine showed the highest level of protein concentration (4 μM), followed by Cherry (1.4 μM), Cerulean (1.3 μM), eGFP (1.1 μM) and Apple (1.0 μM). Citrine was also, by far, the brightest FP variant, with a normalized actual brightness of 5.53 (defined as the product of concentration × extinction coefficient × quantum yield) compared with eGFP (1.00). Citrine was followed in brightness by Apple (0.91), Cerulean (0.60) and Cherry (0.52). Differences in relative brightness between all FP variants were statistically significant, with the exception of the difference between Apple and eGFP. It is interesting to note that Citrine is 10.4-fold brighter than the dimmest color variant, Cherry, and approximately sixfold brighter than the remaining color variants. An assessment of the monomeric red FPs revealed that Apple was 1.7-fold brighter than Cherry.
Table 1.
Physical properties of the FPs used in this study
| Protein (color) | Excitation peak (nm) | Emission peak (nm) | Brightnessa | Reference |
|---|---|---|---|---|
| Cerulean (cyan) | 433/445 | 475/503 | 27/24 | Rizzo et al., 2004 |
| EGFP (green) | 488 | 507 | 34 | Cormack et al., 1996 |
| Citrine (yellow) | 516 | 529 | 59 | Griesbeck et al., 2001 |
| Apple (red) | 568 | 592 | 37 | Shaner et al., 2008 |
| Cherry (red) | 587 | 610 | 16 | Shaner et al., 2004 |
Protein names and property values are based on literature.
Product of molar extinction coefficient and quantum yield (mM × cm)−1.
Fig. 4.
Comparison of intrinsic brightness of different color variants of FPs. (A) Confocal images of ESC clones with alleles that express GFP (EN.GFP), Citrine (EN.YFP), Apple (EN.Apple) and Cherry (EN.Cherry). Calibration bars indicate the fluorescence intensity level that corresponds to one color shade in the image; intensity is expressed in arbitrary units. (B) Quantitative confocal microscopy measurements of FP concentration (black line) and relative actual expressed brightness (concentration × extinction coefficient × quantum yield; colored bars). The mean intensities for two clones, with ten images acquired for each clone, were averaged together for each data point. **P<0.01 for all comparisons except between EN.GFP and EN.Apple. Bars indicate s.e.m. (n=20 each). (C,D) Quantification of mean fluorescence intensities of EN.GFP and EN.YFP by FACS using a 488 nm excitation laser (C), and EN.Apple and EN.Cherry using a 532 nm excitation laser (D). Bars indicate s.e.m. (n=2 each).
Again, FACS analysis was used to confirm the differences between different fluorescent color variants. It was not possible to quantitatively compare the FPs by FACS using the same excitation wavelengths used in confocal microscopy. However, the same FACS excitation wavelengths were used for YFP and eGFP, as well as for Cherry and Apple. Thus, it was possible to quantitatively compare fluorescence mean intensity between YFP and eGFP (Fig. 4C), and between Cherry and Apple (Fig. 4D). In this setting, Apple exhibited 1.4-fold greater fluorescence intensity than Cherry. By contrast, Citrine and eGFP showed no significant difference.
Generation of polychrome fluorescent embryos and confirmation of germline competency of the mESCs
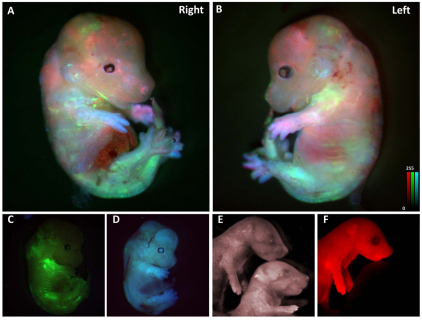
To confirm the pluripotency of mutant mESC lines after gene targeting and RMCE, and to validate the expression of three of the FPs in vivo, we performed blastocyst injections of mESCs expressing cyan (EN.CFP), green (EN.GFP) and red (EN.Cherry) FPs. Four ESCs of each clone were simultaneously injected per blastocyst then transferred into pseudopregnant females. Seven out of nine transfers resulted in pregnancies. Embryos were harvested from two of the pregnant animals at E15.5 and two at E17.5 for fluorescent imaging. In total, 21 of 34 embryos exhibited varying degrees of fluorescence, with the majority of the embryos showing one or two colors (Fig. 5A–D). However, one high-percentage chimeric embryo displayed polychrome fluorescence, indicating that it was derived from all of the different mESC clones (Fig. 5A,B). The remaining pregnant females were brought to term, and several high-degree chimeric males were bred to C57Bl/6J females to test for germline transmission. Although it was not our goal to generate a mouse for every different FP tested, one agouti mouse from these matings that exhibited bright red fluorescence (Fig. 5E,F) was used to establish a ROSA26Cherry mouse line.
Fig. 5.
Polychrome mouse embryos. Four mESCs expressing cyan (EN.CFP), green (EN.GFP) and red (EN.Cherry) FPs were microinjected into 3-day-old blastocysts and implanted into pseudopregnant female mice. Embryos were harvested at E15.5 and E17.5, and fluorescent images obtained. (A,B) This high-degree chimeric embryo displayed cyan, green and red fluorescence in different tissues, indicating that it was derived from the three different mESC types. (A,B) Right and left perspectives, respectively. (C,D) In most embryos, one FP dominated expression, e.g. green (C) or blue (D). (E,F) Offspring of a polychrome chimera show germline transmission of the EN.Cherry allele. Images of two littermates showing one with the EN.Cherry allele and one with a wild-type ROSA26 allele under white (E) and fluorescence excitation (F). Calibration bars indicate the fluorescence intensity level that corresponds to one color shade in the image; intensity is expressed in arbitrary units.
Analysis of Cherry expression in ROSA26Cherry mice
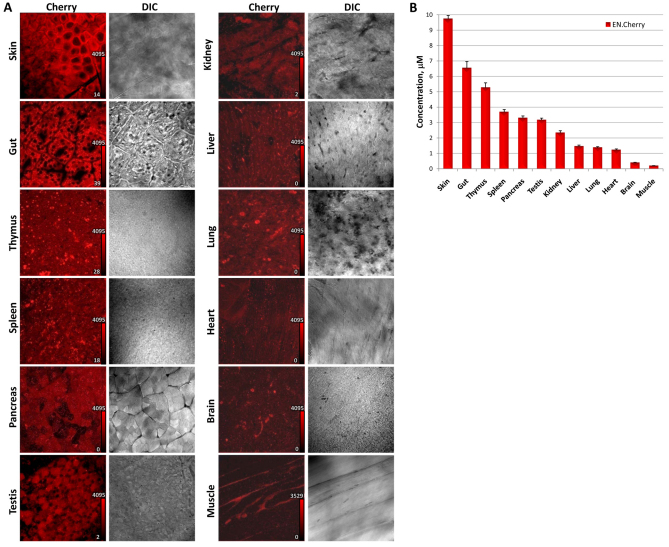
To assess Cherry expression from the ROSA26 locus in vivo, we examined the fluorescence of 12 different organs and tissues using the ROSA26Cherry animals (Fig. 6A). In each case, red fluorescence was easily visible, but fluorescent intensity varied by tissue type. Thus, to quantify the differences in Cherry expression, we performed quantitative confocal microscopy using whole-mount tissues. The punctate pattern of Cherry fluorescence observed in mESCs was also present in all of the examined tissues (Fig. 6A). Interestingly, the amount of Cherry in different organs was highly variable, with approximately a 30-fold difference between skin and muscle, which were the highest and lowest expressing tissues, respectively (Fig. 6B).
Fig. 6.
Cherry expression in different tissues. (A) Confocal images of different tissues from ROSA26Cherry mice. Calibration bars indicate the fluorescence intensity level that corresponds to one color shade in the image; intensity is expressed in arbitrary units. (B) Quantitative confocal microscopy measurements of Cherry concentration. The mean intensities for two animals, with ten images acquired for each tissue per animal, were averaged together for each data point. Bars indicate s.e.m. (n=20 for each; except n=10 for thymus and testis).
DISCUSSION
In this study, we generated a ROSA26LCA allele and used it to create an allelic series of mESCs to quantitatively assess both intrinsic and extrinsic variables that affect the relative brightness of FPs when expressed in mammalian cells and tissues. The ROSA26 locus was chosen for this study because it is widely thought to be expressed at a consistent level in mouse cells, and its use has been proven in numerous studies examining the effects of protein misexpression and as a Cre-inducible genetic tag (Zambrowicz et al., 1997; Kohlhepp et al., 2001; Safran et al., 2003).
Several features of how the ROSA26LCA allele was generated merit discussion. First, the plasmids used to generate this allele can be used to generate other LCA alleles. These plasmids are designed to enable the use of BAC recombineering, thereby simplifying the assembly of both gene targeting and RMCE vectors. Second, the allele contains puΔtk, a fusion protein that can be used to perform both positive and negative selection. This fusion protein also enables mESCs containing the LCA allele to be tested for germline competency without the problematic Sertoli cell toxicity of the wild-type HSV-TK (Chen and Bradley, 2000). Third, the combination of mutant loxP sites present in the LCA allele and exchange vector enable efficient Cre-driven cassette exchange (Araki et al., 2002). Indeed, we found that the use of the tandem lox66/71-lox2272 site strategy is much more efficient than the loxP/inverted-loxP combination that we previously reported for both the Gck and Ptf1a alleles (Long et al., 2004; Burlison et al., 2008). Moreover, because recombination between the lox66 and lox71 sites results in a doubly mutant lox site that can no longer efficiently bind Cre or undergo recombination, the post-RMCE allele is predicted to remain stable in the presence of Cre (Araki et al., 1997).
Using the ROSA26LCA allele, we generated a series of FP-expressing alleles by RMCE, thereby enabling us to perform a side-by-side comparison of specific design variations and to accurately assess the impact of both extrinsic and intrinsic variables on FP expression and relative brightness. This approach, which results in the insertion of a single copy transgene into a defined genetic location, circumvents confounding issues, such as gene copy number and location, that have previously limited the quantitative assessment of different transgenes or design features that influence FP expression or, for that matter, expression of any other gene. As a result, we were able to accurately assess the impact of several different allele design features.
First, we examined the effect of a translational enhancer on FP expression and efficiency. Previously, it was shown that the 5′-UTR of the Xenopus β-globin gene, along with an optimized translational initiation site (ACCAUGG), increased translational efficiency by 10- to 300-fold (Falcone and Andrews, 1991). When this sequence was inserted in the 5′-UTR, we observed only a modest 1.26-fold increase in FP concentration and/or brightness, with no change in mRNA expression. Both EN.CFP and nonEN.CFP contain the optimized initiation site (ACCAUGG), but only EN.CFP contains the Xenopus β-globin 5′-UTR leader sequence. Therefore, although the effect is modest, inclusion of the 5′-UTR of the Xenopus β-globin gene improves FP expression in mammalian cells by enhancing translational efficiency.
Second, we examined three different 3′-UTR configurations. Several prior studies have reported that intervening intron sequences can have a major role on gene expression, both in vitro (Buchman and Berg, 1988; Nott et al., 2004) and in vivo (Choi et al., 1991). For instance, Buchman et al. showed that including an intron in the transcription unit of a recombinant SV40 virus carrying a rabbit β-globin gene sequence resulted in up to a 400-fold increase in mRNA expression. Similarly, Choi et al. showed that insertion of intervening sequences in a transgene caused a 5-to 300-fold increase in expression (Choi et al., 1991). Other studies have shown that a β-globin polyadenylation sequence results in greater mRNA accumulation than a SV40 polyadenylation site (Westwood et al., 1993). However, the effect on expression of placing these different 3′-UTR sequences into a given transgene or allele remains poorly quantitated. In this study, we observed at least a twofold increase in FP expression and a 1.75-fold increase in translation when using a β-globin splice and polyadenylation sequence compared with an allele that only contains a β-globin or SV40 polyadenylation signal sequence. Thus, the cassette containing both the Xenopus 5′-UTR translational enhancer sequence and the intron-containing rabbit β-globin polyadenylation sequence provided the highest FP expression among the design variants that we tested.
Third, we assessed the relative brightness of five different FP variants when placed into the identical genetic context of a mammalian cell. Quantitative microscopic imaging enabled us to determine that Citrine was both the brightest and highest expressed FP, followed by Apple, Cerulean, eGFP and, finally, Cherry. These results differ significantly from prior measurements of relative brightness of purified FPs. However, the differences can be understood when one considers that our in vivo measurements also reflect differences in protein accumulation within living mammalian cells. Our measurements also reflect the excitation wavelength used, as well as the portion of the emission spectra that was measured. For instance, in our confocal measurements of FP brightness, we observed that Citrine that was excited at 514 nm was over fivefold brighter than eGFP that was excited at 488 nm, but there was no difference when brightness was assessed using a 488 nm laser in FACS. This difference reflects the attenuation of Citrine absorbance caused by the off-peak excitation by the 488 nm laser used in the FACS analysis.
We also directly compared Apple with Cherry and found that Apple is 1.7-fold brighter by confocal microscopy analysis and 1.4-fold brighter by FACS analysis. The slight differences observed are probably due to the different excitation wavelengths and filter sets used for the two analyses. Interestingly, Cherry displayed a punctate cellular labeling pattern, whereas Apple, like eGFP, Cerulean and Citrine, was uniformly expressed throughout the cells. The observed punctate Cherry pattern might be due to protein misfolding that results in cellular aggregates. Similar aggregates were observed when Cherry was overexpressed in the Phoenix Eco cell line (Shcherbo et al., 2007). Although our findings indicate that Cherry is not as bright as Apple, the farther-red-shifted emission of Cherry might offer an advantage for experiments that involve multiple FPs. Because cellular autofluorescence is low in the red region, the minor expression differences between Cherry and Apple are unlikely to limit either for in vivo use. Indeed, even though Cherry was the least bright FP when assessed in cells, Cherry fluorescence from the ROSA26Cherry allele was easily visualized in 12 different tissues.
The ROSA26Cherry mouse also allowed us to assess the pattern and levels of Cherry expression in different tissues. We found that Cherry displayed punctate labeling in all tissues tested, similar to that observed in mESCs. Interestingly, although Cherry was detected in all tissues examined, expression varied up to 30-fold, with the highest expression being in skin and the lowest in muscle. The reason for these expression differences was not explored but could either reflect differences in ROSA26 promoter activity in different tissues in adult mice, or different FP turnover rates.
Although our studies were performed using only a single gene locus, they have implications for the design and use of other FP-containing transgenes and alleles. Although we did not design nor test alleles that were deliberately engineered for poor expression, on the basis of these data we can extrapolate that a poorly designed allele might be more than threefold lower in expression than the most highly expressed allele used in this study. The design of transgenes and/or alleles is highly variable in terms of sequences placed on the 5′ and 3′ sides of a coding sequence, such as those for an FP. Moreover, splice and polyadenylation sequences from endogenously expressed genes might intentionally be retained when gene targeting is used to insert an FP into a gene, even though the impact of these sequences on RNA stability is almost never known. Although a threefold difference in expression is seemingly small, in some settings it might mean the difference between experimental success and failure, especially if the excitation wavelengths available for use result in off-peak excitation or if the FP emission spectra overlaps with other proteins that exhibit significant autofluorescence.
METHODS
Plasmids
Four general use plasmids were made using standard cloning techniques and confirmed by restriction enzyme analysis and sequencing. The following four plasmids were made: (1) pLCA.71/2272, an insertion vector for gene targeting, contains tandem lox71 and lox2272 sites (Araki et al., 2002) flanking a fusion protein between puromycin N-acetyltransferase and a truncated form of HSV1 thymidine kinase (puΔtk), under control of a mouse phosphoglycerol kinase (PGK) promoter, as well as an EM7-driven neomycin resistance (neoR) cassette [from PL452 (Liu et al., 2003)]. The puΔtk fusion gene does not cause male infertility in transgenic animals (Chen and Bradley, 2000), thereby allowing LCA-containing mESCs to be tested for germline competence. (2) pMCS.DT-A, a retrieval vector for gene targeting, contains an MC1-driven diphtheria toxin A sequence and multiple cloning site (MCS). (3) pMCS.66/2272 contains an MCS flanked by tandem lox66 and lox2272 sites that recombine with those in pLCA.71/2272 and can therefore be used to generate exchange vectors for use in RMCE. (4) pMCS.66/2272.Hygro is a variant of pMCS.66/2272 that contains a PGK-driven hygromycin resistance (hygroR) gene flanked with tandem FLP recognition target (FRT) sites. These vectors have been deposited into Addgene (www.addgene.org).
Gene targeting
The pROSA26.LCA targeting vector was made by a two-step BAC recombineering procedure (Copeland et al., 2001). First, two inner homology regions from the ROSA26 gene locus were PCR-amplified from a mouse ROSA26 BAC (clone number: 58-D17, RPCI-22 library) and cloned into pLCA.71/2272. The resulting construct was used to insert the lox71-, PGK-puΔtk-, EM7-neoR-and lox2272-containing cassette into the BAC. Second, two outer homology regions were cloned into pMCS.DT-A, and the resulting plasmid was used to retrieve a longer segment of DNA containing the loxed PGK-puΔtk and EM7-neoR cassette, thereby generating the pROSA26.LCA gene targeting vector. 200 μg of this plasmid was linearized with NotI and used to electroporate 3.5×107 TL-1 ESCs (Labosky et al., 1994) at 240 V and a capacitance setting of 500 μFD. After selection using 1.5 μg/ml puromycin (Sigma) for 7–8 days, surviving colonies were isolated for DNA analysis by Southern blot.
FP exchange vectors
pROSA26.Ex, which was made from pMCS.66/2272 and contains a 4.081 kb fragment of the mouse ROSA26 promoter that was removed from the gene during the generation of the ROSA26LCA allele, served as the starting point for assembling the FP-containing exchange vectors. CFP (Cerulean) was obtained from pCerulean-C1 vector (Rizzo et al., 2004), eGFP from pEGFP-N1 vector (Clontech), YFP (Citrine) from pmCit-N3 vector (Rizzo et al., 2004), Cherry from pmCherry-C1 vector (Clontech), and Apple from pmApple-N1 vector (Clontech). Five of the exchange cassettes, EN.CFP, EN.GFP, EN.YFP, EN.Cherry and EN.Apple, contain the 5′-UTR from the Xenopus β-globin gene, which has been reported to function as a translational enhancer (Falcone and Andrews, 1991). In addition, these exchange plasmids contain an 886 bp DNA fragment from pBI-LG (Clontech) containing the 3′ end of exon 2, intron 2, exon 3 and polyadenylation sequences of the rabbit β-globin gene. The nonEN.CFP exchange vector lacks the translational enhancer sequence but is identical in other respects. In EN.CFP.bgpA, CFP is followed by a 540 bp intronless polyadenylation sequence from the rabbit β-globin gene, also cloned from pBI-LG. EN.CFP.SV40 contains an SV40 polyadenylation sequence cloned from pTet-on-Advance (Clontech). Each exchange vector contains a PGK/EM7-neoR cassette flanked by FRT sites. Annotated sequences for these FP-containing exchange vectors were deposited into GenBank (accession numbers from HM771696 to HM771703). The vectors are available upon request.
Recombinase-mediated cassette exchange
RMCE was performed using a positive-negative selection strategy previously described (Long et al., 2004). In brief, 5.6×106 mESCs containing the ROSA26LCA allele were co-electroporated with 40 μg of an exchange plasmid and 40 μg of pBS185, a Cre-expression plasmid (Sauer and Henderson, 1989). 200 μg/ml neomycin (Invitrogen) was used for positive selection and 8 μM gancyclovir (Sigma, St Louis, MO) for negative selection. Surviving clones were screened by DNA PCR using the following primers: (1) 5′-AGACT-TATCTACCTCATAGGTG-3′, (2) 5′-GTGAGAACAGAGTACC-TACAT-3′, (3) 5′-GAGGATCATAATCAGCCATACC-3′, (4) 5′-TCACAAGCAATAATAACCTGTAGT-3′. Primers 1 and 2 amplify a 537 bp DNA fragment from an exchanged allele and 503 bp DNA fragment from the wild-type ROSA26 locus. Primers 3 and 4 detect a 565 bp sequence only in an exchanged allele. Properly exchanged clones were expanded from the master plate and stocked for future use.
Blastocyst microinjections
All procedures performed on mice were approved by the Vanderbilt University Animal Care and Use Committee. Chimeric mice were generated by the microinjection of three mutant mESCs: R26.EN.CFP clone 5B9:1C3, R26.EN.Cherry clone 5B9:1C11 and R26.EN.GFP clone 5B9:1D10. Four cells for each clone were injected into each blastocyst then transferred into pseudopregnant females. After 15.5 and 17.5 days of gestation, pregnant females were euthanized and chimeric embryos were imaged by fluorescent microscopy. Several post-injection litters were brought to term, and three chimeras were mated with C57BL/6 females to test for germline transmission. Confirmation of germline transmission of the EN.Cherry allele and genotyping of established ROSA26Cherry mice were carried out by PCR.
Quantitative reverse-transcription PCR
mESC clones were grown to 70% confluency and total RNA isolated using the RNeasy mini kit (Qiagen). cDNA was synthesized from 2 μg RNA with Omniscript reverse transcriptase and oligo(dT) primers (Applied Biosystems). Quantitative reverse-transcription (RT)-PCR was performed using a 7900HT Real-time PCR System (Applied Biosystems) with 2 ng of template and Power SYBR Green PCR Mastermix (Applied Biosystems). The primers used to detect CFP sequences were 5′-TGCTGCCCGACAACC-ACTAC-3′ and 5′-CGGTCACGAACTCCAGCAGG-3′. β-actin cDNA was detected with 5′-ACGATGCTCCCCGGGCTGTA-TTC-3′ and 5′-TCTCTTGCTCTGGCCTCGTCACC-3′. The ΔΔCt method was used to determine relative expression. Data was normalized to β-actin and expressed relative to the ROSA26LCA allele.
Quantitative confocal microscopy
For analysis of mESCs, cells were grown on glass-bottom dishes (MatTek) on a monolayer of irradiated MEFs. For analysis of mouse tissues, different organs from two mice (6-weeks old and 20-weeks old) were dissected in PBS, cut into thin slices with a razor blade and mounted under coverslips in Aqua Poly/Mount (Polysciences) on MatTek dishes. Confocal microscopy was performed on a LSM710 (Zeiss) using a Fluor 40× 1.3 NA oil-immersion objective. The following excitation wavelengths and emission bandpass filters were used for FP detection: eGFP: 488 nm and 494–650 nm bandpass; Citrine: 514 nm and 520–640 nm bandpass; Cerulean: 458 nm and 465–601 nm bandpass; Cherry and Apple: 561 nm and 572–708 nm bandpass. All images were acquired using 512×512, 0.415 μm diameter pixels and 12-bit gray levels. DIC images were acquired simultaneously with the fluorescence images. For each clone, ten fields of view per dish were measured, and detector gain was set to avoid saturation. After each imaging experiment, serial dilutions of the respective purified protein solution were quantified using the same settings to establish a calibration curve for the conversion of gray level values into protein concentration values. Image analysis was performed using ImageJ 1.43s software (http://rsb.info.nih.gov/ij/). DIC images were used to create regions of interest (ROI) encompassing each cell. The ROI was used to determine mean fluorescence intensity of each cell, and the area outside the ROI was used to determine background to be subtracted from the fluorescence intensity. Intensity values were converted to concentration values by interpolation with the calibration curve.
FACs
Cells were grown to confluence on gelatin-coated plates without MEFs, harvested with 0.25% trypsin/EDTA and resuspended in phenol-red-free DMEM (Gibco). Fluorescence was analyzed on a 5-laser BD LSRII in the Vanderbilt Flow Cytometry Core, and fluorescent intensity was calculated using FACSDiva software. 7-AAD (Sigma) was used to assess viability, and ROSA26LCA mESCs were used as negative controls. CFP fluorescence was analyzed using a 405 nm laser and a 450/50 bandpass filter; GFP and YFP were analyzed using a 488 nm laser, 505 nm LP mirror and 530/30 bandpass filter; and mApple and mCherry were analyzed with a 532 nm laser, 600 nm LP mirror and 610/20 bandpass filter. SPHERO Ultra Rainbow calibration beads (Spherotech) were used to calibrate the instrument for more accurate quantification of fluorescent intensities.
RESOURCE IMPACT.
Background
Fluorescent proteins (FPs) are widely used to monitor gene expression and study cellular dynamics in cells and tissues. However, many variables can significantly affect experimental outcomes when FPs are expressed in the context of transgenes or gene targeting vectors in cell lines and mice. Variables that are intrinsic to the FP itself include the excitation and emission wavelengths, as well as the quantum yield of the FP. Variables extrinsic to the FP include the transcriptional activity of the specific promoter used to drive expression of the FP, the translational efficiency of the mRNA generated and the stability of the mRNA. Although many of the intrinsic factors have been precisely characterized, the extrinsic factors have been more difficult to accurately assess. Specifically, the variability that occurs owing to transgene copy number and insertion site has been a major impediment in quantifying the genetic factors that influence FP reporter gene expression in cells and mice.
Results
The authors of this study overcome this difficulty by using recombinase-mediated cassette exchange (RMCE) to generate an allelic series in mouse embryonic stem cells (mESCs), thereby allowing the quantitative assessment of intrinsic and extrinsic variables that influence FP expression in cells and mice. Eight different ROSA26 alleles are generated and compared to one another to assess the effects of intrinsic and extrinsic variables on FP protein expression in cells, as well as the relative brightness of different color variants of FPs. The authors show that the combination of genetic elements in an expression cassette influences FP expression up to threefold, and that relative expressed brightness of individual FPs varied up to tenfold in relation to one another. Among the five FPs examined, Citrine was the brightest, followed by Apple, eGFP, Cerulean and finally Cherry. In vivo expression of Cherry in ROSA26Cherry mice was observed in all organs, yet varied up to 30-fold between different tissues.
Implications and future directions
These results should help investigators to make more informed choices regarding the design of transgenes or vectors for expressing FPs and other proteins in cells and mice. In addition, the basal plasmids used in this study have design features that enable their use in BAC recombineering strategies and that simplify other aspects related to the design of gene targeting and RMCE vectors. Overall, these resources will simplify the generation of new loxed cassette acceptor (LCA) alleles, gene targeting and RMCE vectors, and mouse models, especially those containing FPs.
Fluorescence microscopy
Chimeric embryos were dissected in PBS and imaged using a Leica MZ 16 FA stereo dissecting fluorescent microscope. Filter sets used were as follows: CFP: excitation 436/20 nm, dichroic 455 nm, barrier 480/40 nm; GFP: excitation 480/40 nm, dichroic 505 nm, barrier 510/21 nm; and Cherry: excitation 540/25 nm, dichroic 565 nm, barrier 605/55. Images were taken using a QImaging RETIGA 4000R camera and processed using Photoshop CS4 software (Adobe Systems).
Acknowledgments
We thank Lila Solnica-Krezel and Kendal Broadie for helpful suggestions; Ray MacDonald for suggestions on use of the translational enhancer element; Gert-Jan Kremers, Jill Lindner and Changqing Zhang for technical assistance; and Eunyoung Choi for help performing the real-time PCR analysis. We also thank the staff of the Vanderbilt Transgenic Mouse/ESC Shared Resource for their expert performance of the blastocyst microinjections; the Vanderbilt Flow Cytometry Core for assistance performing FACS analysis; and the Vanderbilt Cell Imaging Resource for help performing confocal microscopy and analysis. These studies were supported by NIH grants DK042502, rDK72473, DK58404, DK20593, DK53434 and DK85064.
Footnotes
COMPETING INTERESTS
The authors declare that they do not have any competing or financial interests.
AUTHOR CONTRIBUTIONS
S.X.C., A.B.O., W.Y., L.A.P., D.W.P. and M.A.M. were involved in conception and design of the experiments and co-wrote the manuscript; S.X.C. performed most of the experiments and analyzed the data; W.Y. and R.G. performed DNA plasmid construction; R.G. performed embryo dissections and real-time PCR analysis; S.H. developed and optimized the RMCE protocol; A.U. performed all of the quantitative confocal microscopy experiments and data analysis; A.B.O. performed fluorescent embryo imaging and mouse tissue preparations for confocal microscopy. All authors read and commented on the manuscript.
REFERENCES
- Araki K., Araki M., Yamamura K. (1997). Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic. Acids Res. 25, 868–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K., Araki M., Yamamura K. (2002). Site-directed integration of the cre gene mediated by Cre recombinase using a combination of mutant lox sites. Nucleic. Acids Res. 30, e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Berg P. (1988). Comparison of intron-dependent and intron-independent gene expression. Mol. Cell Biol. 8, 4395–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlison J. S., Long Q., Fujitani Y., Wright C. V., Magnuson M. A. (2008). Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev. Biol. 316, 74–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. T., Bradley A. (2000). A new positive/negative selectable marker, puDeltatk, for use in embryonic stem cells. Genesis 28, 31–35 [DOI] [PubMed] [Google Scholar]
- Choi T., Huang M., Gorman C., Jaenisch R. (1991). A generic intron increases gene expression in transgenic mice. Mol. Cell Biol. 11, 3070–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland N. G., Jenkins N. A., Court D. L. (2001). Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2, 769–779 [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Valdivia R. H., Falkow S. (1996). FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38 [DOI] [PubMed] [Google Scholar]
- Falcone D., Andrews D. W. (1991). Both the 5′ untranslated region and the sequences surrounding the start site contribute to efficient initiation of translation in vitro. Mol. Cell Biol. 11, 2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck O., Baird G. S., Campbell R. E., Zacharias D. A., Tsien R. Y. (2001). Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 276, 29188–29194 [DOI] [PubMed] [Google Scholar]
- Heim R., Prasher D. C., Tsien R. Y. (1994). Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl Acad Sci USA 91, 12501–12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhepp R. L., Hegge L. F., Moser A. R. (2001). The ROSA26 LacZ-neo(R) insertion confers resistance to mammary tumors in Apc(Min/+) mice. Mamm. Genome 12, 606–611 [DOI] [PubMed] [Google Scholar]
- Labosky P. A., Barlow D. P., Hogan B. L. (1994). Mouse embryonic germ (EG) cell lines: transmission through the germline and differences in the methylation imprint of insulin-like growth factor 2 receptor (Igf2r) gene compared with embryonic stem (ES) cell lines. Development 120, 3197–3204 [DOI] [PubMed] [Google Scholar]
- Liu P., Jenkins N. A., Copeland N. G. (2003). A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13, 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q., Shelton K. D., Lindner J., Jones J. R., Magnuson M. A. (2004). Efficient DNA cassette exchange in mouse embryonic stem cells by staggered positive-negative selection. Genesis 39, 256–262 [DOI] [PubMed] [Google Scholar]
- Niswender K. D., Blackman S. M., Rohde L., Magnuson M. A., Piston D. W. (1995). Quantitative imaging of green fluorescent protein in cultured cells: comparison of microscopic techniques, use in fusion proteins and detection limits. J. Microsc.. 180, 109–116 [DOI] [PubMed] [Google Scholar]
- Nott A., Le Hir H., Moore M. J. (2004). Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 18, 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyabi O., Naessens M., Haigh K., Gembarska A., Goossens S., Maetens M., De Clercq S., Drogat B., Haenebalcke L., Bartunkova S., et al. (2009). Efficient mouse transgenesis using Gateway-compatible ROSA26 locus targeting vectors and F1 hybrid ES cells. Nucleic Acids Res. 37, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olenych S. G., Claxton N. S., Ottenberg G. K., Davidson M. W. (2007). The fluorescent protein color palette. Curr. Protoc. Cell Biol. Chapter 21, Unit 21 25. [DOI] [PubMed]
- Patterson G. H., Knobel S. M., Sharif W. D., Kain S. R., Piston D. W. (1997). Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 73, 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo M. A., Springer G. H., Granada B., Piston D. W. (2004). An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol 22, 445–449 [DOI] [PubMed] [Google Scholar]
- Rizzo M. A., Davidson M. W., Piston D. W. (2009). Fluorescent protein tracking and detection: applications using fluorescent proteins in living cells. Cold Spring Harb. Protoc.. 2009, pdb top64. [DOI] [PubMed] [Google Scholar]
- Safran M., Kim W. Y., Kung A. L., Horner J. W., DePinho R. A., Kaelin W. G., Jr (2003). Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol. Imaging 2, 297–302 [DOI] [PubMed] [Google Scholar]
- Sauer B., Henderson N. (1989). Cre-stimulated recombination at loxP-containing DNA sequences placed into the mammalian genome. Nucleic Acids Res. 17, 147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol.. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Lin M. Z., McKeown M. R., Steinbach P. A., Hazelwood K. L., Davidson M. W., Tsien R. Y. (2008). Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5, 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbo D., Merzlyak E. M., Chepurnykh T. V., Fradkov A. F., Ermakova G. V., Solovieva E. A., Lukyanov K. A., Bogdanova E. A., Zaraisky A. G., Lukyanov S., et al. (2007). Bright far-red fluorescent protein for whole-body imaging. Nat. Methods 4, 741–746 [DOI] [PubMed] [Google Scholar]
- Tsien R. Y. (1989). Fluorescent probes of cell signaling. Annu. Rev. Neurosci. 12, 227–253 [DOI] [PubMed] [Google Scholar]
- Tsien R. Y., Miyawaki A. (1998). Seeing the machinery of live cells. Science 280, 1954–1955 [DOI] [PubMed] [Google Scholar]
- Westwood J. A., Jones I. M., Bishop D. H. (1993). Analyses of alternative poly(A) signals for use in baculovirus expression vectors. Virology 195, 90–99 [DOI] [PubMed] [Google Scholar]
- Yew N. S., Wysokenski D. M., Wang K. X., Ziegler R. J., Marshall J., McNeilly D., Cherry M., Osburn W., Cheng S. H. (1997). Optimization of plasmid vectors for high-level expression in lung epithelial cells. Hum. Gene Ther. 8, 575–584 [DOI] [PubMed] [Google Scholar]
- Zambrowicz B. P., Imamoto A., Fiering S., Herzenberg L. A., Kerr W. G., Soriano P. (1997). Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. USA 94, 3789–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]