Abstract
Glioblastoma multiforme, which represents 80% of malignant gliomas, is characterized by aggressiveness and high recurrence rates. Despite therapeutic advances, patients with glioblastoma multiforme show a poor survival, and identification of novel markers and molecular targets for therapy is needed. A role for BAG3, a member of the BAG family of HSC/HSP70 co-chaperones, in promoting tumor cell growth in vivo has recently been described. We analyzed BAG3 levels by IHC in specimens from patients affected by brain tumors and we found that BAG3, although negative in normal brain tissues, was highly expressed in astrocytic tumors and increasingly expressed in more aggressive types of cancer; it was particularly high in glioblastomas. Down-regulating BAG3 both in vitro and in vivo in a rat glioblastoma model resulted in increased sensitivity to apoptosis, suggesting that BAG3 is a potential target for novel therapies. Finally, we determined that the underlying molecular mechanism requires the formation of a complex of BAG3, HSP70, and BAX that prevents BAX translocation to mitochondria, thus protecting tumor cells from apoptosis. Our data identify BAG3 as a potential marker of glial brain tumor sensitivity to therapy and thus also an attractive candidate for new molecular therapies.
Glioblastoma multiforme (GBM) is the most aggressive and most common tumor of the brain, accounting for approximately 25% of all brain tumors, 50% to 60% of all astrocytic tumors, and 80% of all malignant gliomas.1,2 Poorly circumscribed margins, microinvasion, and the infiltrating nature of astrocytes are contributing factors for the notorious aggressiveness and high rates of recurrence of glioblastomas; moreover, the severe neurological dysfunctions that accompany this tumor compromise both quality of life and survival. Despite significant advances in neurosurgical techniques, including the introduction of gamma knife surgery, and aggressive multimodal treatments, the median survival time for GBM is approximately 56 weeks.3,4 Recent studies elucidating some of the molecular abnormalities underlying the pathogenesis of glioblastomas are contributing to the development of novel therapeutic approaches.5–7 Therapies targeting a single pathway, however, seem to lack clinical benefits,8 and therefore future directions in the treatment of these devastating neoplasms call for the characterization of novel targets and the design of multiple target approaches.9,10
Among proteins that sustain cell survival and promote in vivo growth of several tumors, a role has recently been described for BAG3.11,12 This is a 74-kDa cytoplasmic protein belonging to the BAG family of co-chaperones. All family members share a conserved domain of 110 to 124 amino acids (BAG domain),13,14 through which they bind to the heat shock protein 70 (HSP70) ATPase domain.13,15 In addition, BAG3 contains a WW domain and a proline-rich repeat (PXXP), through which it interacts with other proteins.16 BAG3 expression is induced in leukocytes and other normal cell types in response to stress.17–19 Notably, however, BAG3 is constitutively expressed in several tumors, including leukemia, lymphoma, myeloma, pancreas and thyroid carcinomas, and melanomas.18,20–23 Several lines of evidence indicate that BAG3 plays a role in tumor cell survival. Indeed down-regulation of BAG3 in primary samples of B-cell chronic lymphocytic leukemia and acute lymphoblastic leukemia results in increased basal as well as drug-induced apoptosis.22,23 Furthermore, in thyroid carcinomas, BAG3 down-regulation sensitizes cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-dependent apoptosis.21 The role of BAG3 in cell survival is, at least in part, mediated by the regulation of HSC70/HSP70 function.11 We have recently shown that BAG3 sustains melanoma cell survival by interfering with the binding of HSP70 to the IKK-γ subunit of the NF-κB-activating IKK complex, thus favoring IKK complex formation and preventing the proteasomal degradation of IKK-γ and finally enhancing NF-κB activation.12 It is likely that in different tumors BAG3 can interfere with the binding of HSP70 with other partners known to sustain cell survival, preventing their degradation.
Here we show that BAG3 is robustly expressed in a large proportion of astrocytomas and glioblastomas and that its expression increases with tumor grade. Furthermore, we demonstrate that BAG3 promotes the binding of HSC/HSP70 to the proapoptotic BCL2 family member BAX, preventing its translocation to mitochondria and protecting glioblastoma cells from apoptosis, and that silencing of BAG3 results in a dramatic decrease in cell proliferation in vitro and in vivo.
Materials and Methods
Antibodies
Antibodies used to detect BAG3 protein were a mouse monoclonal antibody (AC-1) and a rabbit polyclonal antibody (TOS-2) distributed by Enzo Life Sciences (Lausen, Switzerland; Farmingdale, NY) and a mouse monoclonal antibody clone AC-2 produced in our laboratories. Polyclonal antibodies recognizing BAX, cleaved caspase-3 (Asp175), and BAD were obtained from Cell Signaling Technology (Danvers, MA). An anti-α-tubulin monoclonal antibody was obtained from Sigma-Aldrich (St. Louis, MO). Secondary antibodies were obtained from Pierce (Thermo Fisher Scientific, Rockford, IL). Anti-GAPDH and -HSP60 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). An anti-HSC/HSP70 polyclonal antibody was obtained from Stressgen (Victoria, BC, Canada; now Enzo Life Sciences).
Cell Cultures and Reagents
The rat malignant glial tumor cell line C6 and the human glioblastoma cell lines A172, T98G, and DBTRG-05MG were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The C6 and A172 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS); the T98G cells were grown in Eagle's minimal essential medium supplemented with 10% FBS, 1% Na-pyruvate, and 1% nonessential amino acids; the DBTRG-05MG cells were grown in Roswell Park Memorial Institute growth medium (RPMI-1640) supplemented with 10% FBS. All of the cell lines were maintained in 5% CO2 at 37°C in a humidified incubator. All of the growth media and supplements were obtained from Gibco-Invitrogen (Carlsbad, CA). Cisplatin [cis-diamminedichloroplatinum(II) (CDDP)] was obtained from Sigma-Aldrich.
Cell Viability, Apoptosis Assay, and Mitochondrial Membrane Potential Measurement
Cell viability was measured by Trypan Blue exclusion using a Bürker counting chamber. Apoptosis was analyzed by propidium iodide incorporation in permeabilized cells and flow cytometry was performed as described previously.12 Mitochondrial membrane potential was assessed by flow cytometry using tetramethylrhodamine ethyl ester as described previously.24 Briefly, cells were exposed to tetramethylrhodamine ethyl ester (Molecular Probes, Eugene, OR) for 1 hour at 37°C. Changes in dye fluorescence were analyzed with a FACScan flow cytometer (BD Biosciences, San Jose, CA).
Co-Immunoprecipitation
For co-immunoprecipitation assays, cells were lysed in HNT buffer (HEPES 20 mmol/L pH 7.5, NaCl 150 mmol/L, Triton 0.1%) supplemented with a protease inhibitor cocktail (Sigma-Aldrich) on ice for 20 minutes. After 5 cycles of freeze and thaw, cell membranes were centrifuged at 15,000 × g. Next, 500 μg of soluble proteins were subjected to immunoprecipitation with 1 μg of polyclonal anti-BAX antibody or 1 μg of rabbit IgG as control in HNTG buffer (HNT buffer, 10% glycerol), overnight at 4°C. Protein A-Sepharose was then added (45 minutes at 4°C), and immunocomplexes were precipitated by centrifugation at 15,000 × g. Next, immunoprecipitated proteins were washed with a low-salt buffer (0.1% SDS, 1% Triton, 20 mmol/L EDTA, 20 mmol/L Tris pH 8.0, 150 mmol/L NaCl), a high-salt buffer (0.1% SDS, 1% Triton, 20 mmol/L EDTA, 20 mmol/L Tris pH 8.0, 500 mmol/L NaCl), and a LiCl buffer (0.25 mol/L LiCl, 1% NP-40, 1% deoxycholate, 1 mmol/L EDTA, 10 mmol/L Tris pH 8.0), followed by two final washes with HNT buffer. Obtained proteins were then loaded onto SDS-PAGE gels and were analyzed for presence of HSC/HSP70 and BAG3 protein by Western blotting.
Densitometry
Scanning densitometry of the bands was performed with image scanning software (SnapScan 1212; Agfa-Gevaert, Mortsel, Belgium). The area under the curve related to each band was determined using Gimp 2 software version 2.6 (available at http://www.gimp.org). Background was subtracted from the calculated values. Results are expressed as means of at least three separate experiments.
IHC
A brain tumor microarray was obtained from US Biomax (Rockville, MD). The TMA represented 151 cases: 13 grade I glial tumors, 85 low-grade diffuse astrocytomas (grade II), 17 anaplastic astrocytomas (grade III), and 36 glioblastomas (grade IV), as well as 8 samples of normal brain adjacent to a brain tumor and 8 normal brain tissues, along with pathology diagnosis and tumor grade data. Immunohistochemistry (IHC) was performed using the avidin-biotin-peroxidase system, according to the manufacturer's instructions (Leica Microsystems, Bannockburn, IL). Our modified protocol included deparaffination in xylene, rehydration through descending degrees of alcohol up to water, nonenzymatic antigen retrieval in citrate buffer, pH 6.0, for 30 minutes at 95°C, and endogenous peroxidase quenching with H2O2 in methanol for 20 minutes. After rinsing with PBS, the samples were blocked with 5% normal horse serum in 0.1% PBS/bovine serum albumin.
To detect BAG3, samples were incubated for 1 hour at room temperature with the monoclonal antibody AC-1 (1:100 dilution, Enzo Life Sciences). After a thorough washing with PBS, sections were incubated with a biotinylated secondary anti-mouse IgG for 20 minutes, then were rinsed, incubated with avidin-biotin-peroxidase (ABC) complexes, and developed with diaminobenzidine (Sigma-Aldrich). Finally, the sections were counterstained with hematoxylin, dehydrated in alcohol, cleared in xylene, and mounted with Permount (Thermo Fisher Scientific, Waltham, MA). Ten nonoverlapping high-power fields (×400) were evaluated, and a labeling index of tumor cells was calculated for each specimen. The labeling index was defined as the percentage of positive cells out of the total number of cells counted in all fields. Only neoplastic cells were counted; to avoid nontumoral cells, margins and areas of infiltration into the brain were excluded. The number of positive cells were divided into three groups: BAG3 negative (<5% positive cells), low expression (5% to 40% positive cells), and high expression (>40% positive cells). The results from the IHC experiments were evaluated separately by two observers (R.F. and L.D.V.), blinded to the histological diagnosis and grading of the tumors.
siRNAs and Transfections
A specific small interfering RNA (siRNA) (5′-AAGGUUCAGACCAUCUUGGAA-3′) targeting bag3 mRNA and a control, nontargeted (NT) RNA (5′-CAGUCGCGUUUGCGACUGG-3′) were obtained from Dharmacon (Thermo Fisher Scientific, La Fayette, CO). Glioma cell lines were transfected with siRNAs at final concentration of 100 nmol/L using TransIT-TKO reagent (Mirus Bio, Madison, WI). Cells were harvested at indicated time points.
Statistical Analysis
Results are expressed as means ± SD or ± SE. Data were analyzed by Student's t-test or χ2 test using GraphPad Prism statistical software version 4.01 (La Jolla, CA). P values from 0.01 to 0.05 were considered significant, P values from 0.001 to <0.01 were considered very significant, and P values of <0.001 were considered highly significant.
Stereotactic Surgeries, C6 Cell Implants, and siRNA Treatments
Rats were housed in an animal facility and were maintained in a temperature-controlled and light-controlled environment with an alternating 12-hour light/dark cycle. All protocols were approved by the local Ethical Committee (DiFarma). For the surgical procedures, all instruments were sterilized beforehand and sterile small-animal surgical techniques were used. The rats were allowed to feed and drink freely until the time of the surgery. Animals were anesthetized by intraperitoneal injection with a ketamine/xylazine solution (200 mg ketamine and 20 mg xylazine in 10 mL of saline solution) at a dosage of 0.15 mg per 10 g body weight. Once the rats were anesthetized, the head of the animals was shaved and positioned in a stereotactic frame with a rat teeth adaptor (World Precision Instruments, Sarasota, FL). The skin was prepared with povidone-iodine 10% and alcohol and a 2- to 3-mm incision was made at the midline and anterior to the interaural line, for clear identification of the bregma and lambda sutures. A burr hole was drilled in the skull at 1.4 mm anterior to bregma and 2.5 mm lateral to the midsagittal suture (the coordinates for the caudate putamen), as described previously.25 A guide screw with an 0.5-mm channel (Plastics One, Roanoke, VA) was inserted into the drilled hole. The top of the screw was approximately 1 mm above the skull surface, and its shaft protruded through the dura and into the brain surface. For glioma formation, 1 × 106 C6 cells suspended in 10 μL of PBS were placed in a 26-gauge Hamilton syringe and inoculated through the guide screw, 4.5 mm down from the surface, to reach the caudate putamen, with an automated microinfuser pump (World Precision Instruments). Finally, a cross-shaped stylet was placed in the central hole of the guide screw, to prevent neoplastic cells from growing into the guide screw hole.
After 2 weeks, the animals were separated into two groups for treatment, with one group receiving BAG3 siRNA and the second group a nontargeted (NT) siRNA as control. The animals were again anesthetized and placed into the stereotactic frame as described above. After removal of the stylet, siRNA was delivered in a Hamilton syringe through the hole in the screw. The guide screw ensured that the tip of the needle penetrated the glioma cells implant and that, as a result, the siRNAs were delivered directly into the tumor. Treatments were performed three times per week for 3 weeks, after which the animals were euthanized. Rat brains were removed from the cranial cavity, bisected coronally at the injection site, fixed in 10% formalin for 3 days, and embedded in paraffin; sections (4 μm thick) were stained with H&E for routine histological evaluation. The maximum cross-sectional area of the intracranial glioblastomas was used to determine tumor area by computer-assisted image analysis (Olympus cellSens version 1.4).
Subcellular Fractionation and Western Blot Analysis
Cells were harvested and washed twice with PBS 1× solution (Mediatech, Herndon, VA). Total proteins were extracted in Tris 250 mmol/L, pH 7.6, supplemented with a protease inhibitor cocktail (Sigma-Aldrich), using five cycles of freeze and thaw. The lysates were centrifuged at 15,000 × g at 4°C and the soluble fractions were collected. Protein concentrations were measured using a Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA). Equal amounts of total protein (30 μg) from each sample were separated electrophoretically in a 12% SDS-PAGE and were blotted on a nitrocellulose membrane (Hybond; Amersham Life Sciences, St Louis, MO). Immunodetection was performed using an enzymatic chemiluminescence kit (ECL Plus; Amersham Biosciences, Piscataway, NJ) according to the protocol provided by the manufacturer. Cell cytosolic and mitochondrial fractions were generated by a digitonin-based subcellular fractionation technique as described previously.26
TUNEL Assay
Labeling of apoptotic cells from the tumor glioma cells implanted in the rat brain was performed using a TUNEL ApopTag peroxidase in situ apoptosis detection kit following the manufacturer's instructions (Intergen, Purchase, NY). Briefly, deparaffinized sections were pretreated with 30 μg/mL of proteinase K (Roche Diagnostics, Indianapolis, IN). After endogenous peroxidase quenching, sections were treated with digoxigenin nucleotide-containing reaction buffer, terminal deoxynucleotidyl transferase, and the modified nucleotides were then detected by immunolabeling with anti-digoxigenin antibody. Finally, sections were developed with diaminobenzidine (Dako, Carpinteria, CA) and counterstained with hematoxylin. A breast carcinoma sample provided by the manufacturer was used as a positive control for apoptosis. Incubation of the sections with reaction buffer, but without terminal deoxynucleotidyl transferase, was performed as a negative control.
Results
Using a BAG3-specific monoclonal antibody, we investigated BAG3 expression in patients affected by gliomas of various grades, taking advantage of a TMA representing 151 cases of tumors (13 grade I astrocytomas, 85 grade II astrocytomas, 17 malignant astrocytomas, and 36 glioblastomas), along with 8 cancer-adjacent normal tissue samples and 8 normal brain tissues. Positivity to BAG3 staining increased in more aggressive tumors, whereas normal brain samples were completely BAG3 negative (Figure 1A). The number of BAG3 positive cells within the tumor samples increased with tumor aggressiveness, and a statistically significant difference was observed when comparing glioblastomas to low grade astrocytomas (P = 0.0003, versus grade I; P = 0.017, versus grade II) (Figure 1B). We defined a scoring system to analyze biopsy samples based on the percentage of positive cells: BAG3 negative (<5% positive cells), low expression (5% to 40% positive cells), and high expression (>40% positive cells). Distribution of BAG3 positivity based on these criteria is summarized in Table 1. Over all tumor samples studied, 81% were BAG3 positive; the percentage of positive cells increased in more aggressive tumors, reaching 63.9% of high-positive biopsy samples in the GBM group (Table 1). A χ2 test confirmed significance of the differences among tumor groups.
Figure 1.
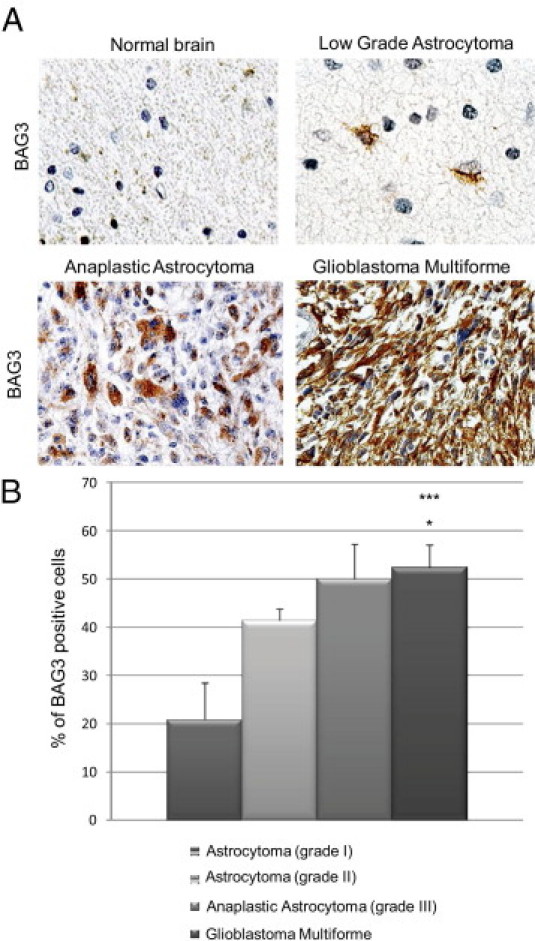
IHC detection of BAG3 in human glial brain tumors. A: BAG3 is detected in the cytoplasm of neoplastic astrocytes, and its expression significantly increases according to the tumor grade, but is barely detectable in the normal brain. Original magnification, ×400 (all images). B: The percentages of BAG3-positive cells were evaluated in all specimens from tumors of the same grade and the mean percentage ± SE was calculated. *P < 0.05 versus grade I; ***P < 0.001 versus grade II.
Table 1.
Clinicodemographic and Pathology Data, with Distribution of BAG3 Positivity
| Age (years ± SD) | M/F (no.) | Samples, no. | BAG3 negative, no. (%) | BAG3 positive, no. (%) |
|||
|---|---|---|---|---|---|---|---|
| BAG3+ | low BAG3+ | high BAG3+ | |||||
| Tissue type | |||||||
| NBT | 31.6 ± 16.3 | 1/7 | 8 | 8 (100) | 0 (0) | ||
| NBT adjacent to brain tumor | 45.5 ± 11.2 | 6/2 | 8 | 8 (100) | 0 (0) | ||
| Brain tumor tissue (combined N = 151) | 46.2 ± 10.4 | 77/74 | 151 | 14 (9) | 137 (91) | ||
| Astrocytoma grade I | 13 | 3 (23) | 6 (46) | 4 (31) | |||
| Astrocytoma grade II | 85 | 6 (7) | 43 (50.6) | 36 (42.4) | |||
| Anaplastic astrocytoma (grade III) | 17 | 1 (5.9) | 6 (35.3) | 10 (58.8) | |||
| Glioblastoma multiforme | 36 | 4 (11.1) | 9 (25) | 23 (63.9)⁎ | |||
BAG3 negative, <5% positive cells; low expression, 5% to 40% positive cells; high expression, >40% positive cells.
NBT, normal brain tissue.
P = 0.013. 3×3 contingency table for BAG3 levels distribution between Glioblastoma Multiforme, Anaplastic astrocytoma, and Astrocytoma grade II groups.
Because BAG3 has a well-established antiapoptotic effect,11,12 we analyzed its influence on glioblastoma cell survival by reducing its levels using a BAG3-specific siRNA. To this end, the rat glioblastoma cell line C6 was transfected with a BAG3-specific siRNA or a nontargeted control sequence (NT siRNA) and cell death was measured in cells grown in 1% FBS. BAG3 was strongly expressed in C6 cells, and its levels were significantly reduced on treatment with a specific siRNA after 48 hours (Figure 2A). The number of viable cells was reduced by >40% in BAG3 siRNA-treated cells, compared with control (nontransfected) and NT siRNA-treated cells. Furthermore, a marked increase of caspase-3 cleaved form in BAG3 siRNA-treated cells indicates that these cells were undergoing apoptotic death. C6 cells were also grown in 10% FBS and transfected with BAG3 siRNA for 48 hours; no change in cells viability was observed, compared with control cells (data not shown). These data indicate that loss of BAG3 sensitizes glioblastoma cells to apoptosis. This effect is not restricted to this particular cell line; A172 cells also showed increased apoptosis in low serum on BAG3 reduction (Figure 2B). Moreover, down-regulation of BAG3 sensitizes cells to different death inducers, as shown by increased sensitivity to cisplatin treatment of two different glioblastoma cell lines (Figure 2C). Of note, in the T98G cells the reduction of BAG3 levels was sufficient to induce death, in the absence of any additional stimulus.
Figure 2.
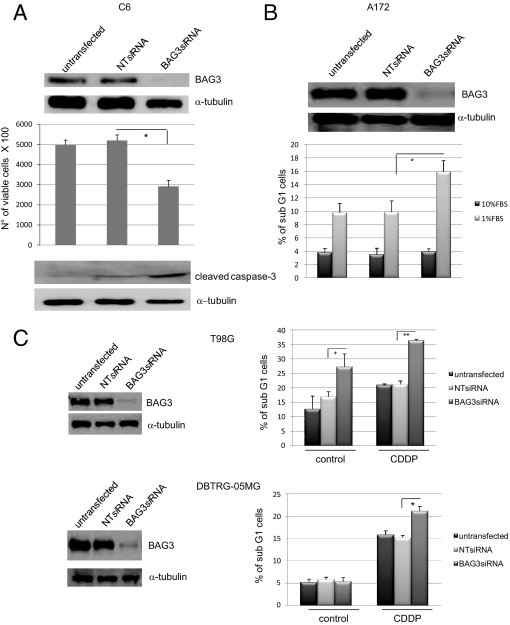
Effect of BAG3 siRNA on glioblastoma cells in vitro. A: C6 cells were plated at 30% of confluence, transfected with a BAG3 siRNA or a nontargeted (NT) siRNA (100 nmol/L), and maintained in medium supplemented with 1% FBS for at least 48 hours. Cell total extracts were obtained and BAG3 levels were analyzed by Western blot. Cell viability and caspase-3 cleavage were analyzed by Trypan Blue dye exclusion and by Western blotting, respectively. Graph depicts the mean percentage ± SD. B: A172 cells were treated as described for C6 cell line. The percentage of sub-G1 cells was assessed by flow cytometry analysis of propidium iodide-stained, fixed cells. Graph depicts the mean percentage ± SD. C: T98G and DBTRG-05MG cells were plated at 30% of confluence, transfected with a BAG3 siRNA or a nontargeted (NT) siRNA (100 nmol/L), and maintained in normal culture conditions or treated with cisplatin (CDDP; 5 μg/mL) for 72 hours. Total cell extracts were obtained and BAG3 levels were analyzed by Western blot. The percentage of sub-G1 cells was assessed by flow cytometry analysis of propidium iodide stained, fixed cells. Graph depicts the mean percentage ± SD. Data are representative of at least three independent experiments.
We then analyzed the molecular mechanism underlying the proapoptotic effects of BAG3 down-regulation. Antiapoptotic proteins of the BCL-2 family, and particularly BAX, are known to regulate glioblastoma cell survival/death balance.27–29 We therefore analyzed BAD and BAX protein levels in C6 cells cultured in high or low serum (10% or 1% FBS). Although the levels of BAD protein remained unaltered, those of BAX increased in cells grown in 1% FBS (Figure 3A). Despite increased BAX levels on serum starvation, no changes in cell viability were observed in these cells unless BAG3 was also down-regulated, indicating that BAG3 can interfere with BAX signaling. Because in the apoptotic process BAX translocates to the mitochondria and induces the depolarization of mitochondrial membrane,30 we determined whether BAG3 siRNA affected these events. Cell treatment with BAG3 siRNA did not result in a significant regulation of total levels of BAX and BAD proteins (data not shown), although down-regulation of BAG3 levels promoted BAX localization to the mitochondrial membrane (Figure 3B). BAD localization did not change. In accord with these results, depolarization of the mitochondrial membrane was significantly enhanced in BAG3 siRNA-treated cells (Figure 3C).
Figure 3.
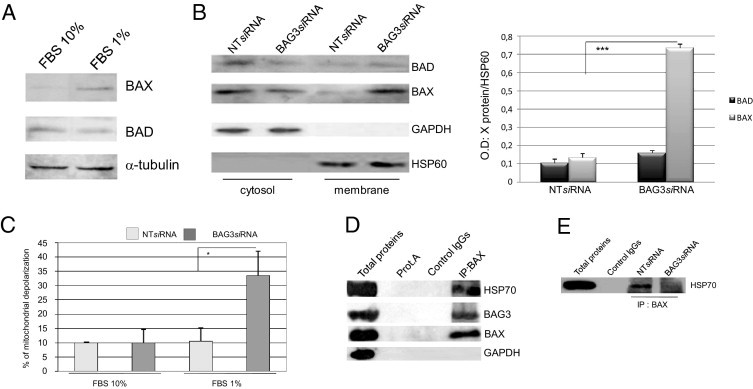
Effect of BAG3 knockdown on BAX translocation to mitochondria. A: Western blot analysis of BAX and BAD proteins in cells cultured for 48 hours in normal (FBS 10%) or reduced (FBS 1%) serum conditions. B: C6 cells were treated as described for A; subcellular fractions were obtained and fraction proteins were analyzed by Western blot using anti-BAD and -BAX antibodies. Antibodies raised against HSP60 and GAPDH were used to monitor equal loading conditions of the mitochondrial and cytosolic fractions. Densitometry analysis of the obtained bands and statistical significance was calculated from measurements of three separate experiments. ***P < 0.001. BAX levels in NT siRNA treated cells versus BAG3 siRNA treated cells. C: C6 cells were plated at 30% confluence, transfected with BAG3 siRNA or a nontargeted (NT) siRNA and cultured in medium containing 10% or 1% FBS for 48 hours. Mitochondrial membrane depolarization was measured by tetramethylrhodamine ethyl ester inclusion and expressed as mean percentage ± SD. *P < 0.05. NT siRNA versus BAG3 siRNA treated cells maintained in 1% FBS medium. D: Proteins (500 μg) from C6 cells maintained for 48 hours in 1% FBS medium were immunoprecipitated using a BAX-specific antibody. Co-immunoprecipitation of BAG3, HSP70, and BAX proteins was assessed by Western blot. An anti-GAPDH antibody was used as control. E: C6 cells were maintained for 48 hours in 1% FBS medium and treated with BAG3 siRNA or NT siRNA. Cell extracts were obtained and 200 μg of proteins were subjected to immunoprecipitation using a BAX-specific antibody. Presence of HSP70 protein was assessed by Western blot.
Next, we explored the mechanism through which reduction of BAG3 levels promotes BAX translocation to mitochondria. HSP70 binds to BAX, preventing its translocation to mitochondria and thus promoting cell survival.31 We therefore investigated whether, in glioblastoma cells, BAX is complexed with HSP70 in the cytosol and whether BAG3 participates in the complex. To this end, we immunoprecipitated BAX protein from lysates of C6 cells maintained for 48 hours in medium containing 1% FBS. Both HSP70 and BAG3 were found to co-immunoprecipitate with BAX (Figure 3D). We then analyzed the effect of BAG3 siRNA on the formation of BAX/HSP70 complex. HSP70 protein did not co-immunoprecipitate with BAX from lysates of cells in which BAG3 was down-regulated (Figure 3E); co-immunoprecipitation was clearly detectable in lysates from cells treated with a control NT siRNA. These findings indicate that BAG3 is required for HSP70 binding to BAX, preventing its translocation to the mitochondria. Reducing BAG3 levels disrupts the interaction among these three proteins, frees BAX, and sensitizes cells to apoptosis.
To investigate the possibility that interfering with this pathway may result in tumor cell death in vivo, C6 cells were stereotactically implanted into the caudate putamen of immunocompetent rats. After 2 weeks, the animals were separated into two groups for treatment, with one group receiving the BAG3 siRNA and the second group receiving the NT siRNA as control. The siRNAs were delivered directly into the tumor via a guided screw system. Treatments were performed three times per week for 3 weeks, after which the animals were euthanized and the brain was extracted and analyzed. Control animals showed larger tumors, occupying the entire basal ganglia, whereas animals treated with BAG3 siRNA showed smaller tumors (Figure 4A). Indeed, BAG3 siRNA-treated animals showed at least an 80% reduction in tumor size, compared with control animals. Histologically, the tumors of control animals were characterized by abundant, large neoplastic cells with pleomorphic nuclei and eosinophilic cytoplasm; BAG3 siRNA-treated tumors showed a reduced cytoplasm and abundant pycnotic nuclei, characteristic of cells undergoing apoptosis (Figure 4B). A terminal deoxynucleotidyl transferase dUTP nick end labeling assay revealed numerous positive cells in BAG3 siRNA-treated tumors, confirming that they are undergoing apoptosis (Figure 4B). These findings serve as proof of principle, that a therapy aimed at reducing BAG3 levels or targeting its interaction with other proteins could be effective in treating these tumors.
Figure 4.
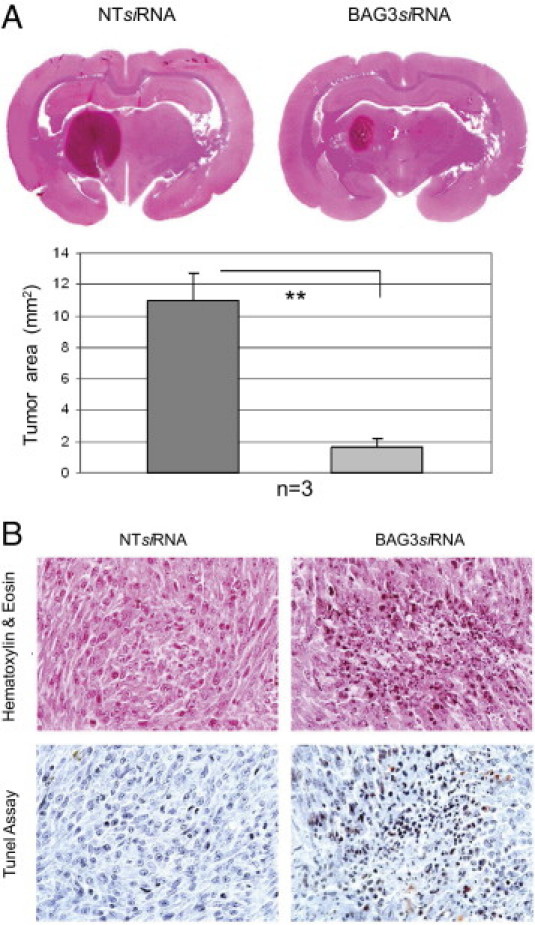
Effect of BAG3 siRNA on C6 rat glioblastoma cells in vivo. A: H&E-stained sections of rat brain show significant reduction of glioma tumors implanted into the caudate putamen of BAG3 siRNA-treated rats, compared with the larger tumors in NT siRNA-treated animals. The difference in tumor area (reported as means ± SD) was significant (**P < 0.01). B: Histological evaluation of the tumors reveals abundant areas of cell death and pycnotic nuclei in the BAG3 siRNA-treated tumors compared with solid sheaths of neoplastic cells in the NT siRNA-treated animals. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay revealed a large number of cells undergoing apoptosis in the BAG3 siRNA-treated tumors.
Discussion
Successful therapy of high-grade brain tumors is likely to require a combination of therapeutic approaches, because GBM cells are notoriously infiltrating and are resistant to single conventional radio and chemotherapeutic treatments. A deeper understanding of glioblastoma biology and physiopathology is necessary to identify new molecular targets for a rational drug design. Here we have reported increased expression of BAG3 in astrocytomas and glioblastomas, compared with normal brain tissues, with a particularly significant increased expression in high-grade glioblastomas. A lower percentage of BAG3-positive cells was also detected in lower grade astrocytomas. Caution is advised when analyzing expression of BAG3 in these particular tumors, because trapped non-neoplastic elements can be abundant in many of these neoplasms, and neoplastic versus non-neoplastic cells are sometimes not reliably distinguishable even to an expert eye, especially in the case of single neoplastic cells infiltrating the adjacent normal brain parenchyma. A proportion of BAG3-negative cells identified as malignant could instead be normal glial cells, leading to underestimation of the number of positive tumor cells, and BAG3-expressing cells in the normal brain could constitute infiltrating single GBM cells. Nonetheless, we think that BAG3 might still prove to be a helpful marker, despite this bias (which could lead to an overestimation of the differences in positivity across different grade lesions). Further studies are needed to analyze the relevance of BAG3 as a prognostic marker in these tumors, correlating its expression to tumor recurrence after resection, response to standard therapies, and patient survival.
More importantly, our results point to BAG3 as a potential candidate target for therapy, because its reduction in vitro sensitizes cells to death. As a proof of principle, we showed that BAG3 knockdown by local siRNA delivery induces apoptosis in a rat glioblastoma model in vivo. In contrast to cancers in other locations, GBM is an attractive target for local gene therapy because of its restricted anatomical location and absence of metastases outside the central nervous system. This allows delivery of vectors directly to the desired site, with only a small risk of systemic toxicity,32 thus supporting the idea of developing a BAG3-targeting strategy, eventually in combination with other drugs. Even though our in vivo data suggest that reduction of BAG3 alone results in tumor cells death, our in vitro data indicate that this may not be sufficient. Moreover, a complete eradication of the tumor might require combinational therapies. These might include triggering other antiapoptotic proteins that might synergize with BAG3, as we have shown before in other systems.11 Investigating potential therapeutic combinations that can synergize with BAG3 is a focus for future studies planned in our laboratory.
We also addressed the molecular mechanism underlying the antiapoptotic activity of BAG3 in glioblastoma, which appears to rely on BAG3 requirement for BAX retention by HSP70 in the cytosol, thus preventing the translocation to the mitochondria that is required for apoptosis induction. Down-regulation of BAG3 would free BAX, allowing its activation in response to a number of proapoptotic stimuli. In some cells in which a latent killing signaling (eg, oncogenic stress) is present, this might be sufficient to activate the death program in the absence of additional stimuli. This is in agreement with the co-chaperone role of BAG3, which binds the heat shock protein and cooperates in several of its functions,33 and with the relevant role of BAX in regulating glioblastoma cell survival/death balance.27–29 BAG3-dependent retention of BAX in the cytosol represents a novel mechanism through which BAX action can be regulated in tumors (Figure 5).
Figure 5.
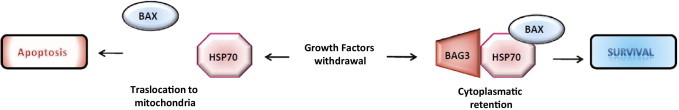
Proposed mechanism for BAG3-mediated regulation of BAX activity. Under stress conditions, the HSP70/BAG3 chaperone pair is able to prevent BAX translocation to the mitochondria. When BAG3 is down-regulated, BAX is able to translocate and promote mitochondrial membrane depolarization.
In conclusion, the present study identified a novel function for BAG3 and its involvement in the biology of malignant central nervous system tumors. Moreover, in addition to establishing BAG3 as a potential marker for astrocytomas and glioblastomas, our findings strongly suggest that BAG3 could be a target for new therapies.
Footnotes
Supported by the U.S. NIH (R01 MH086358-01A1) and the Italian Ministry for Research.
References
- 1.Central Brain Tumor Registry of the United States (CBTRUS) CBTRUS; Hinsdale, IL: 2010. 2009–2010 CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2004–2006. [Google Scholar]
- 2.Ohgaki H., Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 3.Grossman S.A., Ye X., Piantadosi S., Desideri S., Nabors L.B., Rosenfeld M., Fisher J., NABTT CNS Consortium Survival of patients with newly diagnosed glioblastoma treated with radiation and temozolomide in research studies in the United States. Clin Cancer Res. 2010;16:2443–2449. doi: 10.1158/1078-0432.CCR-09-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke J., Butowski N., Chang S. Recent advances in therapy for glioblastoma. Arch Neurol. 2010;67:279–283. doi: 10.1001/archneurol.2010.5. [DOI] [PubMed] [Google Scholar]
- 5.Zheng H., Ying H., Wiedemeyer R., Yan H., Quayle S.N., Ivanova E.V., Paik J.H., Zhang H., Xiao Y., Perry S.R., Hu J., Vinjamoori A., Gan B., Sahin E., Chheda M.G., Brennan C., Wang Y.A., Hahn W.C., Chin L., DePinho R.A. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell. 2010;17:497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chautard E., Loubeau G., Tchirkov A., Chassagne J., Vermot-Desroches C., Morel L., Verrelle P. Akt signaling pathway: a target for radiosensitizing human malignant glioma. Neuro Oncol. 2010;12:434–443. doi: 10.1093/neuonc/nop059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bose D., Meric-Bernstam F., Hofstetter W., Reardon D.A., Flaherty K.T., Ellis L.M. Vascular endothelial growth factor targeted therapy in the perioperative setting: implications for patient care. Lancet Oncol. 2010;11:373–382. doi: 10.1016/S1470-2045(09)70341-9. [DOI] [PubMed] [Google Scholar]
- 8.Krakstad C., Chekenya M. Survival signaling and apoptosis resistance in glioblastomas: opportunities for targeted therapeutics. Mol Cancer. 2010;9:135. doi: 10.1186/1476-4598-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayden E.C. Genomics boosts brain-cancer work. Nature. 2010;463:278. doi: 10.1038/463278a. [DOI] [PubMed] [Google Scholar]
- 10.Bredel M. Translating biological insights into clinical endpoints in neuro-oncology. Lancet Oncol. 2009;10:928–929. doi: 10.1016/S1470-2045(09)70192-5. [DOI] [PubMed] [Google Scholar]
- 11.Rosati A., Ammirante M., Gentilella A., Basile A., Festa M., Pascale M., Marzullo L., Belisario M.A., Tosco A., Franceschelli S., Moltedo O., Pagliuca G., Lerose R., Turco M.C. Apoptosis inhibition in cancer cells: a novel molecular pathway that involves BAG3 protein. Int J Biochem Cell Biol. 2007;39:1337–1342. doi: 10.1016/j.biocel.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Ammirante M., Rosati A., Arra C., Basile A., Falco A., Festa M., Pascale M., d'Avenia M., Marzullo L., Belisario M.A., De Marco M., Barbieri A., Giudice A., Chiappetta G., Vuttariello E., Monaco M., Bonelli P., Salvatore G., Di Benedetto M., Deshmane S.L., Khalili K., Turco M.C., Leone A. IKK{gamma} protein is a target of BAG3 regulatory activity in human tumor growth. Proc Natl Acad Sci USA. 2010;107:7497–7502. doi: 10.1073/pnas.0907696107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briknarová K., Takayama S., Brive L., Havert M.L., Knee D.A., Velasco J., Homma S., Cabezas E., Stuart J., Hoyt D.W., Satterthwait A.C., Llinás M., Reed J.C., Ely K.R. Structural analysis of BAG1 co-chaperone and its interactions with Hsc70 heat shock protein. Nat Struct Biol. 2001;8:349–352. doi: 10.1038/86236. [DOI] [PubMed] [Google Scholar]
- 14.Sondermann H., Scheufler C., Schneider C., Hohfeld J., Hartl F.U., Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 15.Takayama S., Xie Z., Reed J.C. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 16.Doong H., Price J., Kim Y.S., Gasbarre C., Probst J., Liotta L.A., Blanchette J., Rizzo K., Kohn E. CAIR-1/BAG-3 forms an EGF-regulated ternary complex with phospholipase c-gamma and hsp70/hsc70. Oncogene. 2000;19:4385–4395. doi: 10.1038/sj.onc.1203797. [DOI] [PubMed] [Google Scholar]
- 17.Pagliuca M.G., Lerose R., Cigliano S., Leone A. Regulation by heavy metals and temperature of the human BAG-3 gene, a modulator of Hsp70 activity. FEBS Lett. 2003;541:11–15. doi: 10.1016/s0014-5793(03)00274-6. [DOI] [PubMed] [Google Scholar]
- 18.Bonelli P., Petrella A., Rosati A., Romano M.F., Lerose R., Pagliuca M.G., Amelio T., Festa M., Martire G., Venuta S., Turco M.C., Leone A. BAG3 protein regulates stress-induced apoptosis in normal and neoplastic leukocytes. Leukemia. 2006;18:358–360. doi: 10.1038/sj.leu.2403219. [DOI] [PubMed] [Google Scholar]
- 19.Rosati A., Leone A., Del Valle L., Amini S., Khalili K., Turco M.C. Evidence for BAG3 modulation of HIV-1 gene transcription. J Cell Physiol. 2007;210:676–683. doi: 10.1002/jcp.20865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao Q., Ozawa F., Friess H., Zimmermann A., Takayama S., Reed J.C., Kleeff J., Büchler M.W. The anti-apoptotic protein BAG-3 is overexpressed in pancreatic cancer and induced by heat stress in pancreatic cancer cell lines. FEBS Lett. 2001;503:151–157. doi: 10.1016/s0014-5793(01)02728-4. [DOI] [PubMed] [Google Scholar]
- 21.Chiappetta G., Ammirante M., Basile A., Rosati A., Festa M., Monaco M., Vuttariello E., Pasquinelli R., Arra C., Zerilli M., Todaro M., Stassi G., Pezzullo L., Gentilella A., Tosco A., Pascale M., Marzullo L., Belisario M.A., Turco M.C., Leone A. The antiapoptotic protein BAG3 is expressed in thyroid carcinomas and modulates apoptosis mediated by tumor necrosis factor-related apoptosis-inducing ligand. J Clin Endocrinol Metab. 2007;92:1159–1163. doi: 10.1210/jc.2006-1712. [DOI] [PubMed] [Google Scholar]
- 22.Romano M.F., Festa M., Pagliuca G., Lerose R., Bisogni R., Chiurazzi F., Storti G., Volpe S., Venuta S., Turco M.C., Leone A. BAG3 protein controls B-chronic lymphocytic leukemia cell apoptosis. Cell Death Differ. 2003;10:383–385. doi: 10.1038/sj.cdd.4401167. [DOI] [PubMed] [Google Scholar]
- 23.Romano M.F., Festa M., Petrella A., Rosati A., Pascale M., Bisogni R., Poggi V., Kohn E.C., Venuta S., Turco M.C., Leone A. BAG3 protein regulates cell survival in childhood acute lymphoblastic leukemia cells. Cancer Biol Ther. 2003;2:508–510. doi: 10.4161/cbt.2.5.524. [DOI] [PubMed] [Google Scholar]
- 24.Rosati A., Quaranta E., Ammirante M., Turco M.C., Leone A., De Feo V. Quassinoids can induce mitochondrial membrane depolarisation and caspase 3 activation in human cells. Cell Death Differ. 2004;11(Suppl 2):S216–S218. doi: 10.1038/sj.cdd.4401534. [DOI] [PubMed] [Google Scholar]
- 25.Lal S., Lacroix M., Tofilon P., Fuller G.N., Sawaya R., Lang F.F. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg. 2000;92:326–333. doi: 10.3171/jns.2000.92.2.0326. [DOI] [PubMed] [Google Scholar]
- 26.Sun X.M., Bratton S.B., Butterworth M., MacFarlane M., Cohen G.M. Bcl-2 and Bcl-XL inhibit CD95-mediated apoptosis by preventing mitochondrial release of Smac/DIABLO and subsequent inactivation of X-linked inhibitor-of-apoptosis protein. J Biol Chem. 2002;277:11345–11351. doi: 10.1074/jbc.M109893200. [DOI] [PubMed] [Google Scholar]
- 27.Ozawa T., Hu J.L., Hu L.J., Kong E.L., Bollen A.W., Lamborn K.R., Deen D.F. Functionality of hypoxia-induced BAX expression in a human glioblastoma xenograft model. Cancer Gene Ther. 2005;12:449–455. doi: 10.1038/sj.cgt.7700814. [DOI] [PubMed] [Google Scholar]
- 28.Tagscherer K.E., Fassl A., Campos B., Farhadi M., Kraemer A., Böck B.C., Macher-Goeppinger S., Radlwimmer B., Wiestler O.D., Herold-Mende C., Roth W. Apoptosis-based treatment of glioblastomas with ABT-737, a novel small molecule inhibitor of Bcl-2 family proteins. Oncogene. 2008;27:6646–6656. doi: 10.1038/onc.2008.259. [DOI] [PubMed] [Google Scholar]
- 29.Giam M., Huang D.C., Bouillet P. BH3-only proteins and their roles in programmed cell death. Oncogene. 2008;27(Suppl 1):S128–S136. doi: 10.1038/onc.2009.50. [DOI] [PubMed] [Google Scholar]
- 30.Knudson C.M., Brown N.M. Mitochondria potential, bax “activation,” and programmed cell death. Methods Mol Biol. 2008;414:95–108. doi: 10.1007/978-1-59745-339-4_9. [DOI] [PubMed] [Google Scholar]
- 31.Gotoh T., Terada K., Oyadomari S., Mori M. hsp70-DnaJ chaperone pair prevents nitric oxide- and CHOP-induced apoptosis by inhibiting translocation of Bax to mitochondria. Cell Death Differ. 2004;11:390–402. doi: 10.1038/sj.cdd.4401369. [DOI] [PubMed] [Google Scholar]
- 32.La Rocca R.V., Mehdorn H.M. Localized BCNU chemotherapy and the multimodal management of malignant glioma. Curr Med Res Opin. 2009;25:149–160. doi: 10.1185/03007990802611935. [DOI] [PubMed] [Google Scholar]
- 33.Meimaridou E., Gooljar S.B., Chapple J.P. From hatching to dispatching: the multiple cellular roles of the Hsp70 molecular chaperone machinery. J Mol Endocrinol. 2009;42:1–9. doi: 10.1677/JME-08-0116. [DOI] [PubMed] [Google Scholar]


