Abstract
Structural crystallography and Nuclear Magnetic Resonance (NMR) spectroscopy are the predominant techniques for understanding the biological world on a molecular level. Crystallography is constrained by the ability to form a crystal that diffracts well and NMR is constrained to smaller proteins. While powerful techniques they leave many soluble, purified protein samples structurally uncharacterized. Small Angle X-ray Scattering (SAXS) is a solution technique that provides data on the size and multiple conformations of a sample, and can be used to reconstruct a low resolution molecular envelope of a macromolecule. In this study SAXS has been used in a high-throughput manner on a subset of 28 proteins where structural information is available from crystallographic and/or NMR techniques. These crystallographic and NMR structures were used to validate the accuracy of molecular envelopes reconstructed from SAXS data on a statistical level, to compare and highlight complementary structural information that SAXS provides, and to leverage biological information derived by crystallographers and spectroscopists from their structures. All of the ab initio molecular envelopes calculated from the SAXS data agree well with the available structural information. SAXS is a powerful albeit low-resolution technique that can provide additional structural information in a high-throughput and complementary manner to improve the functional interpretation of high-resolution structures.
1. Introduction
Structural biology aims to understand life on an atomic scale by using structural information to discern a molecule’s functional attributes. To date over 60,000 macromolecular structures have been deposited in the Protein Data Bank (PDB)1. Of these approximately 86% were determined with single crystal X-ray diffraction methods. The importance of this method is reflected in the number of Nobel prizes that have been associated with it including the determination of the structure of DNA2, the structure of vitamin B123, the structure of the photosynthetic reaction center4, the enzymatic mechanism underlying the synthesis of adenosine triphosphate5, the structure of potassium channels6, the molecular basis of eukaryotic transcription7 and most recently the ribosome8. Unfortunately, most proteins do not readily produce diffraction-quality crystals. Where failure as well as success has been rigorously tracked only 34% of expressed and purified targets provide a crystal and out of that only 12% result in a structure deposited in the PDB9. Crystallographic structures require crystals, while crystallization remains fundamentally a hit-or-miss proposition.
The Hauptman-Woodward Medical Research Institute provides a high-throughput crystallization screening service to the structural genomics and biological crystallography community. Macromolecular samples are screened against 1536 chemically diverse cocktails10 using the microbatch-under-oil technique11. This service has been in operation for over 10 years and to date has screened 12,500 proteins for over 1,000 laboratories worldwide. The screening laboratory has worked in close collaboration with the Northeast Structural Genomics (NESG) consortium, screening their samples for crystallization leads. In this effort approximately 50% of soluble proteins that enter the screening laboratory provide promising crystallization lead conditions; ~45% of these have been successfully optimized by NESG resulting in a PDB deposition. While this success rate is relatively good in the structural genomics field (providing evidence of good initial sample preparation and crystallization methods), this means that 78% of the soluble, purified proteins do not result in crystallographic structures. NMR techniques can provide structural information for samples recalcitrant to crystallization. In the NESG case, approximately 44% of the structural depositions result from NMR methods. For the Protein Structure Initiative (PSI) as a whole, only ~14% of the soluble purified targets make it to a PDB deposition, 21% of which were determined by NMR. To put this into perspective, there are greater than 30,000 soluble, purified samples from the US PSI that failed to provide structures; this number is almost half of the current structural information in the PDB. Even low-resolution structural information from these samples would significantly enhance the understanding of the biological world.
Small Angle X-ray Scattering (SAXS) is a technique that can provide low-resolution structural information, a molecular envelope from a solution of the sample12; a crystal is not required. In this study SAXS has been used in a high-throughput manner as a technique that is complementary to crystallography and NMR. The remainder of solutions from samples provided by NESG for high-throughput crystallization screening have been used for SAXS analysis. To date, this has been carried out for over five hundred samples. In this paper the information obtained from SAXS on a subset of 28 samples where either crystallographic, NMR, or a combination of both of these structures are available is described. We demonstrate how information from SAXS can complement and enhance high-resolution structural information. Based upon these observations it is proposed that SAXS should be adopted as a routine complementary analysis technique in structural biology that can be used to resolve and improve the interpretation of biological function from structural information.
2. Materials and Methods
2.1 Samples
In this study complementary SAXS data were collected from 28 different protein samples where structural information is available through crystallography, NMR or a combination of both techniques. The protein samples are NESG targets that represent large protein domain families, biomedical themes, and targets nominated by the biomedical community. The NESG biomedical themes focus on eukaryotic proteins, particularly human proteins involved in cancer biology, protein-protein interaction networks, specific biochemical pathways, and proteins implicated in other human diseases. The protocols for selection, cloning, expression, purification and crystallization of each sample is described elsewhere42–44. After purification each sample is concentrated to between 5–10 mg/ml using an Amicon (Millipore, Billerica, MA) centrifugal filtration unit with a 5 kDa molecular weight cutoff membrane. SDS-PAGE and mass spectrometry analysis are used to confirm purity and molecular weight respectively. Anlaytical gel filtration with static light scattering detection is used to screen for aggregation and determine the oligomeric state of each sample.
These 28 protein samples are summarized in Table 1 and can be divided into four sets. The first set encompasses 13 proteins where a crystallographic structure is available. These range in molecular weight from 9.5 kDa to 48.5 kDa. The second set consists of two proteins where two constructs were studied for each protein target. Two crystallographic structures were available from different constructs for the first and a single crystallographic structure for the second. The third set consists of nine proteins for which there is an NMR structure; the fourth set includes two protein targets where both NMR and crystallographic structures are available.
Table 1.
Samples used for the SAXS analysis are divided into four sets. The first set (1–13) contains 13 proteins, each having crystallographic structures. The second set (14–17) contains 2 proteins with two different constructs of the first having two crystallographic structures and the second a single structure. The third set (18–26) contains 9 proteins, each having an NMR structure. The fourth set (27–28) contains two proteins where both NMR and crystallographic structures are available. The sample name, ID, PDB identifier, reference, the oligomeric state in solution characterized on preparation by light scattering and gel filtration, initial concentration (mg/ml), molecular weight (Da) and number of residues are listed. The oligomeric state in solution is defined in the table as M (monomer), D (dimer), Tri (trimer), T (tetramer), Hep (Heptamer) or a combination. While all the samples have structures deposited in the PDB the majority are as yet unpublished. We are grateful to the authors in the references for the ability to use this structural data at this early stage
| # | Name | NESG ID | PDB | Ref | State | Conc | MW | Res |
|---|---|---|---|---|---|---|---|---|
| Samples where crystallographic structures were available | ||||||||
| 1 | Domain of Unknown Function | DhR2A | 3HZ7 | 13 | M | 6.9 | 9523 | 87 |
| 2 | Diguanylate cyclase with PAS/PAC sensor | MqR66C | 3H9W | 14 | D | 8.2 | 13611 | 210 |
| 3 | Nmul_A1745 protein from Nitrosospira multiformis | NmR72 | 3LMF | 15 | T | 6.9 | 14069 | 484 |
| 4 | Domain of Unknown Functiion | DhR85C | 3MJQ | 16 | D | 10.7 | 14609 | 252 |
| 5 | Sensory box/GGDEF family protein | SoR288B | 3MFX | 17 | D | 9.1 | 14779 | 258 |
| 6 | MucBP domain of the adhesion protein PEPE_0118 | PtR41A | 3LYY | 18 | M | 9.5 | 14300 | 131 |
| 7 | Sensory box/GGDEF domain protein | CsR222B | 3LYX | 19 | D | 12.7 | 15341 | 248 |
| 8 | HIT family hydrolase | VfR176 | 3I24 | 20 | D | 11.0 | 17089 | 298 |
| 9 | EAL/GGDEF domain protein | McR174C | 3ICL | 21 | M | 5.0 | 18738 | 171 |
| 10 | Diguanylate cyclase | MqR89A | 3IGN | 22 | M | 7.5 | 20256 | 177 |
| 11 | Putative NADPH-quinone reductase | PtR24A | 3HA2 | 23 | D | 9.5 | 20509 | 354 |
| 12 | MmoQ (Response regulator) | McR175G | 3LJX | 24 | M | 8.8 | 32032 | 288 |
| 13 | Putative uncharacterized protein | DhR18 | 3HXL | 25 | M | 9.6 | 48519 | 446 |
| Samples where multiple constructs and crystallographic structures were available | ||||||||
| 14 | Putative hydrogenase | PfR246A (78–226) | 3LRX | 26 | D | 11.4 | 17701 | 316 |
| 15 | PfR246A (83–218) | 3LYU | 27 | D | 8.4 | 16321 | 284 | |
| 16 | Alr3790 protein | NsR437I | 3HIX | 28 | M | 5.3 | 11760 | 105 |
| 17 | NsR437H | 3HIX | 28 | M | 6.5 | 15700 | 141 | |
| Samples where NMR structures were available | ||||||||
| 18 | MKL/myocardin-like protein 1 | HR4547E | 2KW9 (NMR) | 29 | D | 10.4 | 8276 | 75 |
| 19 | MKL/myocardin-like protein 1 | HR4547E | 2KVU (NMR) | 30 | D | 10.4 | 8276 | 75 |
| 20 | Putative peptidoglycan bound protein (LPXTG motif) | LmR64B | 2KVZ (NMR) | 31 | M | 5.0 | 9712 | 85 |
| 21 | E3 ubiquitin-protein ligase Praja1 | HR4710B | 2L0B (NMR) | 32 | M/D | 5.6 | 10297 | 91 |
| 22 | Transcription factor NF-E2 45 kDa subunit | HR4653B | 2KZ5 (NMR) | 33 | M | 10.0 | 10623 | 91 |
| 23 | YlbL protein | GtR34C | 2KL1 (NMR) | 34 | M | 11.0 | 10661 | 94 |
| 24 | Cell surface protein | MvR254A | 2L0D (NMR) | 35 | Tri | 5.9 | 12385 | 114 |
| 25 | Domain of Unknown Function | MaR143A | 2KZW (NMR) | 36 | M | 6.6 | 16312 | 145 |
| 26 | N-terminal domain of protein PG_0361 from P.gingivalis | PgR37A | 2KW7 (NMR) | 37 | M | 12.9 | 17485 | 157 |
| Samples where both crystallographic and NMR structures were available | ||||||||
| 27 | GTP pyrophosphokinase | CtR148A | 2KO1 (NMR) | 38 | D | 8.0 | 10042 | 176 |
| 3IBW | 39 | T | 8.0 | 10042 | 176 | |||
| 28 | Lin0431 protein | LkR112 | 2KPP (NMR) | 40 | M/Hep | 6.3 | 12747 | 114 |
| 3LD7 | 41 | M | 6.3 | 12747 | 100 | |||
There are a high percentage of crystallographic structures that have residues missing in the coordinates deposited in the PDB, compared to the total number of residues in the protein sequence. Indeed, it is estimated that conformational flexibility results in unstructured regions of 40 amino acids or more in length in 50% of eukaryotic proteins45. Although some efforts were made in construct design to eliminate large disordered N- and C-terminal segments42, in many cases disordered ends and disordered internal loops are observed in these protein structures. Since dynamics and conformational changes are crucial for the function of many macromolecular complexes and enzymes46, even low resolution information about conformational distributions of these residues is useful.
2.2 Crystallization
Each sample (450 μl at ~5–13 mg/ml concentration) was shipped to the Hauptman-Woodward Medical Research Institute’s high-throughput screening laboratory on dry ice, thawed upon arrival (typically within one day of receipt), and set up in 1536 crystallization plates10. Each of the 1536 experiments were imaged immediately after the sample was added, and then in weekly intervals for six weeks (as well as a control imaging before the sample was added to the cocktails). For all of the NESG samples, each image was manually inspected and classified as ‘crystal’ or ‘no crystal’. These classifications and the images were then communicated to NESG scientists for crystallization optimization and structural data collection.
2.3 X-ray Crystallographic and Solution NMR Structure Determination
The crystallographic and NMR structures used in this study were all solved by NESG staff scientists, and have been deposited in the Protein Data Bank1. In cases where a paper on the structure is not currently available the Digital Object Identifier (DOI) for the PDB deposition is provided in the reference list accompanied by the authors involved. The crystallographic asymmetric unit is not necessarily the biological oligomer. This oligomer was predicted using a theoretical analysis of binding energy and entropy of dissociation with the Protein Interfaces, Surfaces and Assemblies (PISA) service at the European Bioinformatics Institute47.
2.4. SAXS data
SAXS data were collected at beamline 4–248 of the Stanford Synchrotron Radiation Lightsource (SSRL). The SAXS preparations used samples that were re-frozen after crystallization screening; all of the SAXS samples underwent two freeze/thaw cycles. Typically, a minimum sample volume of 60 μl was used. The sample was diluted with its matching sample buffer to prepare 3 solutions of known concentrations. At the beamline an automated sample loader (manuscript in preparation), compatible with the PCR tubes, was used to collect data on as many as 96 experiments without user intervention, or the need to open the hutch. A wavelength of 1.3 Å was used for eight consecutive two-second exposures collected at each of the 3 sample concentrations. Each sample was oscillated back and forth in a quartz capillary cell during data collection to minimize radiation damage effects. All of these samples had an identical matching buffer. The samples were loaded in 8 well PCR strips such that a buffer blank was recorded followed by three concentrations of each of the two samples and then a final buffer blank with a wash cycle between each. The original concentration was diluted in 2:1, 1:2 and 1:5 ratios of sample and buffer blank. Using the 96-well capacity of the beamline sample loader, a series of 24 proteins was studied in each automated run. Typical time for a single sample concentration series was approximately 15 minutes with the majority of that time spent on liquid handling, e.g. sample loading and washing the fluid apparatus between each concentration. The data were processed and azimuthally integrated with SASTool (manuscript in preparation) and then visually examined with Primus49. Each of the eight exposures were compared for similarity to ensure no radiation damage took place and were averaged using SASTool to increase the signal to noise ratio. The SAXS data for different protein concentrations were assessed with Kratky plots and screened for aggregation using Guinier plots50. Guinier regions and Radius of gyration (Rg) estimates were derived by the Guinier approximation I(q) = I(0) exp(−q2Rg2/3) with qRg<1.3 using the AutoRg function of Primus where q = 4π sin θ/λ. The highest quality estimate as determined by AutoRg was used to select which of the three concentrations would go on to further processing. Zero extrapolated curves were not used because the examination of the concentration series showed no evidence of aggregation or repulsion in the higher concentration, stronger signal data. AutoGNOM51 was used to compute the pair distribution functions, P(R), for each sample and to determine the maximum particle dimension, Dmax, and these values were compared with those determined manually by GNOM to ensure consistency. A molecular weight was estimated from the program AutoPOROD of the ATSAS package52. Five ab initio shape reconstructions (molecular envelopes) were generated by DAMMIF53 and averaged with DAMAVER54. The program CRYSOL55 was used to calculate the scattering intensity from deposited crystallographic and NMR structures and estimate an Rg and fit the data by minimizing the discrepancy, χ, according to:
where Ie is the experimental scattering, Ic is the calculated scattering, and σ is the experimental error as determined by SASTool48. Other variables are given elsewhere55. In our case the experimental errors are underestimated as the detector is treated as an ideal photon counter; χ values here should therefore be regarded as a relative indicator of goodness of fit. Volume fractions for cases of oligomeric mixtures were estimated using the program OLIGOMER49. In these cases estimates for χ and the convoluted Rg of the mixture in solution and were taken from OLIGOMER.
2.5 Comparison of structural data
Rg and Dmax were calculated from the crystallographic and NMR structures using the program CRYSOL55. These values were compared with those derived from the SAXS data with constant subtraction enabled. For visualization purposes envelopes produced by the SAXS data were automatically overlaid with structures derived from X-ray crystallographic and NMR techniques using the program SUPCOMB, and were followed by manual adjustments using the program PyMOL.
3. Results
Table 2 summarizes and compares data calculated from crystallographic and NMR structural information with that measured from the SAXS data. In general, experimentally determined Rg values are consistent with those calculated from the structural information. In most cases the SAXS calculated Dmax are somewhat larger than those for the crystallographic cases, but smaller than those for the NMR structures. This might be expected due to missing residues in the crystallographic case. On the other hand, these NMR structure ensembles may overestimate the breadth of the true conformational distribution, since the set of 20 conformers deposited in the PDB does not account for the population distribution across the ensemble. The Porod calculated molecular weights are, again for the most part, integer multiples of the measured molecular weight. The SAXS determined oligomer is shown along with the relationship to that seen in the crystallographic structure. The SAXS data are recorded under conditions diluted from the initial sample preparation and the crystallographic structures are necessarily determined under different biochemical conditions. Details for each group, and in particular deviations from the known crystallographic structure, are described below. The observed data and structural fit to the observed data (continuous line) are shown with the structures and ab initio envelopes calculated for each group in Figures 1 to 4 and for those where a mixture was observed, Figure 5. In the majority of cases the fit to the experimental data is good.
Table 2.
A summary of structural (crystallography and NMR) and SAXS results. The sample # refers to the identical number in Table 1. The number of unresolved residues in the structure (mainly crystallographic) is listed together with the Rg and Dmax (in Å) determined from the available structure. The Rg and Dmax from the SAXS data are shown together with the difference from the available structural information. The molecular weight (in Da) calculated from a Porod analysis is listed along with the ratio of this weight with that derived initially from mass spectrometry in table 1. Finally the SAXS determined oligomer, (Monomer, Dimer or Tetramer), the relationship to the available structure and the χ of the fit are listed. A special case is described below for samples 16 and 17. Further details are given in the text
| # | Residues observed |
# Res missing |
Rg structure |
Dmax structure |
Rg SAXS |
ΔRg | Dmax SAXS |
Δ dmax | Porod MW |
MW Ratio |
SAXS oligomer1 |
Oligomer Assign. |
SAXS fit (χ) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Samples where crystallographic structures were available | |||||||||||||
| 1 | 74 | 13 | 13.7 | 42.0 | 14.9 | 1.2 | 53.2 | 11.2 | 7827 | 0.8 | M | 4.2 | |
| 2 | 198 | 12 | 16.6 | 67.0 | 19.8 | 3.2 | 67.4 | 0.4 | 24555 | 1.8 | D | sym | 2.6 |
| 3 | 436 | 48 | 22.4 | 62.3 | 23.2 | 0.8 | 75.3 | 13.0 | 50064 | 3.6 | T | sym | 1.6 |
| 4 | 214 | 38 | 23.3 | 81.2 | 23.6 | 0.3 | 82.7 | 1.5 | 37348 | 2.6 | D/T* | PDB | 2.6 |
| 5 | 224 | 34 | 19.9 | 57.6 | 19.8 | −0.1 | 64.2 | 6.6 | 28828 | 2.0 | D | PDB | 2.2 |
| 6 | 107 | 24 | 19.6 | 76.3 | 21.5 | 1.9 | 82.0 | 5.7 | 11085 | 0.8 | M | 6.1 | |
| 7 | 236 | 12 | 21.4 | 64.7 | 22.2 | 0.8 | 76.8 | 12.1 | 31410 | 2.0 | D | PDB | 3.8 |
| 8 | 286 | 12 | 20.5 | 63.1 | 21.1 | 0.6 | 71.4 | 8.3 | 34786 | 2.0 | D | PDB | 2.0 |
| 9 | 162 | 9 | 17.6 | 54.0 | 18.7 | 1.1 | 65.5 | 11.5 | 20468 | 1.1 | M | 3.7 | |
| 10 | 165 | 12 | 17.5 | 58.0 | 18.5 | 1.0 | 65.8 | 7.8 | 19069 | 0.9 | M | 4.2 | |
| 11 | 336 | 18 | 26.1 | 80.8 | 26.0 | −0.1 | 89.7 | 8.9 | 59937 | 2.9 | D/T* | PDB/sym | 1.4 |
| 12 | 252 | 36 | 21.3 | 61.5 | 22.5 | 1.2 | 81.9 | 20.4 | 37254 | 1.2 | M | 2.9 | |
| 13 | 416 | 30 | 28.5 | 95.0 | 27.6 | −0.9 | 98.5 | 3.5 | 40027 | 0.8 | M | 1.4 | |
| Samples where multiple constructs and crystallographic structures were available | |||||||||||||
| 14 | 272 | 44 | 20.8 | 59.6 | 21.1 | 0.3 | 69.2 | 9.6 | 30670 | 1.9 | D | PDB | 1.9 |
| 15 | 258 | 26 | 21.1 | 61.8 | 22.0 | 0.9 | 79.7 | 17.9 | 32657 | 2.0 | D | PDB | 1.8 |
| 16 | 93 | 12 | 18.0 | 59.5 | 18.2 | 0.2 | 64.7 | 5.2 | 15875 | 1.3 | D2 | PDB | 1.7 |
| 17 | 93 | 48 | 20.4 | 75.0 | 20.8 | 0.4 | 73.0 | −2.0 | 15920 | 1.0 | D1 | PDB | 2.5 |
| Samples where NMR structures were available | |||||||||||||
| 18 | 75 | 0 | 22.5 | 122.4 | 16.8 | −0.9 | 58.4 | −64.0 | 6771 | 0.8 | M | 4.7 | |
| 19 | 75 | 0 | 17.7 | 94.4 | 16.5 | −1.2 | 58.4 | −36.0 | 6771 | 0.8 | M | 1.4 | |
| 20 | 85 | 0 | 19.0 | 80.8 | 18.7 | −0.3 | 68.0 | −12.8 | 9724 | 1.0 | M | 1.7 | |
| 21 | 91 | 0 | 16.4 | 71.0 | 15.9 | −0.5 | 59.6 | −11.4 | 7862 | 0.8 | M | 1.5 | |
| 22 | 91 | 0 | 22.3 | 123.1 | 19.6 | −2.7 | 68.0 | −55.1 | 10762 | 1.0 | M | 1.6 | |
| 23 | 87 | 7 | 14.3 | 55.8 | 14.5 | 0.2 | 49.7 | −6.1 | 8479 | 0.8 | M | 1.4 | |
| 24 | 114 | 0 | 16.5 | 67.8 | 19.6 | 3.1 | 66.6 | −1.2 | 12609 | 1.0 | M | 5.9 | |
| 25 | 145 | 0 | 49.0 | 325.5 | 26.6 | −22.4 | 94.7 | −230.8 | 15386 | 0.9 | M | 7.4 | |
| 26 | 157 | 0 | 19.8 | 67.5 | 17.5 | −2.3 | 60.6 | −6.9 | 15238 | 0.9 | M | 2.1 | |
| Samples where both crystallographic and NMR structures were available | |||||||||||||
| 27* | 176 | 0 | 18.0 | 66.7 | 19.1 | 1.1 | 68.3 | 1.6 | 22589 | 2.2 | D | PDB | 2.5 |
| 158 | 18 | 18.1 | 52.5 | 19.0 | 0.9 | 68.3 | 15.8 | PDB | 2.4 | ||||
| 28* | 114 | 0 | 18.5 | 104.4 | 18.5 | 0.0 | 68.2 | −36.2 | 10721 | 0.8 | M | 2.3 | |
| 87 | 13 | 14.8 | 44.1 | 18.4 | 3.6 | 68.2 | 24.1 | 7.4 | |||||
The SAXS data compared to the NMR structure is listed above with the crystallographic comparison below as in Table 1.
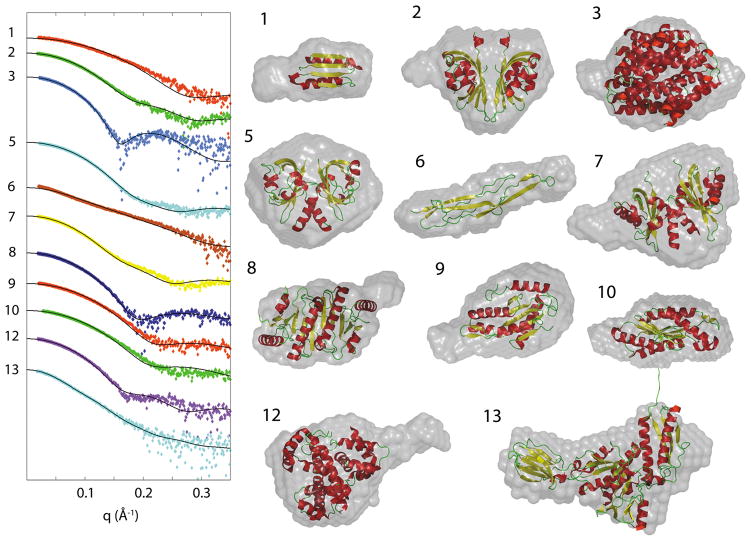
Figure 1.
The observed SAXS data and structural fit to the observed data (continuous line) for samples with crystallographic structure. The ab initio SAXS derived envelopes overlaid with crystallographic structure are also shown illustrating the agreement between the techniques. The samples are numbered as in Tables 1 and 2. Samples 4 and 11 contained a mixture of oligomers and are shown below.
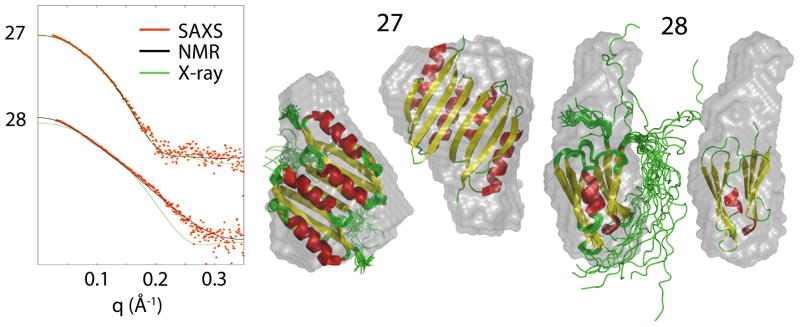
Figure 4.
Ab initio SAXS-derived envelopes overlaid with NMR and crystallographic structure to show the agreement between the different structural methods. The figures are shown to approximate scale and illustrate multiple conformations determined from the NMR data.
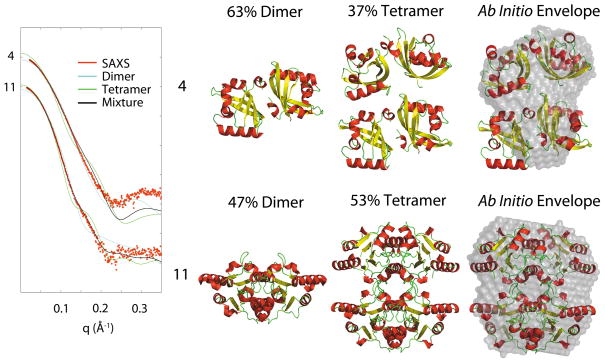
Figure 5.
Structures of oligomers based on analysis of the SAXS data and known monomer structure. The ab initio SAXS-derived envelopes are shown assuming a monodisperse solution.
3.1 Crystallographic and SAXS comparison
For the 13 samples in the set of crystallographically determined structures, there was relatively good agreement between the Rg of the model and that calculated from the SAXS data with an average deviation of < 1 Å, Table 2. The difference in Dmax between the crystallographic and SAXS envelope is greater, having no correlation with the percentages of missing residues in the crystallographic structure. This is not surprising given that missing residues may contribute to the Dmax if missing from the longest axis or may have little to no contribution if predominately missing from a shorter axis.
The observed data and structural fit to the observed data (continuous line) with crystallographic structures and ab initio molecular envelopes are shown in Figure 1 with the exception of samples 4 and 11 where a mixture of oligomers is indicated (see below). Outliers clearly visible by eye occur in samples 1 and 6; however there is a good correlation with all the envelopes and the known structure. The known crystallographic structure is represented in a ribbon form for clarity but in reality occupies more space when side chains are taken into account. For a number of cases the molecular envelope clearly extends beyond the known structure, extending further than can be explained by side chains on the backbone. These instances are consistently located in areas with residues missing from the crystallographic structure, but could also be attributed to slight undetected aggregation artificially enhancing the calculated Dmax. The highest χ values are observed for samples 6 (χ=6.1), 1 (χ=4.2), and 10 (χ=4.2). The crystallographic structures for these samples are missing 22%, 18% and 7% of the residues respectively which could contribute to this, although better χ values are observed for samples 4 and 12 which are both missing 14% of their residues in the crystallographic structures. In sample 1, a total of 13 residues are unresolved in the crystallographic structure. The molecular envelope reconstruction suggests evidence of these on the left hand side of the envelope. Sample 2 is a dimer in solution and 12 residues are unresolved in the crystallographic structure. When reconstructing the molecular envelope it is possible to add known symmetry information to the reconstruction and averaging (as in the case of an oligomer) but in our case ab initio modeling and averaging without symmetry constraints were used. A similar effect is seen for sample 7 where 12 residues were missing from the crystallographic structure. The molecular envelope accounts for missing residues in samples 8, 9 and 12, with 12, 9 and 36 unresolved residues, respectively. In each case the portion of envelope unexplained by the available crystallographic structure is positioned adjacent to the point where residues become unresolved in the structure. In sample 5 there are 34 residues missing but in this case it is not clear where those residues reside. These samples are structurally diverse yet in all the cases, the molecular envelopes show good agreement (at the resolution of the technique) with the known structures.
Where the molecular weight calculated from the Porod volume indicated an oligomer, different oligomers were compared with the experimental scattering. In Table 2 the oligomer assignment is noted as either “PDB”, where the chosen oligomer is present within the asymmetric structure provided by the PDB, or as “sym” where the oligomer in solution is not present in the PDB, but is chosen based on the crystal symmetry operator. In two cases, the oligomer seen in solution using SAXS was not the oligomer indicated by the asymmetric unit in the PDB, but was a smaller unit present within it. In case 5, the PDB contains a trimer, whereas the SAXS data not only favored a dimer, but was also able to clearly distinguish which dimer from the two possibilities. In case 9, the PDB oligomer is a dimer, whereas the SAXS selects the monomer. In both of these cases, the SAXS-selected oligomer agrees with the oligomer observed by gel filtration. For the majority of cases, with the exceptions of samples 6 and 9, the PISA prediction was in good agreement with the SAXS derived oligomer. In sample 6, PISA predicted an elongated dimer and in sample 9 three dimers in solution were predicted. Neither of these cases were supported by the SAXS data. In certain cases no oligomer provided by the PDB or via symmetry operation was found to be consistent with the SAXS data when comparing the Rg, Dmax, and overall fit to the curve. In this study, since the atomic structure is already known the results can be analyzed as a mixture of oligomers. Samples 4 and 11 showed clear evidence of oligomer mixtures with sample 4 consisting of 63% dimer and 37% tetramer and sample 11 consisting of 47% dimer and 53% tetramer. These are discussed below.
3.2 Sensitivity to different constructs
Alternate constructs were available for two samples. The first sample, a putative hydrogenase, had crystallographic structures for both constructs where SAXS data was collected, samples 14 and 15 with 316 and 290 residues, respectively. Interestingly, for sample 15, the construct with fewer residues, a significantly larger Dmax, (79.7 Å) for the construct with fewer residues, compared to the Dmax (69.2 Å) for the construct with more residues. The corresponding crystallographic structure for sample 15 shows a dimer in the PDB where one monomer has two fewer residues in the electron density than the adjacent monomer. In the adjacent monomer, these two residues appear to form a beta strand secondary structural element while in the adjacent monomer, lacking these two residues, this element is not present. This may reflect a higher level of disorder for these and neighboring residues in solution and subsequently for the five additional residues absent from this terminus. The SAXS envelope for this sample fits well to the overall crystallographic structure with the exception of an additional region present on only one side of the dimer. The disordered residues present in one monomer may be occupying this area. However, SAXS is a technique sensitive to aggregation and this extension of the SAXS envelope by an additional 10 Å compared to the similar construct may result from minor levels of aggregation present in solution that has escaped detection via static light scattering and Guinier analysis of multiple concentrations. Without further data it is not possible to distinguish the source of this difference.
The second example of multiple constructs, the protein Alr3790, has a single crystallographic structure (PDB id 3HIX), the Alr3790 protein, for the two constructs, whereas SAXS data for each construct, samples 16 and 17, are clearly different. These constructs had 105 residues (providing the crystallographic structure) and 141 residues respectively (out of 151 in the protein). The 3HIX structural model shows a trimer in the asymmetric unit. This trimer did not fit the SAXS data for either construct. Breaking the trimer into two separate dimers, D1 and D2, showed that each construct forms a structurally distinct dimer in solution. Sample 16 contains 36 fewer residues than sample 17 and these extra residues are located precisely at the D1 dimer interface. A possible explanation for the two solution states is that these extra residues impede D1 dimer formation in sample 17, but not being present in sample 16, allow the formation of dimer D2. The comparison of the calculated scattering for each possible dimer configuration with the experimental SAXS data clearly distinguishes the correct dimer formation for each construct. The envelope for sample 17 appears to underestimate the volume of the entire D1 dimer. Given that analytical gel filtration data and Porod molecular weight indicated a monomer in solution, it is possible that the monomer form may have a significant population in solution. A mixture analysis using both monomer and dimer only gave marginal improvements to the fit (χ = 2.1 to 2.5 respectively), and no improvement to the size parameters (data not shown). If a monomer population is also present at a low concentration it does not appear to greatly affect the SAXS curve. The PISA analysis predicted a stable hexamer consisting of a trimer of dimers for samples 14 and 15. While the hexamer is not shown to exist from the SAXS data, the dimer is present. For samples 16 and 17, the PISA analysis predicts both dimers to be equally stable. The SAXS data for sample 16 shows one dimer, while for sample 17 the SAXS data shows the other. For both examples the observed SAXS data, structural fit to the observed data and the envelopes with overlaid crystallographic structure are shown in Figure 2. Again the globular region of these proteins is well represented by the SAXS-derived ab initio molecular envelope.
Figure 2.
The observed SAXS data and structural fit to the observed data (continuous line) for samples with crystallographic structure and multiple constructs. Ab initio SAXS-derived envelopes are overlaid with crystallographic structure. Sample 17 contained a mixture shown below. The figures are shown to the same approximate scale as those in Figure 1 and the remaining structural representations in Figures 3 and 4.
3.3. NMR and SAXS comparison
The SAXS Dmax was consistently smaller than that derived from the corresponding solution NMR structure. In each case the NMR structural data consists of the 20 lowest energy conformers from 100 that were calculated. The Dmax in the NMR case is calculated from the maximum dimension of the total envelope of all 20 conformers. As such it can lead to an overestimate of Dmax as it measures the extremes within this set of conformers and does not take into account relative populations of these conformers or their dynamics. Although it is possible to obtain such population distributions from NMR studies, this information is not available from these NMR structural ensembles. For all the samples except sample 25, the calculated and measured Rg’s are similar. The Rg is defined as the root mean square distance of the atoms in the molecule from their common center of gravity. As such it is less sensitive to extremes within the population of conformers derived from the NMR data. Of note are samples 18 and 19, where the same SAXS data is compared against two NMR structures. The first, sample 18, was compared to a structure with no residual dipolar coupling information and the second, sample 19, was compared to a structure making use of residual dipolar coupling in the refinement process. Although these two NMR structures are similar, with backbone rmsd between the mean coordinates of each ensemble of 4.6 Å (1.5 Å for the well defined residues, 20–75), the fit of the SAXS data is significantly better to the latter. Figure 3 shows the SAXS data, structural fit to the data, and the NMR structures overlaid on each SAXS envelope. For samples 20 through 24, results similar to those observed in comparing SAXS data to the crystallographic structures are seen. The SAXS envelope accurately contains the globular portion of the NMR model; where the SAXS and NMR model diverge is consistent with the expected location of disordered residues. An exception to this appears to occur in the case of sample 21, where the NMR model indicates disordered structure extending away from the top right of the ordered portion but the SAXS envelope indicates that structural envelope is predominately to the right of the ordered portion of the molecule. Samples 22 and 25 both show large structurally-disordered regions. SAXS is a technique that is sensitive to the time- and ensemble-averaged volume occupied by a protein, but there will be a case where the amount of time a protein molecule is in a particular position or the percentage of molecules in that position is too small to produce a signal that can be interpreted as the envelope and not noise. Though NMR can be used to characterize distributions of conformations in disordered regions by interpreting the data as arising from ensemble averaging, these methods were not used for these NMR structures and the distributions of conformations in disordered regions cannot be interpreted as representative of the true conformational distributions in solution. In this case limitations of each technique must be realized and a balance needs to be made between the limitations of both techniques.
Figure 3.
The observed SAXS data and structural fit to the observed data (continuous line) for samples with NMR structures. The ab initio SAXS-derived envelopes are overlaid with the NMR structures to show the agreement between the data. The figures are shown to approximate scale as in the other structural illustrations and illustrate multiple conformations determined from the NMR data.
The SAXS data shows that samples 18, 19 and 24 are in the monomeric state, which is in disagreement with the oligomeric state determined by analytical gel filtration. NMR 1D N15 T1/T2 measurements are a more accurate technique than gel filtration for oligomer determination. NMR 1D N15 T1/T2 measurements on samples 18, 19 and 24 confirm the monomeric state, in agreement with the SAXS data.
3.4. The combination of Crystallography and NMR with SAXS
For two samples, 27 and 28, both crystallographic and NMR structures were available. In each of these cases, the SAXS envelopes were in good agreement with the crystallographic and NMR structures. For sample 27, the Rg and Dmax from the SAXS data were each within ~1 Å of the NMR structure. The Rg was within ~1 Å of the crystallographic structure, but the Dmax measures 15.8 Å greater when SAXS data is compared to the crystallographic structure. This is consistent with the crystallographic structure having 18 missing residues, ~10% of the structure, while NMR accounted for all of the residues. For sample 28, the Rg for the NMR data was in exact agreement with the SAXS data but the Dmax from the SAXS data was ~36 Å less. The crystallographic structure had 13 missing residues, 13% of the structure, which accounts for a smaller Rg and Dmax when compared to the SAXS values. The difference in Dmax from the NMR structure and SAXS data is discussed above.
The observed SAXS data and structural fit to the observed data and ab initio envelopes with structures overlaid are shown in Figure 4. The NMR and crystallographic structures are similar and fit well into the SAXS-derived molecular envelopes. In the case of sample 27, no residues are missing from the NMR structure (a) and 18 are missing from the crystallographic structure (b). For the NMR structure, regions of structural disorder are consistent with envelope regions otherwise not explained by the NMR structure. Similarly, in the X-ray structural case, missing residues are represented by the molecular envelope density consistent with the position and number of those residues. Sample 28 is missing 13 residues in the crystallographic structure (a), but can be positioned using the SAXS envelope. The NMR data (b), while fitting the experimental SAXS data better than the crystallographic (accounting for these missing residues), appears to place the bulk of the disordered region in a different location than the SAXS envelope suggests. When comparing these two examples, 18 missing residues can make little difference to the overall calculated curve in cases such as sample 27, while a similar number of missing residues can have a great impact on the calculated curve, as seen in sample 28. This may be due to location of the missing residues as well as their size compared to the size of the particle as a whole. The PISA analysis predicted the same dimer organization for sample 27 as determined by the SAXS data. However, the dimer predicted by PISA for sample 28 was not seen in the SAXS data.
3.5. Mixtures
This study has been used to determine how well SAXS ab initio molecular envelope reconstructions represent known structures and from this gain an idea of the accuracy of cases where no structural information is present. However, having this structural information also allows us to analyze samples as mixtures. Samples 4 and 11 were determined to be mixtures of oligomers from the SAXS data, Figure 5.
In these examples, scattering from each oligomer was calculated and estimates of volume fractions present in solution were carried out. For sample 4, the fit to the curve improved from χ = 7.8 for the dimer (not shown) to χ = 2.6 for the dimer-tetramer mixture. This is seen primarily in the improvement of the low q-region of the curve, corresponding to the overall size of particles in solution. This Dmax value reflect this as well, 58.7 Å for the dimer (not shown), 81.2 Å for the tetramer, in good agreement with the SAXS estimated value of 82.7 Å. The 28 residues of the dimer, and 56 residues of the tetramer that were missing in the crystallographic structure, may explain the poor fit beyond about 0.13 Å−1 for the mixture. For sample 11, the fit improved dramatically from χ = 13.5 for the dimer and χ = 10.4 for the tetramer (not shown) to χ = 1.4 for the mixture. Similarly, the Dmax for the dimer is only 71.0 Å, but for the tetramer it increases to 80.8 Å, closer to the SAXS derived Dmax of 89.7 Å. The PISA analysis indicated that both dimers were in stable oligomeric states but did not identify either tetramer.
Without knowledge of the structure, one is unable to determine volume fractions of oligomers in solution. Ab initio reconstructions for mixtures should not be carried out because most algorithms, including that used in the DAMMIF reconstructions in this study, assume a monodisperse solution and are not suited for polydisperse mixtures53. Attempts at reconstructing ab initio envelopes when samples are known to be polydisperse are shown in Figure 5. It is readily seen that in some cases, e.g. sample 4, the envelope is a poor representation of either oligomer in solution, whereas in sample 11, the envelope appears to be able to accommodate most of the tetramer. This illustrates that while an ab initio model may be constructed, it is not reliable for either oligomer if the solution is polydisperse. In a polydisperse solution containing multiple oligomers of the same basic quaternary unit the intramolecular distances within the basic quaternary unit will be similar for each oligomer and thus contribute similarly to the intensity profile as a monodisperse solution. Only the additional intramolecular distances present in the larger oligomer that are not present in the basic quaternary unit will contribute to the scattering differently than the monodisperse solution. This highlights the importance of oligomer screening prior to SAXS data collection, or the use of biophysical or biochemical separation techniques to ensure a single monodisperse oligomer population distribution, especially when no other structural information is available. If prior structural information exists, this illustrates the strength of the application of SAXS for mixed-oligomer analysis to characterize solutions containing mixtures of quaternary structures.
4. Discussion
SAXS is not a new biophysical technique but it has only recently been applied to high-throughput structural biology56. In this paper, SAXS has been approached from a different perspective, that of a high-throughput crystallization screening laboratory. SAXS has been used to characterize remnants of samples that remained after crystallization screening. Over five hundred different proteins from this group have been characterized to date. From these samples we have presented a subset of cases where crystallographic and/or NMR structural information was available. In some cases this was known prior to SAXS, in other cases it became known subsequently. In all cases, SAXS studies using minimal amounts of sample at multiple concentrations but a single buffer condition, produced molecular envelopes that were consistent with crystallographic and NMR based structural knowledge. We acknowledge the limitations of SAXS; for example, disordered regions may be averaged to a single area that is not representative of the actual molecular structure. Similarly, the SAXS envelope may not be completely sensitive to highly dynamic regions of a structure and in extreme cases could insufficiently sample and subsequently incorrectly represent the volume occupied by the flexible portion of the molecule. SAXS experiments can be performed on all of the greater than 30,000 soluble, purified samples produced by the US PSI. SAXS could be used to structurally characterize the majority of these samples. We have demonstrated that these envelopes appear to be highly consistent with known structural information. If these samples could be characterized structurally, albeit at low resolution, they would significantly increase the amount of structural knowledge that is currently available.
The fact that our envelopes are in good agreement with known structures does not imply that envelopes for samples recalcitrant to crystallization will necessarily be representative of the structures of these samples. There could be significant biochemical, biophysical, or structural reasons for failure to crystallize. However, SAXS is a powerful technique for characterizing samples in solution. It can distinguish between natively unfolded samples, those with flexible disordered regions and those that may have multiple globular regions with flexible linkers. We can identify these problem cases and limit our analysis to those samples that are well-behaving. In doing so we can have reasonable confidence that the molecular envelope produced from SAXS data reflects the molecular structure. However, reasonably confident is not completely confident. Without complementary structural, or biochemical knowledge we can never be 100% certain of the accuracy of the envelope. We have to remain wary and have to settle for the fact that most of what we see from envelope reconstructions is correct, but this is not always going to be the case.
An important note in this study is the observation of two cases of mixtures. We have a limited sample set that has been well characterized on preparation but then cycled through freeze thaw cycles both before and after crystallization trials prior to SAXS analysis. Samples should be as fresh as possible and homogeneous. One approach that is clearly recommended is the use of size-exclusion chromatography and light scattering techniques immediately before SAXS data collection to monitor monodispersity57.
SAXS is clearly complementary to high-resolution structural techniques such as crystallography and NMR spectroscopy. We have demonstrated that it provides unique quaternary structural information from the solution state that can be leveraged into biological knowledge that is not determined using independent methodologies. This is exemplified by the identification of oligomer organizations for samples 2, 3, 4, 5, 6, 9, 11, 16 and 17 that are alternatives to those seen in the crystallographic structures. Binding energy and entropy of dissociation can be calculated where structural information is present enabling prediction of the biological oligomer with services such as PISA47. This approach has been shown to be successful in 80–90% of cases, a similar success rate seen with our data. However, SAXS can directly identify these oligomers to support or challenge the prediction.
In the case of NMR, special data collection and analysis methods are required to determine the correct representation of highly disordered regions. In such disordered regions, SAXS data indicates a more compact structure than that indicated by the reported NMR conformational ensemble. To some extent, this is an issue with the calculation of Dmax from an ensemble of conformers, but there are clear cases where this alone does not fully explain difference in Dmax values. Specific modeling of SAXS sensitivity is needed to resolve this case. There are methods to treat molecules or parts of molecules as ensembles of conformers within the SAXS analysis. The Ensemble Optimization Method (EOM)58 randomly generates conformers, bins them to create ensembles and using a genetic algorithm, optimizes the ensembles by comparing the average scattering profile of their conformers to the experimental data. Using an increasing number of conformers per ensemble, and an analysis of the deviation of experimental data from predicted data, SAXS analysis can be used to study dynamic structural regions. In this study we have not made use of these methods due to the number of samples examined and the computational resources required for each case. On the other hand, the NMR methods used by the NESG consortium are not aimed at accurate representation of conformational distributions in disordered regions, which requires special methods and considerations.
The structural and biochemical data used in this study are publically available. We are happy to provide the SAXS data associated with this study to groups that may use it for further development. We have used SAXS to complement high-throughput crystallization screening and are uniquely positioned with the availability of a large number of well-behaved and well-characterized samples courtesy of the NESG efforts. We have presented a top-level overview of our initial results on a subset of samples where structural information was already available. SAXS data has been useful and provided additional information in these cases.
The strength of SAXS shown by our results causes us to echo the conclusions of Hura et al.56 in adopting the method for high-throughput structural genomic studies and to go one step further in suggesting that it is in fact essential. While X-ray crystallography and NMR are clearly powerful structural techniques, when SAXS analysis is added, the synergistic relationship between the techniques provides a far greater understanding of the biological system as a whole.
Acknowledgments
Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (P41RR001209) and the National Institute of General Medical Sciences. We would like to acknowledge those responsible for the structural information we have used from the PDB and Dr. George DeTitta for access to the high-throughput screening laboratory and remnants of samples left after crystallization screening. The referees are acknowledged for useful comments. This work was supported in part by NIH grants R01 GM088396 to EHS, and Protein Structure Initiative grants U54 GM074958 and U54 GM094597 to GTM, and U54 GM074899 to George DeTitta. Dr. E. Lattman is acknowledged for useful discussions.
References
- 1.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watson JD, Crick FH. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 3.Hodgkin DC, Kamper J, MacKay M, Pickworth JJHR, Shoemaker CB, White JG, Prosen RJ, Trueblood KN. Proc R Soc Lond A. 1957;242:228–263. [Google Scholar]
- 4.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Journal of Molecular Biology. 1984;180:385–398. doi: 10.1016/s0022-2836(84)80011-x. [DOI] [PubMed] [Google Scholar]
- 5.Abrahams JP, Leslie AG, Lutter R, Walker JE. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 6.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 7.Cramer P, Bushnell DA, Kornberg RD. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 8.Schlunzen F, Hansen HA, Thygesen J, Bennett WS, Volkmann N, Levin I, Harms J, Bartels H, Zaytzev-Bashan A, Berkovitch-Yellin Z, et al. Biochem Cell Biol. 1995;73:739–749. doi: 10.1139/o95-082. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Oughtred R, Berman HM, Westbrook J. Bioinformatics. 2004;20:2860–2862. doi: 10.1093/bioinformatics/bth300. [DOI] [PubMed] [Google Scholar]
- 10.Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. J Struct Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 11.Chayen NE, Shaw Stewart PD, Blow DM. J Cyst Growth. 1992;122:176–180. [Google Scholar]
- 12.Putnam CD, Hammel M, Hura GL, Tainer JA. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 13.Forouhar F, Lew S, Seetharaman J, Sahdev S, Xiao R, Ciccosanti C, Maglaqui M, Everett JK, Nair R, Acton TB, Rost B, Montelione GT, Tong L, Hunt JF. 2009 doi: 10.2210/pdb3hz7/pdb. [DOI] [Google Scholar]
- 14.Seetharaman J, Su M, Wang H, Foote EL, Mao L, Nair R, Rost B, Acton TB, Xiao R, Everett JK, Montelione GT, Tong L, Hunt JF. 2009 doi: 10.2210/pdb3h9w/pdb. [DOI] [Google Scholar]
- 15.Forouhar F, Lew S, Seetharaman J, Sahdev S, Xiao R, Ciccosanti C, Lee D, Everett JK, Nair R, Acton TB, Rost B, Montelione GT, Tong L, Hunt JF. 2010 doi: 10.2210/pdb3lmf/pdb. [DOI] [Google Scholar]
- 16.Vorobiev S, Neely H, Seetharaman J, Wang D, Ciccosanti C, Mao L, Xiao R, Acton TB, Everett JK, Montelione GT, Tong L, Hunt JF. 2010 doi: 10.2210/pdb3mjq/pdb. [DOI] [Google Scholar]
- 17.Forouhar F, Abashidze M, Seetharaman J, Mao M, Xiao R, Ciccosanti C, Lee D, Everett JK, Nair R, Acton TB, Rost B, Montelione GT, Tong L, Hunt JF. 2010 doi: 10.2210/pdb3mfx/pdb. [DOI] [Google Scholar]
- 18.Seetharaman J, Lew S, Wang D, Janjua H, Cunningham K, Owens L, Xiao R, Liu J, Baran MC, Acton TB, Montelione GT, Tong L, Hunt JF. 2010 doi: 10.2210/pdb3lyy/pdb. [DOI] [Google Scholar]
- 19.Seetharaman J, Chen Y, Wang D, Janjua H, Cunningham K, Owens L, Xiao R, Liu J, Baran MC, Acton TB, Montelione GT, Tong L, Hunt JF. 2010 doi: 10.2210/pdb3mjq/pdb. [DOI] [Google Scholar]
- 20.Seetharaman J, Abashidze M, Forouhar F, Janjua H, Xiao R, Ciccosanti C, Foote EL, Acton TB, Rost B, Montelione GT, Hunt JF, Tong L. 2009 doi: 10.2210/pdb3i24/pdb. [DOI] [Google Scholar]
- 21.Kuzin A, Chen Y, Seetharaman J, Mao M, Xiao R, Ciccosanti C, Foote EL, Wang H, Everett JK, Nair R, Acton TB, Rost B, Montelione GT, Tong L, Hunt JF. 2009 doi: 10.2210/pdb3icl/pdb. [DOI] [Google Scholar]
- 22.Vorobiev S, Neely H, Seetharaman J, Wang H, Foote EL, Ciccosanti C, Sahdev S, Xiao R, Acton TB, Montelione GT, Tong L, Hunt JF. 2009 doi: 10.2210/pdb3ign/pdb. [DOI] [Google Scholar]
- 23.Kuzin A, Su M, Seetharaman J, Sahdev S, Xiao R, Ciccosanti C, Maglaqui M, Everett JK, Nair R, Acton TB, Rost B, Montelione GT, Hunt JF, Tong L. 2009 doi: 10.2210/pdb3ha2/pdb. [DOI] [Google Scholar]
- 24.Kuzin A, Scott L, Forouhar F, Abashidze M, Seetharaman J, Mao M, Xiao R, Ciccosanti C, Wang H, Everett JK, Nair R, Acton TB, Rost B, Montelione GT, Hunt JF, Tong L. 2010 doi: 10.2210/pdb3ljx/pdb. [DOI] [Google Scholar]
- 25.Forouhar F, Neely H, Seetharaman J, Sahdev S, Xiao R, Ciccosanti C, Maglaqui M, Everett JK, Nair R, Acton TB, Rost B, Montelione GT, Hunt JF, Tong L. 2009 doi: 10.2210/pdb3hxl/pdb. [DOI] [Google Scholar]
- 26.Forouhar F, Abashidze M, Seetharaman J, Mao M, Xiao R, Ciccosanti C, Foote EL, Belote RL, Everett JK, Nair R, Acton TB, Rost B, Montelione GT, Tong L, Hunt JF. 2010 doi: 10.2210/pdb3lrx/pdb. [DOI] [Google Scholar]
- 27.Forouhar F, Abashidze M, Seetharaman J, Sahdev S, Xiao R, Foote EL, Ciccosanti C, Belote RL, Everett JK, Nair R, Acton TB, Rost B, Montelione GT, Tong L, Hunt JF. 2010 doi: 10.2210/pdb3lyu/pdb. [DOI] [Google Scholar]
- 28.Vorobiev S, Chen Y, Forouhar F, Maglaqui M, Ciccosanti C, Mao L, Xiao R, Acton TB, Montelione GT, Tong L, Hunt JF. 2009 doi: 10.2210/pdb3hix/pdb. [DOI] [Google Scholar]
- 29.Liu G, Shastry R, Ciccosanti C, Janjua H, Acton TB, Xiao R, Mao B, Everett JK, Montelione GT. 2010 doi: 10.2210/pdb2kw2/pdb. [DOI] [Google Scholar]
- 30.Liu G, Xiao R, Janjua J, Acton TB, Mao B, Everett J, Montelione GT. 2010 doi: 10.2210/pdb2kvu/pdb. [DOI] [Google Scholar]
- 31.Cort JR, Ramelot TA, Lee D, Ciccosanti C, Janjua H, Acton TB, Xiao R, Everett JK, Montelione GT, Kennedy MA. 2010 doi: 10.2210/pdb2kvz/pdb. [DOI] [Google Scholar]
- 32.Liu G, Tong S, Xiao R, Acton TB, Everett JK, Montelione GT. 2010 doi: 10.2210/pdb2l0b/pdb. [DOI] [Google Scholar]
- 33.Liu G, Janjua H, Xiao R, Ciccosanti C, Shastry R, Acton TB, Tong S, Everett JK, Montelione GT. 2010 doi: 10.2210/pdb2kz5/pdb. [DOI] [Google Scholar]
- 34.Lee H, Montelione GT, Prestegard JH. 2009 doi: 10.2210/pdb2kl1/pdb. [DOI] [Google Scholar]
- 35.Cort J, Lee D, Ciccosanti C, Janjua H, Acton TB, Xiao R, Everett JK, Montelione GT, Kennedy MA. 2010 doi: 10.2210/pdb2l0d/pdb. [DOI] [Google Scholar]
- 36.Mills JL, Eletsky D, Lee C, Lee K, Ciccosanti T, Hamilton R, Acton JB, Xiao G, Everett TK, Prestegard JG, Montelione GT, Szyperski T. 2010 doi: 10.2210/pdb2kzw/pdb. [DOI] [Google Scholar]
- 37.Eletsky A, Mills JL, Lee D, Ciccosanti C, Hamilton K, Acton TB, Xiao R, Everett JK, Montelione GT, Szyperski T. 2010 doi: 10.2210/pdb2kw7/pdb. [DOI] [Google Scholar]
- 38.Eletsky A, Garcia E, Wang H, Ciccosanti C, Jiang M, Nair R, Rost B, Acton TB, Xiao R, Everett JK, Lee H, Prestegard J, Montelione GT, Szyperski T. 2009 doi: 10.2210/pdb2ko1/pdb. [DOI] [Google Scholar]
- 39.Vorobiev S, Su M, Seetharaman J, Janjua J, Xiao R, Ciccosanti C, Wang H, Everett JK, Nair R, Acton TB, Rost B, Montelione GT, Tong L, Hunt JF. 2009 doi: 10.2210/pdb3ibw/pdb. [DOI] [Google Scholar]
- 40.Tang Y, Xiao R, Ciccosanti C, Janjua H, Lee DY, Everett JK, Swapna GV, Acton TB, Rost B, Montelione GT. Proteins. 2010;78:2563–2568. doi: 10.1002/prot.22760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vorobiev S, Chen Y, Lee D, Patel DJ, Ciccosanti C, Sahdev S, Acton TB, Xiao R, Everett JK, Montelione GT, Hunt JF, Tong L. 2010 doi: 10.2210/pdb3ld7/pdb. [DOI] [Google Scholar]
- 42.Xiao R, Anderson S, Aramini J, Belote R, Buchwald WA, Ciccosanti C, Conover K, Everett JK, Hamilton K, Huang YJ, Janjua H, Jiang M, Kornhaber GJ, Lee DY, Locke JY, Ma LC, Maglaqui M, Mao L, Mitra S, Patel D, Rossi P, Sahdev S, Sharma S, Shastry R, Swapna GV, Tong SN, Wang D, Wang H, Zhao L, Montelione GT, Acton TB. J Struct Biol. 2010;172:21–33. doi: 10.1016/j.jsb.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Acton TB, Gunsalus KC, Xiao R, Ma LC, Aramini J, Baran MC, Chiang YW, Climent T, Cooper B, Denissova NG, Douglas SM, Everett JK, Ho CK, Macapagal D, Rajan PK, Shastry R, Shih LY, Swapna GV, Wilson M, Wu M, Gerstein M, Inouye M, Hunt JF, Montelione GT. Methods Enzymol. 2005;394:210–243. doi: 10.1016/S0076-6879(05)94008-1. [DOI] [PubMed] [Google Scholar]
- 44.Acton TB, Xiao R, Anderson S, Aramini JM, Buchwald W, Ciccosanti C, Conover K, Everett JK, Hamilton K, Huang YJ, Janjua H, Kornhaber GJ, Lau J, Lee DY, Liu G, Maglaqui M, Ma L-C, Mao L, Patel D, Rossi P, Sahdev S, Sharma S, Shastry R, Swapna GVT, Tang Y, Tong SN, Wang D, Wang H, Zhao L, Montelione GT. Methods Enzymol. 2011 doi: 10.1016/B978-0-12-381274-2.00002-9. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vucetic S, Brown CJ, Dunker AK, Obradovic Z. Proteins. 2003;52:573–584. doi: 10.1002/prot.10437. [DOI] [PubMed] [Google Scholar]
- 46.Boehr DD, Dyson HJ, Wright PE. Chemical Reviews. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 47.Krissinel E, Henrick K. Journal of Molecular Biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 48.Smolsky IL, Liu P, Niebuhr M, Ito K, Weiss TM, Tsuruta H. Journal of Applied Crystallography. 2007;40:S453–S458. [Google Scholar]
- 49.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- 50.Guinier A, Foumet F. Small Angle Scattering of X-rays. Wiley Interscience; New York: 1955. [Google Scholar]
- 51.Svergun DI. Journal of Applied Crystallography. 1992;25:495–503. [Google Scholar]
- 52.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. Journal of Applied Crystallography. 2007;40:S223–S228. [Google Scholar]
- 53.Franke D, Svergun DI. Journal of Applied Crystallography. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Volkov VV, Svergun DI. Journal of Applied Crystallography. 2003;36:860–864. [Google Scholar]
- 55.Svergun D, Barberato C, Koch MHJ. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- 56.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA. Nature Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rambo RP, Tainer JA. RNA. 16:638–646. doi: 10.1261/rna.1946310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]