Abstract
Currently available bioreactor systems used by tissue engineers permit either direct, high-magnification observation of cell behavior or application of mechanical loads to growing tissue constructs, but not both simultaneously. Further, in most loading bioreactors, the volume of the dead space is not minimized to reduce the cost associated with perfusion media, exogenous stimulatory/inhibitory agents, proteases, and label. We have designed, developed, and tested a bioreactor that simultaneously satisfies the combined requirements of providing (i) controlled tensile mechanical stimulation, (ii) direct high-magnification imaging capability, and (iii) low dead-space volume. This novel mechanostimulatory (uniaxial tensile loading) bioreactor operates on an inverted microscope and permits continuous optical access (up to 600×) to a loaded, growing construct for extended periods of time (weeks). The reactor employs an adjustable reaction chamber in which the dead space can be reduced to <2 mL. The device has been used to cultivate our human primary corneal fibroblast-derived, tissue-engineered system for up to 14 days. Using the instrument we have successfully recorded (i) the process of fibroblasts populating, growing to confluence, and stratifying on different substrates; (ii) recorded complex and organized cell sheet motions; and (iii) recorded the behavior of a subpopulation of what appear to be degradative/catabolic cells within our fibroblast culture. The device is capable of providing detailed, long-term, dynamic images of mechanically stimulated cell/matrix interaction that have not been observed previously.
Introduction
Role of mechanics in the guidance of tissue formation
In spite of early1,2 and more recent3–6 recognition of the role of mechanics in shaping biological structure, we are only just beginning to untangle the details of the mechanically sensitive mechanisms that transduce force or strain signals into load-optimized structures. In vertebrate animals, there is substantial and long-established evidence that properly applied mechanical forces are necessary for the appropriate growth, maintenance, and remodeling of musculoskeletal system components.2,5–10 While there has been a significant research effort focused on mechanobiology of the extracellular matrix,10–20 there is no consensus on how even the simplest connective tissue structures are initially formed, grown, and maintained.21,22 Nonetheless, it has been recognized that the mechanical state of a tissue is a potent regulator of (i) fibroblast differentiation state, (ii) migration direction/speed, and (iii) the local secretome. This has led tissue engineers to include mechanical stimulation in their construct formation protocols.23–33 However, there is little data that elucidates how mechanical forces change the dynamics of fibroblast populations in cell culture systems. This is because most investigations are performed by examining the culture at fixed time points either during or after the mechanical stimulation protocol is complete. Discrete time point studies are likely to miss important dynamic behavioral changes that could provide critical information on how fibroblasts proliferate, organize, and synthesize matrix under loaded conditions. This limitation is due principally to the scarcity of bioreactor systems that permit direct, live, high-magnification observation of fibroblast culture responses to mechanical stimulation. Given the explosive growth in the availability of vital dyes, GFP labels, and both inducible and tranfectable gene expression systems, the availability of such a bioreactor would greatly improve our ability to tease apart complex mechanotransduction pathways that operate over long-time scales or that are sequentially activated.
Previous bioreactor designs
In general, the core design requirements for connective tissue engineering bioreactors include long-term sterility, environmental control, transfusion/conditioning of growth media, and mechanical stimulation in the physiological range. Over the last decade, there has been a rapid series of improvements in bioreactor design.34–37 Perfusion cultures have resulted in higher cell densities by providing a constant flow of fresh media to cell populations.38 Stirred vessels have been used for dynamic seeding of scaffolds resulting from increased collisions.39 Low-gravity devices have yielded better mass transfer obtained by achieving Reynolds numbers more conducive to a minimal boundary layer.40 Axial loading devices have been shown to increase the rate of growth for tissue such as central nervous axons.41 Dynamic compression bioreactors have examined the protein incorporation and synthesis in articular cartilage.42 Wartella and Wayne have designed a biaxial bioreactor that combines tension with the perpendicular application of compression.43 Cyclic traction devices have demonstrated an increase in mechanical properties over nonstimulated samples by ∼150% for collagen-based constructs for tendon repair.44
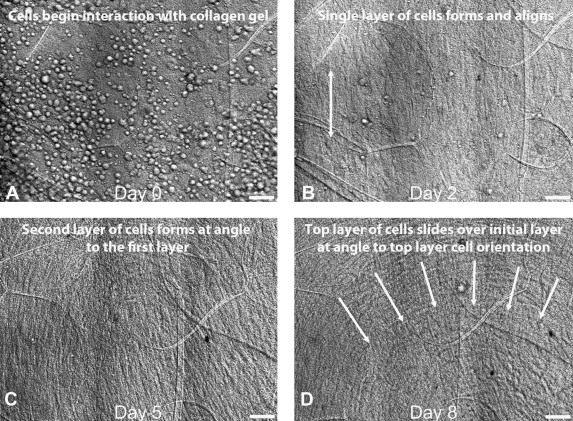
Given this successful proliferation of devices and improvements of outcome, it is clear that bioreactors will play a critical role in the field of tissue engineering. Experimental results from loading bioreactors have shown that mechanical stimulation of tissues and engineered constructs results in increases in cell proliferation, protein production, molecular incorporation, and higher levels of structural organization, in general, leading to enhanced mechanical properties. The ability to examine the detailed dynamics of these phenomena over extended periods should further enable tissue engineers to tune culture systems to optimize the effects of mechanics on construct morphology. For instance, the formation of cell–cell junctions appears to be critical to the production of organized collagen in corneal stem cell cultures.45 However, the lack of time-resolved imaging limits the ability to determine when and how cell–cell connections are produced. Such information and the ability to mechanically manipulate the substrate could lead to methods that promote formation of cell–cell junctions and enhanced organization of the resulting matrix. The direct observation of temporal patterning of cells in engineered cultures could also lead to protocols that better control cell organization and matrix production. We have seen interesting patterning already with our device (Fig. 11 in Results).
FIG. 11.
(A) Optical DIC image sequence (8 days) showing initial seeding of HPCF population, (B) first cell layer formation (cells elongated in direction of arrows), and then (C) second layer formation of cells on a disorganized construct. The last frame (D) depicts a generally organized sheet-motion of one layer of HPCFs sliding over a second layer of cells. Movies of the population dynamics and the cell sheet motion are available in Supplementary Data. One can see numerous phenomena, including the rate of proliferation of cells during the entire observation period.
Overall design goals
In addition to the normally attendant considerations associated with designing a bioreactor (biocompatibility, environmental control, perfusion system, and sterility), our overall goal was to produce a system that simultaneously satisfies three design criteria: (i) load/strain-controlled stimulation of the culture, (ii) direct optical access to the construct at high magnification for extended periods, and (iii) low dead-space volume. In our opinion, the inability to satisfy all three of these design requirements limits the utility of other bioreactors for applications that seek to elucidate real-time culture responses to mechanical stress.
Specific design goals
The device should have the capacity to uniaxially load a specimen to a force range and resolution of 0–10 N±0.01 N. The strain resolution should be ∼0.5% at lowest gage length ∼15 μm. Tissue samples of dimensions 3–16 mm gage length×0.01–2 mm thick×0.1–12 mm wide should be accommodated. For most connective tissues, a force of 0–10 N on a small thin specimen (e.g., area 10 μm×6 mm to 0.6×10−6 m2) should be adequate to produce stresses well into the normal physiological range (e.g., ∼166 MPa). The device should also provide a temperature-controlled aseptic environment (37°C±0.5°C) while providing operational perfusion ports and minimizing the chamber volume dead space. Finally, the device shall mount with minimal modification to a standard inverted microscope stage and provide the ability to perform high magnification transmission optical microscopy (up to 600× with a high numerical aperture [NA] oil immersion objective).
Materials and Methods
General design solution
The bioreactor we have designed (shown in Fig. 1) satisfies our general design criteria in that it can mount onto the stage of an inverted microscope, maintain a cell culture for extended periods, and permit direct 600× observation of a mechanically stimulated tissue sample. The design also includes a chamber that is adjustable to minimize the dead-space volume, thus reducing the cost of media, reagents, and label.
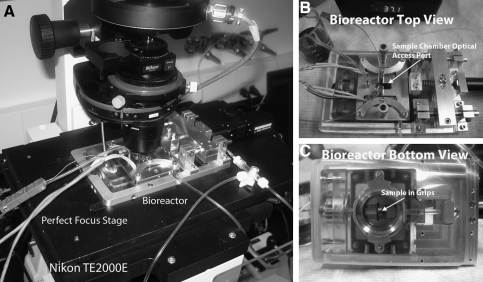
FIG. 1.
General Design Solution Hardware. (A) View of the system in operation on our Nikon TE2000E outfitted with a Perfect Focus stage. (B) Bioreactor top view showing the overall layout of the system. Optical access to the sample chamber is indicated by the arrow. (C) Bioreactor bottom view showing the adjustable sight glass, which provides access for a high-magnification oil-immersion objective. A sample is loaded into the grips in the chamber (indicated by the arrow).
Detailed design solution
Main chamber components
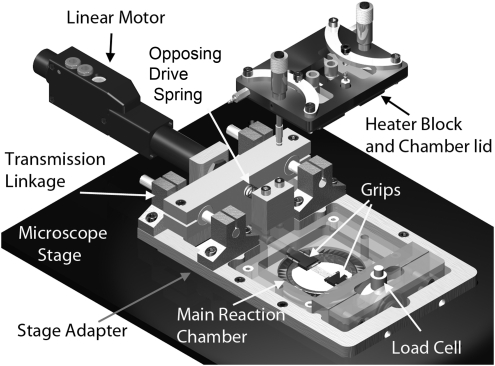
Figure 2 is a rendered drawing showing an exploded view of the main components of the bioreactor. The device is constructed around a central, adjustable-height reaction chamber that is formed by the junction of three individual components. Each of these components performs a different function. The chamber lid is the main temperature control component and provides an optical window for viewing the specimen. The chamber bottom is a vertically adjustable viewport that permits the objective to approach the sample to within the working distance of a high NA 600× objective (∼200 μm above a #1 cover slip). The bioreactor housing, which comprises the sides of the reaction chamber, provides mechanical support for the adjustable viewport, the temperature control block, the grips, and load-sensing components while it also forms a lateral thermal blockade to prevent ambient heat loss and improve thermal control. The main bioreactor housing is held in place by the microscope stage adapter, which also serves to mechanically support the linear motor and grip actuation system.
FIG. 2.
Labeled, rendered image of the bioreactor main components. The device is set into the Perfect Focus stage of the microscope. The grips lie above the adjustable sight glass in the main reaction chamber and, with the sample, form a connection between the linear actuator (through the transmission link) to the load cell in the lower right of the image. The heater block forms the top of the chamber and, with the adjustable sight glass, creates a thin channel in which the grips reside. The system provides ample access to the gripping system, but includes many seals that need to be vigorously maintained.
Material compatibility
To meet the biocompatibility and sterilizability design requirements, any components that interface with the chamber fluid or specimen are constructed from 316-stainless-steel (heater block, grips), glass (adjustable viewport and window in chamber lid), polycarbonate (main bioreactor housing), or silicone rubber (gasket between the chamber lid and the chamber as well as all o-rings). Stainless steel and glass components can be autoclaved and the polycarbonate and silicone rubber can be exposed to 70% ethanol.
Actuation/force-sensing components
Figure 2 shows the force actuation and sensing mechanisms. The grips that hold the specimen are actuated by a spring-opposed, linear motor (KT-NA08A50; Zaber), which operates at a range of velocities from 2.2×10−4 mm/s to 8.0 mm/s. The grip assembly is mounted on high-precision guide rails via precision bearings (INST 2 MS1XL3; Danaher Motion). The motor force is continuously opposed by a 2.9 lb/in tensioning spring to keep the drive system under load, which minimizes backlash. Actuation forces are transmitted to the grips that are set below the force-line produced by the motor. This offset mechanism permits the microscope objective close access to the sample without drastic modification to the microscope stage. However, the design produces some bearing-runout-backlash along the guide rails, which we have found to be acceptable relative to the strain accuracy requirements. To move the grip, the linear motor must overcome a (∼3.0 N) frictional load from the o-ring seal on the moving grip arm, and it must also counter the spring force (max 14.0 N at full throw), which opposes the motor. Thus, the maximum opposition load is 17.0 N, which is within the ability of the chosen linear actuator (50.0 N thrust force). When the grips are at their maximum separation (16.5 mm), the spring still provides 7.0 N, which is sufficient to overcome friction and still achieve a tensile load equal to the full capacity of our load cell. Force sensing is accomplished using a standard load cell (Honeywell Sensotec Model 31 Low), which has a constant form factor for load cells with capacities ranging from 0.5 N to 5.0 N±0.30% full scale. Although the bioreactor has been designed with the intent of low force application, Honeywell also manufactures a second form factor, Model 31 Mid, with capacities ranging from 10 N to 10 lbf (∼44.5 N±0.30% full scale). The Model 31 Mid has the same connection used to hold the gripping assembly, and the changed dimensions are all smaller, permitting the use of an insert, so the Model 31 Mid can be held in the current bioreactor. Force and load control are performed via a PID controller module through LabVIEW.
Temperature control
Figure 2 also shows the bioreactor temperature control module or heater block. Thermal input is provided by two 1/8"-diameter cartridge heaters (C1E14-L36; Watlow) embedded in two copper inserts. The inserts are coated with a thin layer of heat sink compound (#340; Dow Corning) and firmly held in intimate thermal contact with the stainless steel chamber lid module, which in turn contacts the chamber fluid. To permit a varying range of fluid exchange flow rates, incoming fluid is routed through the steel blocks to preheat it before entering the chamber. The temperature of the system is output through two thermocouples, positioned on either side of the sample. A redundant system is used to prevent a failed thermocouple from allowing the bioreactor to grossly overheat and damage the microscope. One thermocouple directly controls the heater controller, and the second provides output to LabVIEW. This provides a recording of the temperature during experimentation, but also controls a relay to the power supply of the heater controller. If the temperature falls outside of 37°C±4°C, the power to the heaters is shut off.
Overall system control
A custom LabVIEW program was created to record relevant operational parameters, to control the linear motor position, and to provide a backup thermal safety lockout. The program provides the ability to run the device in load or strain control while it records forces, motor displacements, and temperature (though temperature is controlled independently via a separate, independent PID system). The user interface also permits calibration of the strain and force-sensing elements. A detailed review of the LABVIEW code is beyond the scope of this article, but can be provided upon request.
Chamber volume
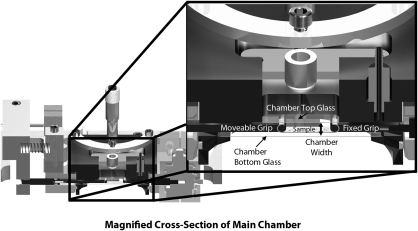
A major design criterion was to minimize the chamber dead space. Attention to this design detail will permit the use of expensive reagents and labels during observation of the cell culture system over long periods. To minimize dead space, an adjustable viewport with a sliding seal was designed in concert with a tissue grip design which ensures that the sample is the lowest object in the chamber. Figure 3 shows how the chamber volume is formed by the assembly of three pieces, one of which is moveable. As can be seen in the image, it is possible to slide the adjustable viewport right up to the tissue. With this design solution, the effective volume can be reduced to the space required to house the grips and the tissue sample (with its intended strain). By properly designing the grips and choosing a sample that is up to 8.5 mm long, it is possible to reduce the chamber volume to ∼2 mL in the current system. The device grips shown are capable of accepting samples with thicknesses of up to 3.5 mm (but this is very thick for the system in general). Our sample substrates are on the order of 30 μm thick.
FIG. 3.
Cross-sectional view of the bioreactor chamber. When the chamber reaction volume is formed by placing together the three main components (heater block, adjustable sight glass, and main reaction chamber assembly), a thin space is formed which just houses the grips. The thinness of the available space can be readily appreciated in the enlarged section view.
Optical access
The feature that most distinguishes this device from previous bioreactors is its ability to run on a high-powered optical microscope in transmission mode at 600× during continuous operation. Figure 4 shows the adjustable viewport that permits the close approach of the objective. The viewport has a piston-like design, utilizing a sliding o-ring seal, with a minimal profile to allow for the rotation the microscope objective turret while the bioreactor remains in place. Additionally, the open design enables full viewing access to the entire width and length of a sample. There is a number 1 cover slip that forms the top of the viewport and permits the use of a 60× (1.45 NA) oil immersion objective (Nikon). Objectives with this power and NA typically have a limited working distance of ∼200 μm above the cover slip. Since every sample is a different thickness (and swelling may occur in situ), the viewport must be mobile and finely tunable. The adjustable viewport is thus designed using 2 μm (Starrett Athol) as vertical adjustment actuators. The micrometers can be used to raise and lower the adjustable viewport as desired, even during operation. It should be noted, however, that for the system to permit high-resolution optical access, the tissue grips must maintain the sample at the lowest point in the chamber.
FIG. 4.
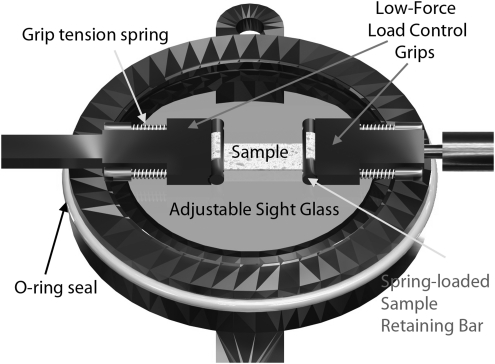
Labeled, rendered image of adjustable chamber bottom with low-force, load control grips. The grips work using a spring-loaded retaining bar that presses the sample into the grip face with a known force. The drawback to this particular grip system is that high forces extend the springs, making encoder-driven strain control inaccurate and potentially leading to slip out. However, for low force applications, they work well.
Gripping samples
To accommodate varying grip designs, the bioreactor provides thin mounting posts onto which different grip attachments may be placed. In Figure 4, one such design (suitable for light-force, load-control experiments) is shown. This particular grip assembly uses four miniature compression springs to clamp the tissue sample in place. The spring loaders thread into the sample retaining bar and when pushed forward, such that the springs are compressed, the sample retaining bar moves apart from the grip, allowing for the tissue to be placed in the grip. The springs are each precompressed to 0.85 N, and with a 200-μm-thick sample, there is a compressive force on the sample of 1.0 N from each spring, or 2.0 N in total. This design is sufficient for testing tissue to a load of the 2.0 N less the minimum force required for frictional clamping. The maximum testing force before slip can be determined by seeding the sample with microbeads and comparing LabVIEW strain measurements with actual strain observed between two beads (further described in Test Plan Design). To test at a higher force, 2-octyl cyanoacrylate, FDA-approved medical-grade glue can be used to adhere the tissue to the grip face.
Test plan design
Position accuracy test
To test the assembled drive train accuracy, we used our ability to image while operating the device. A micropipette was fixed to the grip shaft and its commanded motion (20, 100, 400, and 800 μm steps; forward and reverse) was optically tracked at 200× magnification. The position of the micropipette as reported by the linear motor encoder was compared to that determined by optical tracking of the pipette tip. Experiments were run in triplicate.
Strain control comparison to Bose ELF 3100
To estimate the accuracy of our device relative to a commercial system, we ran a series of identical uniaxial, strain-control tests on our bioreactor and on the Bose ELF 3100 Dynamic Mechanical Analyzer (ElectroForce Systems Group; Bose Corporation). The test specimens were 6-mm-wide×0.11-mm-thick calibrated rubber bands that were cut to 3, 5, or 7 mm in length. Specimens were positioned on the devices and prestretched to 0.01 N load, the resulting strain for which was assumed to be the reference or zero strain. Experimental strains of 1%, 2%, and 4% were commanded, reached with a 0.01 mm/s rate as strain rate, and held fixed at that strain to permit relaxation. Load, grip strain, and optical strain were recorded. The difference in the value of load during dynamic and equilibrium strain was examined as well as the repeatability of the load values. Experiments were run in triplicate.
Strain and load control tests on ersatz tissue construct
The ability of the bioreactor to control the load and strain on a dense collagen specimen under fluid was tested. Type I bovine collagen (Advanced Biomatrix Inc) was polymerized (3 mg/mL) according to the manufacturer's instructions and then densified in a dialysis cassette (Spectrum Laboratories) against 40% PEG (20 kDa; Sigma-Aldrich) overnight. To permit optical tracking of the specimen strain, polystyrene microbeads (3 and 10 μm beads; Polysciences, Inc.) were seeded into the collagen solution before polymerization. A custom-made cutting die was used to generate accurate and repeatable test strips from the original construct sheet. Tensile specimens were cut to 15.0 mm length×6.0 mm width×40.0 μm thickness.
Strain control: The specimen was mounted into the spring grips (shown in Fig. 4) and preloaded to 0.01 N. The initial gage length was taken as the grip-to-grip separation, following a short equilibration period. To demonstrate the bioreactor's ability to control strain, the system was commanded to strain the tissue to 5.0% at a rate of 0.005 mm/s and the actual strain (optically tracked manually by locating the position of a pair of beads near the center of the optical field in each test before and then after application of displacement) was compared to the requested value and the reported valued from the linear motor encoder. In addition, the load relaxation data following the strain application was recorded.
Load control: A similar specimen was fixed into the grips and tested by requesting 0.1 N be maintained. The load reported by the load cell was recorded as was the creep response of the tissue (tracked optically and via the motor encoder). Each experiment was repeated five times.
Temperature control test
To test the bioreactor thermal control system, we examined its ability to achieve and maintain a set temperature during perfusion of the chamber (no specimen, fluid filled). Room temperature phosphate buffered saline was perfused through the reaction chamber at 4 different rates (0, 4, 40, and 400 μL/min). These flow rates produce maximum Reynolds numbers below 10−4, indicating strictly laminar flow. Three runs at each flow rate were performed and the temperature in the center of the chamber was monitored. We set the control point temperature to a value that produced ∼37°C in the chamber center.
Complete functionality testing
Having attained satisfactory mechanical and thermal control, the environmental qualities were investigated to validate that the device is capable of providing an aseptic/sterile chamber, able to support a living cell culture system for extended periods of time. To perform the complete functionality testing, we transferred our established human primary corneal fibroblast (HPCF) tissue engineering construct system46,47 into the bioreactor. Briefly, HPCFs from a human donor were expanded in culture, released in the medium, and seeded into the bioreactor under sterile conditions in a laminar flow hood. Sterilization of the system was achieved by exposure to 70% ethanol and UV. Once seeded into the bioreactor, the cells were fed Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic at a rate of 4 μL/min and warmed to 37°C. The medium flow rate was adequate to exchange the media while keeping the shear stress below 0.1 dyn/cm2. The bioreactor was mounted onto our NIKON TE2000E with a Perfect Focus focal plane stabilization attachment. Four different paradigms for the construct system were tested: (i) direct HPCF plating onto the glass bottom of the chamber (no grips), (ii) direct HPCF plating onto a collagen gel-coated glass bottom of the chamber (no grips), (iii) direct HPCF plating onto a dense, disorganized collagen construct held in tissue grips, and (iv) direct HPCF plating onto a dense, disorganized collagen construct held in tissue grips (with application of 6% strain). For each of these experiments, HPCFs were seeded into the bioreactor onto different substrates and tracked continuously for periods up to 14 days.
Results
Device component testing
A summary of the final design specifications and system operational ranges, accuracy, and repeatability is presented in Tables 1 and 2.
Table 1.
Design Specifications: Physical Parameters and Ranges of the Bioreactor
| Bioreactor physical parameter | Grip separation range | Max sample width | Sample thickness | Chamber volume | Force rangea | Actuation speed |
|---|---|---|---|---|---|---|
| Value | 0.0–16.5 mm | 12 mm | ∼10 μm–2 mm | <2 mL (adjustable) | 0.0–4.0 N | 2.2×10−4−8 mm/s |
Force range is adjustable. Depends on tensioning spring constant, force transducer (which is interchangeable), and motor thrust capacity. Current design is limited by tensioning spring to 4.0 N maximum force, but can be increased significantly if the grips are changed and the load cell is also changed.
Table 2.
Accuracy of the Bioreactor
| Accuracya | Position | Force | Strain | Temperature | Chamber volume |
|---|---|---|---|---|---|
| Value | 6% max errorb | 8% average errorc | 2.85% average errord | 1.2% errore | <2 mL |
Data obtained through repeated observations minimum n=3.
Maximum error of 6% (1.23 μm) was obtained for backward translation over short distances (20 μm). Longer distance translations exhibited much smaller errors (i.e., 0.7% over 400 μm translation–2.7 μm error).
Average error relative to BOSE ELF 3100 measured in strain control mode at equilibrium force on tensile, elastic test strip at 3 different strains (1%, 2%, and 4%), and three different strip lengths (3, 5, and 7 mm). Bioreactor repeatability was on average better than the ELF 3100 in our hands (5.5% vs. 6.8%).
Average error based on a 5% applied strain to 7 mm gage length specimen or ∼10 μm.
Average error was based on deviation from 37°C. Repeatability of the bioreactor was 0.2% (exceedingly stable temperature wise).
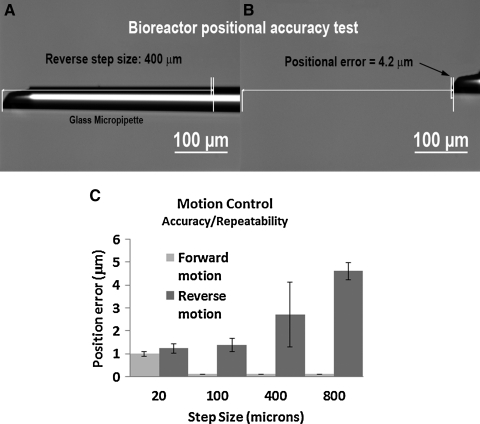
Position accuracy test
Figure 5 shows the accuracy and repeatability of the transmission linkage for forward and backward position commands of 20, 100, 400, and 800 μm. The bar graph (Fig. 5C) compares the optically sampled position error and standard deviation for each commanded step. The device operated well within our design specifications with minimal backlash (maximum of ∼5 μm). The forward positional error was undetectable optically for the three larger commanded moves (<1 μm).
FIG. 5.
Positional accuracy of device. (A) and (B) represent backward motion of the actuation system over a 400 μm commanded-move. The motion commanded is analogous to a 5% strain request for an 8.0-mm-long sample. (A) Initial position of the pipette (tip indicated by arrows) as observed on the microscope. (B) Final position of the pipette following the commanded position change. (C) Graph showing the average positional error for a series of forward and reverse steps of varying distances. Error bars are standard deviations in the positional error. The device was exceedingly accurate in the forward direction (note 0.0 standard deviation and essentially no detectable error for steps 100, 400, and 800). However, the bioreactor exhibited some backlash in the reverse direction and had a maximum average detected positional error of <5 μm.
Strain control comparison to Bose ELF 3100
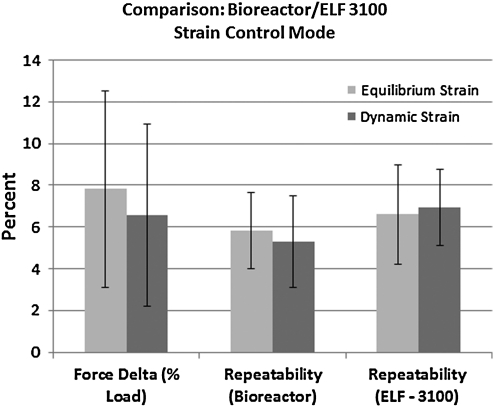
The bar graph in Figure 6 demonstrates how the bioreactor compares to the Bose ELF 3100. A total of 36 tests were run at multiple strains (1%, 2%, and 4%) and with specimen strips of varying length (3, 5, and 7 mm). The graph indicates that the bioreactor and the Bose system were in reasonably good agreement relative to force measurement during both dynamic strain and at equilibrium (aggregate average force value disagreement <8%). In general, the bioreactor seemed to overestimate the load relative to the Bose System (though both units use the same transducer). Our bioreactor was, on average, slightly more repeatable than the ELF-3100, but there was no significant difference detected.
FIG. 6.
Comparative (our bioreactor vs. Bose ELF-3100) results of a series of strain-control experiments (1%, 2%, and 4% strain) run on elastomeric strips cut to three different lengths (3, 5, and 7 mm). The first bar set is a measure of the error in the reported forces between the two systems. The average difference in the reported force values at identical strains under both dynamic and equilibrium conditions was <8%. However, it should be stated that our bioreactor consistently gave a higher force reading. The second and third bar sets provide a measure the force output repeatability (standard deviation/average force value) for the two systems.
Strain and load-control tests
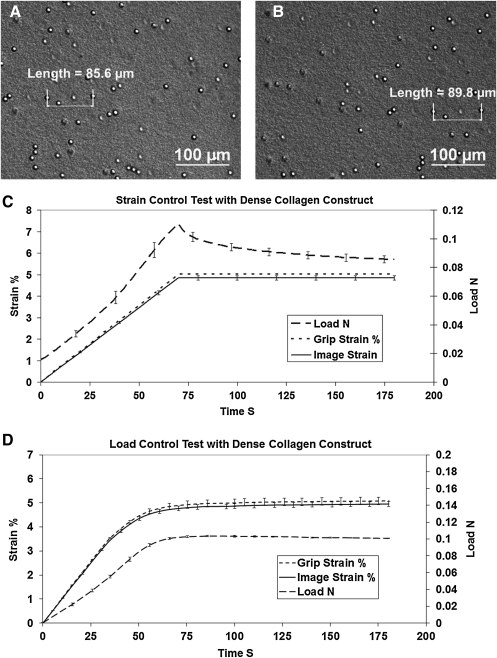
Figure 7 is representative of the kind of data obtained in this series of tests. The image shows the collagen substrate in perfusion fluid before and after the application of a known strain of 5%. Because it is a strain control test, the load evolved with the applied strain.
FIG. 7.
Strain and load control results. (A) and (B) depict the method of extracting optical strain from the sample. (A) Initial position of two beads embedded in the collagen strip. (B) Final position of the beads following applied displacement or force. Distance between the beads was measured optically (manually) by finding the center of the beads on a magnified, calibrated image. (C) Strain-control test aggregate results showing ramp to 5% strain. The image strain generally underestimates the encoder-based grip strain because of the spring-based grip design. Load curve is typical of nonlinear biological samples extended under increasing strain and shows typical relaxation during strain hold. (D) Load-control aggregate results showing ramp to 0.1N load. The load rises smoothly to 0.1 N and remains there. Note compression of error bars in the load curve. Strain curves are consistent. As before, optical strain underestimates grip strain. The dense collagen construct undergoes little creep.
Strain-control: The optically tracked strain is measured and compared to the encoder-reported strain (Fig. 7C). We thus have a true measure to compare to the output of the system. For a commanded 5% strain, the response of the system was 4.85%±0.097%. The resulting error is 2.85% of the commanded value. We suspect that the strain undershoot is due to the type of grips we used, which are spring loaded. Indeed if one looks at the comparison of optical strain to reported grip strain, it can be seen that the difference in the signal increases with applied load and that the optical strain is lower than the reported grip strain. The force data show the classic nonlinear increase in force with strain followed by the expected relaxation of the collagen strip after the strain reaches its setpoint of 5%.
Load-control: Because we could not independently measure load by a second method in the system during the load-control tests (unlike in strain-control where we had optical strain available), we thus have plotted the load and report the optically measured strain values with standard deviations. This should give an idea of the repeatability in load control. Figure 7D shows that our target load of 0.1 N is reached quickly and held stably. The evolved equilibrium strain response the sample (measured optically) reaches a consistent value of 4.95%±0.099%.
Temperature system testing
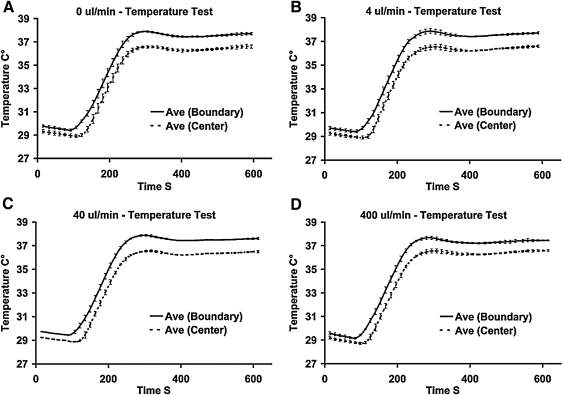
To replicate in vivo thermal conditions, the temperature of the environment inside the device will typically be set to 37°C. Figure 8 shows the transient and steady state temperature control performance of the system. The time to reach ∼37°C from a cold start is <5 min. The device meets our goal of an internal chamber temperature variation of ±0.5°C with a standard deviation as low as ±0.04°C in the center of the chamber with perfusion flow. We found that flowing fluid through the device appeared to stabilize the temperature as the standard deviation dropped substantially during temperature tests where a perfusion flow was applied. Temperature control was generally unaffected by changes in the perfusion flow rate over two orders of magnitude.
FIG. 8.
Four different thermal performance test runs on the bioreactor under conditions of varying flow rate. The target center chamber temperature was 37°C. Solid lines represent the average temperature from the two control thermocouples on either side of the chamber (labeled: Boundary). The dashed line represents the temperature of the center of the bioreactor. All curves represent experimental runs in triplicate. (A) No flow, Center: 36.6°C±0.13°C, Control points: 37.7°C±0.11°C. (B) 4 μL/min, Center: 36.6°C±0.09°C, Control points: 37.7°C±0.07°C. (C) 40 μL/min, Center: 36.5°C±0.08°C, Control points: 37.6°C±0.08°C. (D) 400 μL/min Center: 36.5°C±0.06°C, Control points: 37.4°C±0.04°C.
Complete functionality testing
The bioreactor was used to directly observe the behavior of HPCFs for extended periods of time in culture. Observation periods typically ran continuously from the initial seeding event to the full stratified construct behavior. There were four different long-term experimental runs conducted with the device. Each tested the system under different culture conditions. To serve as a baseline “check,” a set of parallel HPCF cultures were run using a standard incubator-based methodology. The methods and results for the parallel culture runs can be found in the Supplementary Data (Supplementary Data are available online at www.liebertonline.com/tec).
Direct HPCF plating on glass
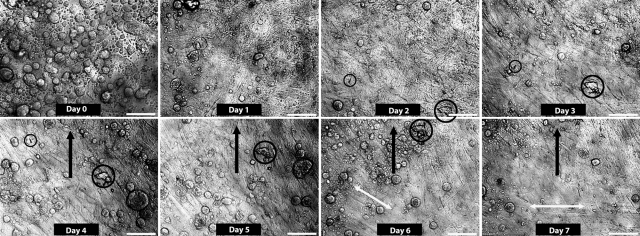
The first test-run entailed simple plating of HPCFs directly onto the chamber bottom viewport cover slip. This test demonstrated the ability of the device to provide an adequate aseptic, perfused, temperature-controlled environment capable of supporting the growth of cells in the device for extended periods. Figure 9 and Supplementary Movie S1 for Figure 9 show a temporal sequence of images of the cell behavior. The ability to image continuously in the same spot (although there are occasionally jumps in the location—see the figure caption for details) permitted us to view the motion of the cells during the entire process of colonization of the cover slip. The cell culture began as individual rounded cells on or near the glass surface and progressed to a mixture of rounded cells in the solution above the glass and elongated cells that had adhered to the glass. Eventually, the cell layer became confluent and initially exhibited random motion. At about day 3, the culture began to show both full cell sheet motion (slow) and the existence of multiple cell layers that displayed different orientations, directions, and speeds. Cells in the system exhibited at least two different general directions of motion (see Supplementary Movie S1 for Figure 9). There was a general slow motion (60 μm/day—tracked by particles/debris on the cell sheet surface) that appeared to involve continuous sheet movement. There was also higher velocity individual cell motion that tended to be at an angle to the slow sheet motion. This first test confirmed our ability to keep the cells alive for extended periods (1 week) in the system, while continuously monitoring their motile behavior.
FIG. 9.
Optical DIC image sequence (7 days) of HPCFs plating directly on chamber glass bottom. On day 0, there are many rounded cells above the glass (as determined by their random motion). By day 1, the glass is coated to confluence by the cells that are randomly oriented. By day 3, there appears to be organized sheet motion of the layer of cells (upward in image). Debris on the culture surface is tracked (black circles) and appears to keep pace with cell movement. In addition to the whole culture sheet motion vertically, there was a general horizontal motion of individual cells, which was faster, variable, and became more marked at days 6 and 7. (Bars 100 μm; see Movie in Supplementary Data for dynamic images of the behavior). Note, occasionally, there are jumps in the movie sequence or shifts in the fluid over the cells; these change could be due to either inadvertent “bumps” of the instrument or failure to relocate after clearing of a bubble. DIC, differential contrast imaging; HPCF, human primary corneal fibroblast.
Direct HPCF plating onto gel-coated glass bottom of chamber
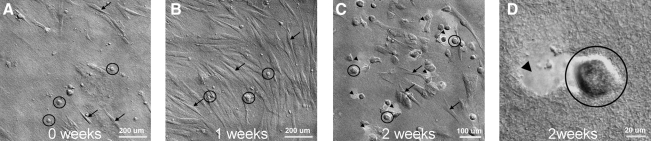
HPCFs were plated onto a standard 3.0 mg/mL atelocollagen-coated cover slip. The cells did not grow to confluence but rather sparsely adhered to the collagen on the chamber bottom. We were able to observe fibroblast behavior for extended periods at relatively high magnification (400×). We were also able to take extensive dynamic panoramic images of the culture, which has the potential to reveal system level behaviors associated with large cell masses. Figure 10 and Supplementary Movies S1 and S2 for Figure 10 show the results of this test for a single location. This test produced our most intriguing finding, which showed that there are two clearly identifiable morphological sets of cells in the culture system. One set of cells demonstrated the typical phenotype and behavior of fibroblasts that entailed an elongated cell profile and extensive filopodial exploration. Cells in the second state were rounded and appeared to be degrading the local collagen gel (as indicated by a widening “hole” within which the cell resided). In the Figure 10 image series, one can see the distribution of the two cell types in the culture system. The movie clips (one high magnification and one panoramic) show the behavior of both cell types in the same field of view. Currently, it is unclear whether the rounded cells are transformed HPCFs or if they are from a subset of cells that were co-cultured from the explant.
FIG. 10.
Optical DIC image sequence (14 days) of HPCFs plated onto a collagen gel-coated cover slip with a low cell density. Two cell types were noted in the culture system at all time points: Rounded (black circles) or Elongated (black arrows). (A) The 0 week time point shows a mix of rounded and elongated HPCFs. (B) At 1 week, elongated HPCFs appear to dominate the population though both types are evident in the culture. (C) At 2 weeks, rounded cells are more prevalent in this region and appear to be eroding the collagen coating (arrowheads). The dynamic imaging suggests that the cells are degrading the collagen gel, but does not confirm this (see Supplementary Movie). (D) The last frame in the sequence shows a higher magnification image of the cell culture. Note that the collagen coating is eroded at multiple locations that coincide with rounded cell positions.
Direct HPCF plating onto dense disorganized collagen held in tissue grips
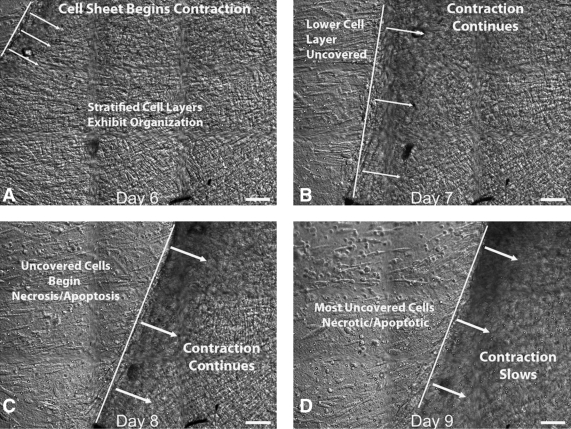
In this test, HPCFs were seeded onto a strip of dense-disorganized collagen (a control for our liquid crystal organized collagen substrate), which was held between the tissue grips. In one experimental series (shown in Fig. 11), we were able to directly observe the initial populating of the collagen substrate by a single aligned layer of HPCF cells (Fig. 11A, B). This was followed by stratification of the cell layers and then organized cell-sheet motion (Fig. 11C, D); See Supplementary Movies S1 and S2 for Figure 11 for initial population and cell sheet motion. We observed the formation of a clearly organized layer of cells on a disorganized substrate. In a second experimental series (shown in Fig. 12) the substrate became densely populated with a multilayered, disorganized colony of HPCFs before organizing. In this experiment, there was no initial formation of a single aligned sheet followed by stratification. Instead, over time (0–6 days–not shown) the cells began to create organization presumably by internally tensioning the cell sheet (Fig. 12A shows organization). At day 6, the HPCF tension became so great that the entire cell sheet was pulled from the substrate over the next 3 days (shown in Fig. 12B, C, and D and Supplementary Movie S1 for Figure 12). We noted that some cells attempted to catch the retreating cell colony, whereas other exposed cells began to die quickly, possibly via apoptosis. This is consistent with our understanding of isolated fibroblast behavior.
FIG. 12.
Optical DIC image sequence (Days 6–9) showing contraction of a multilayered, stratified culture of HPCFs. In the sequence, the cell sheet contracts (in direction of arrows) under internally generated cell force, which begins in image A. Over the next 3 days (B–D) the cell layer peels off the collagen construct. An interesting observation is the apoptotic/necrotic behavior HPCFs that have been exposed by the contracting cell sheet and the “chasing” of the retreating cell sheet by individual fibroblasts (seen in C and D). Movies of the dynamics are available in Supplementary Data.
Direct HPCF plating onto dense disorganized collagen held in tissue grips and subject to uniaxial strain
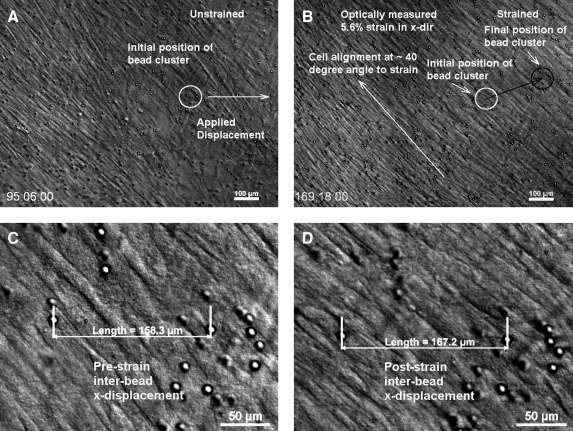
In the final experimental series we plated HPCFs onto the dense disorganized collagen and, after 4 days (reaching of near cell confluence), we applied a uniaxial strain of ∼5.6% (Fig. 13). The Supplementary Movie S1 for Figure 13 shows HPCFs first populating the gel substrate and forming a generally aligned single layer of cells. Upon application of mechanical force, the cells appeared to move with the strain and re-orient at uniform angle to the strain axis (about 40°). For full dynamics of the process, see the Supplementary Movie S1 for Figure 13.
FIG. 13.
Optical DIC image sequence of cells on dense disorganized construct before and after applied strain. (A) Low magnification image of cells on dense collagen construct at 95 h, just before applied strain. A cluster of microbeads is identified that was tracked following strain application. (B) Tracking the motion of the cluster of beads indicates that the mechanical strain applied to the matrix in an x-displacement with a concomitant vertical drift of the construct. The strain appears to alter the alignment of the cells at ∼40° to the horizontal axis. (C) High magnification image of the bead cluster before straining of the construct. NIS elements software (Nikon TE2000E) was used to determine the distance between two microbeads. (D) High magnification image of the bead cluster following application of the strain. The optical strain was calculated by finding the same two beads and measuring the horizontal distance between them. The measured optical strain was 5.6% (6% commanded strain). It should be noted that because DIC has exceedingly good out of plane rejection, some of the beads appear to have disappeared. However, they are still there, but appear as black spots or streaks. See the Supplementary Data for a movie of the dynamics of the system before and after applied construct strain. The circle indicates the position of a cluster of beads before and after strain. The second layer of out-of-focus cells is actually on the backside of the thin collagen strip.
Discussion
The results of the testing/investigation indicate that the bioreactor has met all of the intended design requirements. It is stable thermally, can be run in load or strain-control mode, and permits direct, high-magnification observation of cells in culture. The ability of the device to maintain viable cell culture systems for up to 2 weeks during continuous imaging exceeded our expectations for culture longevity. During the experimental series testing, we found unexpected HPCF behavior almost immediately. This suggests that the ability to observe tissue engineering cell culture systems (such as our HPCF-based system) continuously and at high magnification should lead to significant advances in our understanding of cell behavior under various stimuli (both chemical and mechanical). The availability of live-stains and the ability to use high-magnification optics (transmission, reflectance, and epifluorescence) further broadens the applicability of the device to a multitude of interesting problems. It should be noted that there are some limitations and requirements (some of which we did not realize a priori) that affect the ability to use the device. (i) Because of the closed system design, the media must be preconditioned with CO2 and predegassed to prevent bubble formation. We have not found preconditioning to be a significant limitation but the evolution of bubbles, even though moderate degassing was performed, does cause some imaging difficulty. This could be readily addressed by modification of the top cover glass angle of inclination. (ii) The use of the focal plane stabilization stage attachment was critical to maintaining the focal plane for producing quality movies. Thermal drift of a standard stage or other components leads to poor layer isolation and movie quality is severely compromised. (iii) Data storage capacity and software stability can be a significant problem. Our 5×5 panoramic movie, which was taken for a period of one night (images every 3 min over 15 h), was 16 GB. Storing such large amounts of data can also cause instability in the NIS software (NIS Elements 3.1), which we use to run the system. This should be considered before attempting image intensive runs. (iv) Microscope inaccessibility is also a problem for long experiments. Because we are trying to continuously image the same location, the microscope is not available during experimentation, which has exceeded 2 weeks for some runs.
Conclusions
The bioreactor performed well in both component and complete functionality testing. It has tight thermal and mechanical control, a low, dead-space volume, and good sterility maintenance in spite of the number of seals. The system should provide tissue engineers with the ability to directly observe the short- and long-term effects of mechanical loads on living constructs under a variety of stimuli of interest.
Supplementary Material
Acknowledgments
The bioreactor was initially designed by undergraduates: Aaron Desjarlais, B.S., Jessica Kornfeld, B.S., Michael Lee, B.S., Matthew McGrath, B.S., and Jeffrey Perry, B.S., for their Capstone Senior Design Course at Northeastern University with the support of NEI R01EY015500 and NIBIB R21EB007317. The final device has been significantly modified by the authors with support from NEI R01EY015500. The first author (J.A.P.) has received support from Draper Laboratories during the period over which the device was developed.
Disclosure Statement
No competing financial interests exist.
Authors Contribution
J.A.P. designed and built the bioreactor; R.Z. performed the testing protocols.
References
- 1.Thompson D.W. On Growth and Form. Cambridge: Cambridge University Press; 1917. [Google Scholar]
- 2.Wolff J. Das Gesetz Der Transformation Der Knochen. Berlin: A Hirschwald; 1891. [Google Scholar]
- 3.Guldberg R. Butler D.L. Goldstein S. Guo X.E. Kamm R. Laurencin C.T., et al. The impact of biomechanics in tissue engineering and regenerative medicine. Tissue Eng Part B Rev. 2009;15:477. doi: 10.1089/ten.teb.2009.0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingber D.E. Mechanical control of tissue morphogenesis during embryological development. Int J Dev Biol. 2006;50:255. doi: 10.1387/ijdb.052044di. [DOI] [PubMed] [Google Scholar]
- 5.Carter D. Beaupre G. Skeletal Form and Function: Mechanobiology of Skeletal Development, Aging and Regeneration. Cambridge: Press Syndicate of the University of Cambridge; 2001. [Google Scholar]
- 6.Carter D.R. Van Der Meulen M.C. Beaupre G.S. Mechanical factors in bone growth and development. Bone. 1996;18:5S. doi: 10.1016/8756-3282(95)00373-8. [DOI] [PubMed] [Google Scholar]
- 7.Chamay A. Tschantz P. Mechanical influences in bone remodeling. Experimental research on Wolff's law. J Biomech. 1972;5:173. doi: 10.1016/0021-9290(72)90053-x. [DOI] [PubMed] [Google Scholar]
- 8.Chalmers J. Ray R. The growth of transplanted foetal bones in different immunological environments. J Bone Joint Surg. 1962;44B:149. [Google Scholar]
- 9.Akeson W.H. Amiel D. Abel M.F. Garfin S.R. Woo S.L. Effects of immobilization on joints. Clin Orthop. 1987;(219):28. [PubMed] [Google Scholar]
- 10.Henderson J.H. Carter D.R. Mechanical induction in limb morphogenesis: the role of growth-generated strains and pressures. Bone. 2002;31:645. doi: 10.1016/s8756-3282(02)00911-0. [DOI] [PubMed] [Google Scholar]
- 11.Altman G.H. Horan R.L. Martin I. Farhadi J. Stark P.R. Volloch V., et al. Cell differentiation by mechanical stress. FASEB J. 2002;16:270. doi: 10.1096/fj.01-0656fje. [DOI] [PubMed] [Google Scholar]
- 12.Arokoski J.P. Jurvelin J.S. Vaatainen U. Helminen H.J. Normal and pathological adaptations of articular cartilage to joint loading. Scand J Med Sci Sports. 2000;10:186. doi: 10.1034/j.1600-0838.2000.010004186.x. [DOI] [PubMed] [Google Scholar]
- 13.Blain E.J. Gilbert S.J. Wardale R.J. Capper S.J. Mason D.J. Duance V.C. Up-regulation of matrix metalloproteinase expression and activation following cyclical compressive loading of articular cartilage in vitro. Arch Biochem Biophys. 2001;396:49. doi: 10.1006/abbi.2001.2575. [DOI] [PubMed] [Google Scholar]
- 14.Cowin S.C. The mechanical and stress adaptive properties of bone. Ann Biomed Eng. 1983;11:263. doi: 10.1007/BF02363288. [DOI] [PubMed] [Google Scholar]
- 15.Hannafin J.A. Attia E.A. Henshaw R. Warren R.F. Bhargava M.M. Effect of cyclic strain and plating matrix on cell proliferation and integrin expression by ligament fibroblasts. J Orthop Res. 2006;24:149. doi: 10.1002/jor.20018. [DOI] [PubMed] [Google Scholar]
- 16.He Y. Macarak E.J. Korostoff J.M. Howard P.S. Compression and tension: differential effects on matrix accumulation by periodontal ligament fibroblasts in vitro. Connect Tissue Res. 2004;45:28. doi: 10.1080/03008200490278124. [DOI] [PubMed] [Google Scholar]
- 17.Humphrey J.D. Stress, strain, and mechanotransduction in cells. J Biomech Eng. 2001;123:638. doi: 10.1115/1.1406131. [DOI] [PubMed] [Google Scholar]
- 18.Parsons M. Kessler E. Laurent G.J. Brown R.A. Bishop J.E. Mechanical load enhances procollagen processing in dermal fibroblasts by regulating levels of procollagen C-proteinase. Exp Cell Res. 1999;252:319. doi: 10.1006/excr.1999.4618. [DOI] [PubMed] [Google Scholar]
- 19.Petroll W.M. Vishwanath M. Ma L. Corneal fibroblasts respond rapidly to changes in local mechanical stress. Invest Ophthalmol Vis Sci. 2004;45:3466. doi: 10.1167/iovs.04-0361. [DOI] [PubMed] [Google Scholar]
- 20.Prajapati R.T. Chavally-Mis B. Herbage D. Eastwood M. Brown R.A. Mechanical loading regulates protease production by fibroblasts in three-dimensional collagen substrates. Wound Repair Regen. 2000;8:226. doi: 10.1046/j.1524-475x.2000.00226.x. [DOI] [PubMed] [Google Scholar]
- 21.Cowin S.C. How is a tissue built? J Biomech Eng. 2000;122:553. doi: 10.1115/1.1324665. [DOI] [PubMed] [Google Scholar]
- 22.Cowin S.C. Tissue growth and remodeling. Annu Rev Biomed Eng. 2004;6:77. doi: 10.1146/annurev.bioeng.6.040803.140250. [DOI] [PubMed] [Google Scholar]
- 23.Schulz R.M. Wustneck N. van Donkelaar C.C. Shelton J.C. Bader A. Development and validation of a novel bioreactor system for load- and perfusion-controlled tissue engineering of chondrocyte-constructs. Biotechnol Bioeng. 2008;101:714. doi: 10.1002/bit.21955. [DOI] [PubMed] [Google Scholar]
- 24.Vunjak-Novakovic G. Meinel L. Altman G. Kaplan D. Bioreactor cultivation of osteochondral grafts. Orthod Craniofac Res. 2005;8:209. doi: 10.1111/j.1601-6343.2005.00334.x. [DOI] [PubMed] [Google Scholar]
- 25.Meyer U. Buchter A. Nazer N. Wiesmann H.P. Design and performance of a bioreactor system for mechanically promoted three-dimensional tissue engineering. Br J Oral Maxillofac Surg. 2006;44:134. doi: 10.1016/j.bjoms.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Demarteau O. Jakob M. Schafer D. Heberer M. Martin I. Development and validation of a bioreactor for physical stimulation of engineered cartilage. Biorheology. 2003;40:331. [PubMed] [Google Scholar]
- 27.Saber S. Zhang A.Y. Ki S.H. Lindsey D.P. Smith R.L. Riboh J., et al. Flexor tendon tissue engineering: bioreactor cyclic strain increases construct strength. Tissue Eng Part A. 2010;16:2085. doi: 10.1089/ten.TEA.2010.0032. [DOI] [PubMed] [Google Scholar]
- 28.Riboh J. Chong A.K. Pham H. Longaker M. Jacobs C. Chang J. Optimization of flexor tendon tissue engineering with a cyclic strain bioreactor. J Hand Surg Am. 2008;33:1388. doi: 10.1016/j.jhsa.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 29.Joshi S.D. Webb K. Variation of cyclic strain parameters regulates development of elastic modulus in fibroblast/substrate constructs. J Orthop Res. 2008;26:1105. doi: 10.1002/jor.20626. [DOI] [PubMed] [Google Scholar]
- 30.Feng Z. Tateishi Y. Nomura Y. Kitajima T. Nakamura T. Construction of fibroblast-collagen gels with orientated fibrils induced by static or dynamic stress: toward the fabrication of small tendon grafts. J Artif Organs. 2006;9:220. doi: 10.1007/s10047-006-0354-z. [DOI] [PubMed] [Google Scholar]
- 31.Cao D. Liu W. Wei X. Xu F. Cui L. Cao Y. In vitro tendon engineering with avian tenocytes and polyglycolic acids: a preliminary report. Tissue Eng. 2006;12:1369. doi: 10.1089/ten.2006.12.1369. [DOI] [PubMed] [Google Scholar]
- 32.Webb K. Hitchcock R.W. Smeal R.M. Li W. Gray S.D. Tresco P.A. Cyclic strain increases fibroblast proliferation, matrix accumulation, and elastic modulus of fibroblast-seeded polyurethane constructs. J Biomech. 2006;39:1136. doi: 10.1016/j.jbiomech.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 33.Kall S. Noth U. Reimers K. Choi C.Y. Muehlberger T. Allmeling C., et al. [In vitro fabrication of tendon substitutes using human mesenchymal stem cells and a collagen type I gel] Handchir Mikrochir Plast Chir. 2004;36:205. doi: 10.1055/s-2004-815745. [DOI] [PubMed] [Google Scholar]
- 34.Ellis M. Jarman-Smith M. Chaudhuri J.B. Bioreactor systems for tissue engineering: A four dimensional challenge. In: Chaudhuri J.B., editor; Al-Rubeai M., editor. Bioreactors for Tissue Engineering. The Netherlands: Springer; 2005. [Google Scholar]
- 35.Portner R. Nagel-Heyer S. Goepfert C. Adamietz P. Meenen N.M. Bioreactor design for tissue engineering. J Biosci Bioeng. 2005;100:235. doi: 10.1263/jbb.100.235. [DOI] [PubMed] [Google Scholar]
- 36.Schulz R.M. Bader A. Cartilage tissue engineering and bioreactor systems for the cultivation and stimulation of chondrocytes. Eur Biophys J. 2007;36:539. doi: 10.1007/s00249-007-0139-1. [DOI] [PubMed] [Google Scholar]
- 37.Haj A.J. Hampson K. Gogniat G. Bioreactors for connective tissue engineering: design and monitoring innovations. Adv Biochem Eng Biotechnol. 2009;112:81. doi: 10.1007/978-3-540-69357-4_4. [DOI] [PubMed] [Google Scholar]
- 38.Griffiths B. Perfusion systems for cell cultivation. Bioprocess Technol. 1990;10:217. [PubMed] [Google Scholar]
- 39.Gooch K.J. Kwon J.H. Blunk T. Langer R. Freed L.E. Vunjak-Novakovic G. Effects of mixing intensity on tissue-engineered cartilage. Biotechnol Bioeng. 2001;72:402. doi: 10.1002/1097-0290(20000220)72:4<402::aid-bit1002>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 40.Chaudhuri J. Al-Rubeai M. Bioreactors for tissue Engineeering: Principles, Design and Operation. Dordrecht: Springer; 2005. [Google Scholar]
- 41.Smith D.H. Wolf J.A. Meaney D.F. A new strategy to produce sustained growth of central nervous system axons: continuous mechanical tension. Tissue Eng. 2001;7:131. doi: 10.1089/107632701300062714. [DOI] [PubMed] [Google Scholar]
- 42.Sah R.L. Kim Y.J. Doong J.Y. Grodzinsky A.J. Plaas A.H. Sandy J.D. Biosynthetic response of cartilage explants to dynamic compression. J Orthop Res. 1989;7:619. doi: 10.1002/jor.1100070502. [DOI] [PubMed] [Google Scholar]
- 43.Wartella K.A. Wayne J.S. Bioreactor for biaxial mechanical stimulation to tissue engineered constructs. J Biomech Eng. 2009;131:044501. doi: 10.1115/1.3049859. [DOI] [PubMed] [Google Scholar]
- 44.Juncosa-Melvin N. Shearn J.T. Boivin G.P. Gooch C. Galloway M.T. West J.R., et al. Effects of mechanical stimulation on the biomechanics and histology of stem cell-collagen sponge constructs for rabbit patellar tendon repair. Tissue Eng. 2006;12:2291. doi: 10.1089/ten.2006.12.2291. [DOI] [PubMed] [Google Scholar]
- 45.Du Y. Sundarraj N. Funderburgh M.L. Harvey S.A. Birk D.E. Funderburgh J.L. Secretion and organization of a cornea-like tissue in vitro by stem cells from human corneal stroma. Invest Ophthalmol Vis Sci. 2007;48:5038. doi: 10.1167/iovs.07-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo X. Hutcheon A.E. Melotti S.A. Zieske J.D. Trinkaus-Randall V. Ruberti J.W. Morphologic characterization of organized extracellular matrix deposition by ascorbic acid-stimulated human corneal fibroblasts. Invest Ophthalmol Vis Sci. 2007;48:4050. doi: 10.1167/iovs.06-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren R. Hutcheon A.E. Guo X.Q. Saeidi N. Melotti S.A. Ruberti J.W., et al. Human primary corneal fibroblasts synthesize and deposit proteoglycans in long-term 3-D cultures. Dev Dyn. 2008;237:2705. doi: 10.1002/dvdy.21606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.