Abstract
Human Scribble (Scrib) is an evolutionary-conserved cell polarity protein, but its potential role in human cancer is controversial. Herein, we show that Scrib is nearly universally overexpressed in cultured tumor cell lines and genetically disparate cancer patient series compared with matched normal tissues in vivo. Instead of a membrane association seen in normal epithelia, tumor-associated Scrib is mislocalized and found predominantly in the cytosol. Small-interfering RNA silencing of Scrib in model lung adenocarcinoma A549 cells inhibited cell migration in wound-healing assays, suppressed tumor cell invasion across Matrigel-coated inserts, and down-regulated the expression of cell motility markers and mediators of epithelial-mesenchymal transition. These data uncover a previously unrecognized exploitation of Scrib for aberrant tumor cell motility and invasion, thus potentially contributing to disease progression in humans.
Human Scribble (Scrib) is an evolutionary-conserved member of the LAP (PDZ and LRR) gene family1 that localizes to basolateral aspects of adherent junctions and maintains apical-basal polarity in normal epithelia.2 Experimental evidence, first collected in Drosophila melanogaster3 and later extended to mammalian cells,4 implicates Scrib as a general regulator of directional cell motility, largely via the assembly of multiprotein complexes and the activation of small GTPase signaling at the leading edge of migrating cells.5,6 Accordingly, loss-of-function Scrib mutants7 or depletion of Scrib4 has disrupted apical-basal polarity8 and has cooperated with oncogenic signals9 to enhance tumor cell migration, invasion, and survival.10
Although these data have prompted a model that Scrib may function in an evolutionary-conserved pathway of tumor suppression,1,3 its role in human cancer, in vivo, has remained vehemently debated. Although initial evidence suggested that Scrib expression is lost during human tumorigenesis, other reports have shown that Scrib levels are increased11 and potentially subcellularly mislocalized12 in at least certain human tumors and that overexpressed Scrib may contribute to the progression of hepatocellular carcinoma in humans.13 Similar conflicting results have been reported8,14 for other cell polarity proteins of the Par complex (Par6β and α protein kinase C), which have been associated with both tumor suppression and oncogenic functions.
In this study, we used a global expression strategy to map the distribution of Scrib in human cancer and to test the impact of this pathway on mechanisms of tumor progression.
Materials and Methods
Patients
Representative samples of cancers of the colon, liver, prostate, uterus, thyroid, lung, bladder, breast, ovary, and stomach (10 cases each) were selected for Cancer Universal TMA construction. An equal number of cases of lymphomas, tumors of the central nervous system, and testicular cancer were also included in the Cancer Universal TMA. The clinical-pathological characteristics of the patients used in this study are given in Supplemental Table S1 (available at http://ajp.amjpathol.org). For TMA construction of non–small cell lung cancer (NSCLC), 226 patients who underwent surgical resection for curative purposes from September 1999 to December 2004 were selected. Of these patients, 154 (68%) had adenocarcinomas, 64 (28%) had squamous cell carcinomas (SCCs), 5 (2%) had adenosquamous carcinomas, and 3 (1%) had large-cell carcinomas. Twenty paired NSCLC and normal counterparts from this series were selected for mRNA analysis, including 9 adenocarcinomas (45%), 9 SCCs (45%), 1 adenosquamous carcinoma (5%), and 1 large-cell carcinoma (5%).
The breast cancer series used in these studies included five ductal in situ carcinomas, 38 invasive ductal carcinomas, two papillary carcinomas, one mucinous cancer, and one lobular cancer. Three breast specimens collected from plastic surgery procedures, with no evidence of neoplasia, were used as normal controls.
Biopsy specimens from five cervical dysplasias (four cervical intraepithelial neoplasias II and 1 cervical intraepithelial neoplasia III), one carcinoma in situ, and two infiltrating cervical cancers were used. All patients in this series were positive for human papillomavirus (HPV) infection by PCR amplification using HPV-specific primers15 and direct sequencing.
TMA Construction
Representative tissue blocks from each patient were used to construct TMA, as previously described.16 For Cancer Universal TMA, one tumoral area from each specimen was punched. For NSCLC and breast cancer TMAs, four tumor areas were selected from each tissue block. Samples of nonneoplastic parenchyma, including 16 cores of normal lung and 20 cores of normal mammary glands, were added to each TMA block. For quality control, a 4-μm-thick section was cut from each TMA block, stained with H&E, and analyzed by immunohistochemistry (IHC).
IHC
For antigen retrieval, slides were microwaved for 35 minutes in citrate solution. Sections (4-μm thick) were cut from TMA or full-section blocks and stained with a goat polyclonal antibody specific to Scrib (see Supplemental Figure S1A at http://ajp.amjpathol.org) (1:100, D-8; Santa Cruz Biotechnology, Santa Cruz, CA) for 1 hour at 22°C. IHC was performed using an autostainer (Optimax i6000; Biogenex, San Ramon, CA), with detection of antibody reactivity with a kit (EnVision Detection Kit; Dako, Milan, Italy) using peroxidase-diaminobenzidine as the chromogen. All slides were counterstained with hematoxylin. Immunoreactivity for Scrib in the various samples was evaluated by two pathologists (M.M. and S.B.) and independently scored for membrane-associated or cytoplasmic localization. When discrepancies in Scrib scoring occurred, a consensus interpretation was reached after reexamination. Because Scrib immunoreactivity was either present or absent in all epithelial cells, normal and neoplastic samples were scored as follows (according to immunoreactivity intensity): 0, negative; 1, weak; 2, moderate; or 3, strong.
Cell Lines, Transfections, and Immunoblotting
Human tumor cell lines representative of malignant mesothelioma (MSTO-211H) and cancers of the colon (LoVo and SW1116), breast (MCF-7, MDA-MB231, and SKBR3), lung (A549), stomach (MKN45 and KATO-III), uterus (Hela), prostate (DU145), and liver (HepG2) were purchased from American Type Culture Collection (Manassas, VA) and maintained in culture, as recommended by the supplier. A549 cells (2 × 106 per well in six-well plates) were transfected with 100 pmol of nontargeting or Scrib-directed small-interfering RNA (siRNA) (siGENOME Smart Pool, catalogue numbers M-010500, D-001206; Dharmacon, Lafayette, CO) and 5 μL Lipofectamine 2000 (Invitrogen, Life Technologies Corporation, Carlsbad, CA). In some experiments, transfected A549 cells were harvested after 48 hours and solubilized in 150 μL radioimmunoprecipitation assay buffer plus protease inhibitors (Roche, Basel, Switzerland); aliquots of cell lysates (50 μg) were probed with antibodies (1 μg/μL) to Scrib (Santa Cruz Biotechnology), Snai2 (LifeSpan Biosciences, Seattle, WA), β-catenin (Thermoscientific, Waltham, MA), vimentin (Dako), Zeb-1 (Sigma-Aldrich, Milan, Italy), FAK-Ptk2 (Santa Cruz Biotechnology), or β-actin (Sigma-Aldrich). Reactive bands were visualized with ECL Plus (GE Healthcare, Milan, Italy).
Real-Time RT-PCR
Total RNA, isolated from 20 cases of NSCLC and matched normal tissue using reagent (Trizol; Invitrogen), was reverse transcribed17 and analyzed for changes in Scrib mRNA expression relative to β2-microglobulin using TaqMan gene expression assays (Hs00363005_m1 and Hs99999907_m1, respectively; Applied Biosystems, Life Technologies Corporation, Carlsbad, CA), according to the 2-ΔCt formula.
Cell Migration and Cell Invasion Assays
Monolayers of transfected A549 cells (70% confluency) were analyzed for changes in cell migration using a wound-healing assay during a 24-hour interval. Quantification of migration distance was determined as the reduction in the wound's gap using computer software (NIH ImageJ). In other experiments, transfected A549 cells were analyzed for cell invasion across Matrigel-coated 8.0-μm inserts (Transwell; BD Bioscience, Milan, Italy) using serum as the chemoattractant for 24 hours. The cells on the apical surface of each insert were scraped off; and membranes with invaded cells were fixed in 100% methanol, stained with toluidine blue (Sigma-Aldrich), and mounted on microscope slides. Invading cells were counted after photographing the membranes, and both peripheral and central areas of the insert were analyzed.
Oncomine Analysis
Differential Scrib expression was analyzed in published microarray data sets using a Web-based data-mining platform (Oncomine; available at http://www.oncomine.org). Relative expression data of Scrib were collected from solid malignancies only, using P ≤ 0.001 as the cutoff (t-statistic provided within the data sets in Oncomine).
Statistical Analysis
All cell-based experiments were performed at least three times in duplicate. Differences among sample groups were analyzed using the unpaired Student's t-test function of GraphPad Prism version 4 software. P < 0.05 was considered statistically significant.
Results
All tumor tissues in the Cancer Universal TMA displayed strong Scrib immunoreactivity, in vivo (IHC intensity >1, Figure 1A), except for thyroid cancers (IHC intensity = 0.7, Figure 1A). The highest expression of Scrib was observed in cancers of the colon, bladder, prostate, uterus, and ovaries (IHC intensity >2; Figure 1, A and B), thus independently of tumor histotype (see Supplemental Table S1 at http://ajp.amjpathol.org). Scrib was also prominently expressed in all 11 cultured tumor cell lines examined by IHC (Figure 1C).
Figure 1.
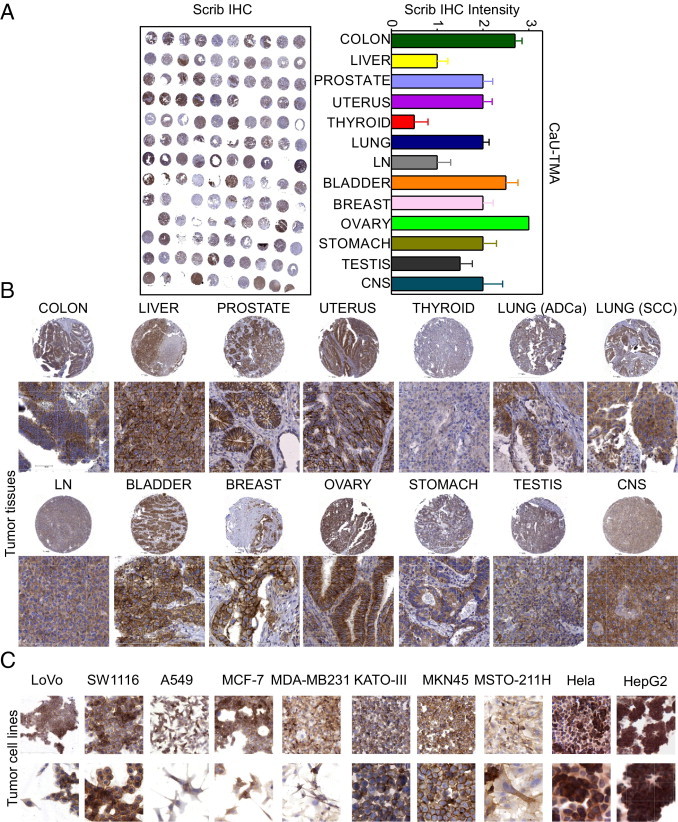
Scrib expression in human cancers. A: Left: Representative image from the Cancer Universal TMA (CaU-TMA). Right: Quantification of IHC intensity. B and C: Scrib immunoreactivity in human tumors (B) or tumor cell lines (C). Original magnification: ×80 (cores); ×200 (cells); ×400 (enlargements). ADCa indicates adenocarcinoma; LN, lymph node; CNS, central nervous system.
Analysis of normal pneumocytes or normal bronchial epithelia revealed undetectable to low levels of Scrib, respectively (Figure 2A). In contrast, Scrib was prominently expressed in tumor tissues of a large series (226 patients) of NSCLCs (IHC average immunoreactivity = 1.34; tumor versus normal, P = 1.87 × 10−8; Figure 2B). For NSCLC subtypes, adenocarcinomas expressed comparatively higher levels of Scrib protein (Figure 2B) and Scrib mRNA (Figure 2C) than SCCs [IHC mean immunoreactivity: adenocarcinoma, 1.54; and SCC, 0.83 (P = 2.9 × 10−9); and fold increase of mRNA levels in adenocarcinoma over SCC, 2.5 (P = 0.0082)]. However, either histotype had higher levels of Scrib compared with normal parenchyma (adenocarcinoma versus normal, P = 2.78 × 10−11 and P = 0.0017 for IHC and gene expression analysis, respectively; and SCC versus normal, P = 0.0002 and P = 0.04 for IHC and gene expression analysis, respectively; Figure 2, B and C).
Figure 2.
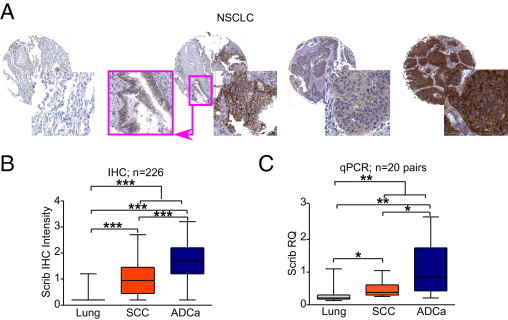
Scrib expression in NSCLCs. A: Representative images of Scrib immunoreactivity in normal lung parenchyma (scored 0, first core), nonneoplastic bronchus (pink inset in the second core) adjacent to cancer (scored 1 and 3, respectively), squamous cell carcinoma (scored 2, third core), and adenocarcinoma (scored 3, forth core) cores. B: Scrib protein expression (IHC intensity) in 226 NSCLCs and 16 normal counterparts. C: Scrib mRNA levels (RQ) in 20 paired histotypes of NSCLC cases and normal parenchyma. *P < 0.05, **P < 0.01, and ***P < 0.0001 (obtained by unpaired t-test). ADCa indicates adenocarcinoma; qPCR, quantitative PCR.
In a series of breast cancers (n = 46; see Supplemental Figure S1B at http://ajp.amjpathol.org), Scrib was ubiquitously overexpressed in tumors compared with normal matched tissues (P = 0.002, P = 0.0007, and P = 0.027; see Supplemental Figure S1C at http://ajp.amjpathol.org), irrespective of hormonal receptor and HER2 status and consistent with the frequent amplification of the Scrib locus in breast18 and ovarian19 cancers. Although Scrib was detectable in normal mammary glands with a membranous distribution at cell-cell junctions, tumor-associated Scrib was subcellularly mislocalized and exhibited a predominantly diffuse cytoplasmic reactivity (see Supplemental Figure S1D at http://ajp.amjpathol.org), similar to earlier reports12 in colon cancer.
Bioinformatic analysis of published microarray data sets (Oncomine) revealed that Scrib was uniformly expressed at high levels in genetically disparate cancer types, compared with matched normal tissues (see Supplemental Figure S2A at http://ajp.amjpathol.org). Although Scrib deregulation cooperates with mutant k-Ras7 or HPV E6 protein20 to enhance tumorigenesis, in our analysis, Scrib levels in human tumors were independent of mutant k-Ras or HPV protein expression (see Supplemental Figure S2, B and C, at http://ajp.amjpathol.org). Accordingly, strong Scrib immunoreactivity was observed in dysplastic or cancerous cervical lesions, compared with moderate positivity detected in matched nontransformed epithelium (n = 8; see Supplemental Figure S3A at http://ajp.amjpathol.org). In this series and similar to the IHC results obtained with breast cancer, Scrib localization in cervical lesions was primarily diffuse and cytosolic, as opposed to its predominant membrane association observed for the normal mucosa (see Supplemental Figure S3B at http://ajp.amjpathol.org).
Transfection of lung adenocarcinoma A549 cells with Scrib-directed siRNA efficiently silenced the intended target, whereas control siRNA had no effect (Figure 3A). Under these conditions, siRNA depletion of Scrib strongly impaired tumor cell migration in a wound closure assay, in vitro (P < 0.0001, Figure 3B); and profoundly inhibited tumor cell invasion across Matrigel-coated inserts (P = 0.02, Figure 3C). In contrast, control nontargeting siRNA had no effect on Scrib levels (Figure 3A) and did not impair tumor cell migration or invasion (Figure 3, B and C). In addition, siRNA depletion of Scrib in A549 cells resulted in reduced expression of cell motility markers and proteins associated with epithelial-mesenchymal transition, including focal adhesion kinase (Ptk2-Fak), β-catenin, and Snai2 (Figure 3D). In contrast, the expression of vimentin or Zeb-1 was only modestly affected; and a control siRNA had no effect on protein levels (Figure 3D).
Figure 3.
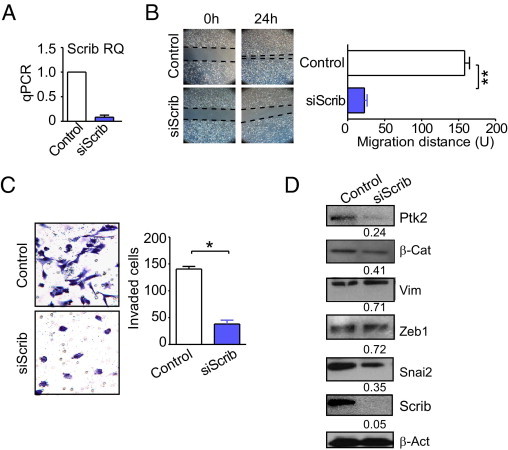
Scrib regulation of tumor cell motility. A: A549 cells were transfected with control nontargeting or Scrib-directed siRNA and analyzed for Scrib mRNA expression. B: Transfected A549 cells were analyzed for cell migration in a wound-healing assay. C: The experimental conditions are as in B, except that transfected A549 cells were analyzed for cell invasion across Matrigel-coated chambers (Transwell). D: Transfected A549 cells were analyzed by Western blotting. β-actin (β-Act) was a loading control. *P < 0.05 and **P < 0.01. qPCR indicates quantitative PCR; siScrib, small-interfering Scrib; Ptk2, Protein tyrosin kinase 2; β-Cat, β-Catenin; Vim, Vimentin.
Discussion
In this study, we showed that Scrib, a pivotal cell polarity module21 with putative tumor suppression functions,1,3 is almost universally overexpressed and subcellularly mislocalized in disparate human cancers, in vivo, independently of oncogenic signaling or viral (ie, HPV) transformation. Nevertheless, Scrib diffusion into the cytoplasm could also be related to saturation of membrane-free positions consequent to Scrib overexpression. Mechanistically, short-term silencing of Scrib in model lung cancer cells profoundly impairs tumor cell migration and invasion and down-regulates several pivotal markers of cell motility and epithelial-mesenchymal transition. These results are at variance with a prevailing model that implicates Scrib in an evolutionary-conserved mechanism of tumor suppression1,3 and suggest that this pathway is broadly exploited during human tumorigenesis, in vivo, enhancing aberrant tumor cell motility and invasion. Thus, the findings potentially contribute to metastatic dissemination and disease progression in humans.22 This alternative model is consistent with other mechanistic results showing that Scrib overexpression in astrocytes perturbed cellular polarity and induced randomly oriented cellular protrusions6; conversely, depletion of Scrib in breast cancer cells inhibited migration,4,5 potentially by disrupting the activation of Rac and PAK motility factors at the leading edge.5 Loss of Scrib at early stages of cellular transformation may disrupt the apical-basal polarity of epithelial cells,2 which is potentially consistent with a putative role in tumor suppression.1,3 Future investigations should address whether Scrib mutations are common events in human cancers or whether mutations are responsible for Scrib delocalization into the cytosol.
However, taken together, our data indicate a more complex scenario, in which aberrant overexpression of Scrib in established tumors and potentially other polarity proteins,23 coupled to their subcellular mislocalization away from cell-cell contacts,8 globally disrupts cell motility pathways, favoring random, as opposed to directional, cell migration.21 This further contributes to the acquisition of a migratory epithelial-mesenchymal transition–like cellular phenotype. In this context, therapeutic targeting of the Scrib pathway may selectively disrupt pivotal mechanisms of random tumor cell motility and invasion22 in tumors, creating concrete new prospects for the development of directed antimetastatic therapies in humans.
Footnotes
Supported by a fellowship of the Doctorate School of Molecular Medicine at Università degli Studi di Milano (A.F.) and in part by grants from the National Institutes of Health (CA140043, CA78810, and CA118005 to D.C.A.) and the Fondazione Berlucchi (S.B.).
D.C.A. and S.B. contributed equally to this work.
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.02.028.
Supplementary data
Scrib protein expression in human breast pathology. A: Validation of Scrib-antibody specificity. Immunohistochemistry with the primary antibody to Scrib was performed in A549 cells transfected with control or Scrib-targeting siRNA. B: Scrib protein expression was evaluated in a series of normal breast glands, ductal carcinoma in situ, and invasive cancers using a tissue microarray platform. C: Quantification of Scrib immunoreactivity (IHC intensity) in normal mammary parenchyma and breast diseases. The number of available areas per tissue category is indicated at the bottom of the bars. D: Representative images of Scrib expression pattern in normal breast glands, ductal carcinoma in situ (DCIS), HR-positive/Her2-negative (HR-POS/Her2-NEG) and HR-negative/Her2-positive (HR- NEG/Her2-POS) invasive cancers. Original magnification x80 (cores) and x400 (enlargments). *P < 0.05; **P < 0.01; ***P < 0.001
Oncomine Cancer Microarray database analysis of Scrib gene expression profiles. Public microarray datasets (Oncomine repository) were queried for expression of Scrib in carcinomas versus normal tissues (A) or in neoplasia with correlated molecular analysis, as k-Ras mutation status (B) or HPV-infection presence (C). Datasets are reported using the name of the involved organ and the name of the author as it appears in Oncomine repository. (ns, not significant; Ca, Carcinoma; Ad-Ca, Adenocarcinoma; SCC, Squamous Cell Carcinoma; SCLC, Small Cell Lung Cancer; HCC, Hepatocellular Carcinoma; AA, Anaplastic Astrocytoma; HOG, Human Oligodendroglioma; GBM, Glioblastoma; DA, Diffuse Astrocytoma; PCA, Prostate cancer; KRAS-WT, k-Ras wild-type; KRAS-Mut, Mutated k-Ras; HPV, Human Papilloma Virus).
A: Scrib expression was evaluated by IHC in cervical lesions and matched normal epithelium in cases of cervical dysplasia (CIN) or invasive carcinomas (SCC) positive for Human Papilloma Virus infection. B: Representative images of cervical lesions and normal epithelium. Original magnification x400. CIN, Cervical intraepithelial neoplasia; ISC, in situ cervical carcinoma; SCC, Squamous cell carcinoma.
References
- 1.Humbert P.O., Grzeschik N.A., Brumby A.M., Galea R., Elsum I., Richardson H.E. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 2008;27:6888–6907. doi: 10.1038/onc.2008.341. [DOI] [PubMed] [Google Scholar]
- 2.Navarro C., Nola S., Audebert S., Santoni M.J., Arsanto J.P., Ginestier C., Marchetto S., Jacquemier J., Isnardon D., Le Bivic A., Birnbaum D., Borg J.P. Junctional recruitment of mammalian Scribble relies on E-cadherin engagement. Oncogene. 2005;24:4330–4339. doi: 10.1038/sj.onc.1208632. [DOI] [PubMed] [Google Scholar]
- 3.Bilder D., Li M., Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 4.Dow L.E., Kauffman J.S., Caddy J., Zarbalis K., Peterson A.S., Jane S.M., Russell S.M., Humbert P.O. The tumour-suppressor Scribble dictates cell polarity during directed epithelial migration: regulation of Rho GTPase recruitment to the leading edge. Oncogene. 2007;26:2272–2282. doi: 10.1038/sj.onc.1210016. [DOI] [PubMed] [Google Scholar]
- 5.Nola S., Sebbagh M., Marchetto S., Osmani N., Nourry C., Audebert S., Navarro C., Rachel R., Montcouquiol M., Sans N., Etienne-Manneville S., Borg J.P., Santoni M.J. Scrib regulates PAK activity during the cell migration process. Hum Mol Genet. 2008;17:3552–3565. doi: 10.1093/hmg/ddn248. [DOI] [PubMed] [Google Scholar]
- 6.Osmani N., Vitale N., Borg J.P., Etienne-Manneville S. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr Biol. 2006;16:2395–2405. doi: 10.1016/j.cub.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Wu M., Pastor-Pareja J.C., Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463:545–548. doi: 10.1038/nature08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aranda V., Nolan M.E., Muthuswamy S.K. Par complex in cancer: a regulator of normal cell polarity joins the dark side. Oncogene. 2008;27:6878–6887. doi: 10.1038/onc.2008.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dow L.E., Elsum I.A., King C.L., Kinross K.M., Richardson H.E., Humbert P.O. Loss of human Scribble cooperates with H-Ras to promote cell invasion through deregulation of MAPK signalling. Oncogene. 2008;27:5988–6001. doi: 10.1038/onc.2008.219. [DOI] [PubMed] [Google Scholar]
- 10.Zhan L., Rosenberg A., Bergami K.C., Yu M., Xuan Z., Jaffe A.B., Allred C., Muthuswamy S.K. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamei Y., Kito K., Takeuchi T., Imai Y., Murase R., Ueda N., Kobayashi N., Abe Y. Human scribble accumulates in colorectal neoplasia in association with an altered distribution of beta-catenin. Hum Pathol. 2007;38:1273–1281. doi: 10.1016/j.humpath.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 12.Gardiol D., Zacchi A., Petrera F., Stanta G., Banks L. Human discs large and scrib are localized at the same regions in colon mucosa and changes in their expression patterns are correlated with loss of tissue architecture during malignant progression. Int J Cancer. 2006;119:1285–1290. doi: 10.1002/ijc.21982. [DOI] [PubMed] [Google Scholar]
- 13.Woo H.G., Park E.S., Lee J.S., Lee Y.H., Ishikawa T., Kim Y.J., Thorgeirsson S.S. Identification of potential driver genes in human liver carcinoma by genomewide screening. Cancer Res. 2009;69:4059–4066. doi: 10.1158/0008-5472.CAN-09-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grifoni D., Garoia F., Bellosta P., Parisi F., De Biase D., Collina G., Strand D., Cavicchi S., Pession A. aPKCzeta cortical loading is associated with Lgl cytoplasmic release and tumor growth in Drosophila and human epithelia. Oncogene. 2007;26:5960–5965. doi: 10.1038/sj.onc.1210389. [DOI] [PubMed] [Google Scholar]
- 15.Fujinaga Y., Shimada M., Okazawa K., Fukushima M., Kato I., Fujinaga K. Simultaneous detection and typing of genital human papillomavirus DNA using the polymerase chain reaction. J Gen Virol. 1991;72:1039–1044. doi: 10.1099/0022-1317-72-5-1039. [DOI] [PubMed] [Google Scholar]
- 16.Barberis M., Pellegrini C., Cannone M., Arizzi C., Coggi G., Bosari S. Quantitative PCR and HER2 testing in breast cancer: a technical and cost-effectiveness analysis. Am J Clin Pathol. 2008;129:563–570. doi: 10.1309/1AKQDQ057PQT9AKX. [DOI] [PubMed] [Google Scholar]
- 17.Romagnoli S., Fasoli E., Vaira V., Falleni M., Pellegrini C., Catania A., Roncalli M., Marchetti A., Santambrogio L., Coggi G., Bosari S. Identification of potential therapeutic targets in malignant mesothelioma using cell-cycle gene expression analysis. Am J Pathol. 2009;174:762–770. doi: 10.2353/ajpath.2009.080721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naylor T.L., Greshock J., Wang Y., Colligon T., Yu Q.C., Clemmer V., Zaks T.Z., Weber B.L. High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005;7:R1186–R1198. doi: 10.1186/bcr1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S.W., Kim J.W., Kim Y.T., Kim J.H., Kim S., Yoon B.S., Nam E.J., Kim H.Y. Analysis of chromosomal changes in serous ovarian carcinoma using high-resolution array comparative genomic hybridization: potential predictive markers of chemoresistant disease. Genes Chromosomes Cancer. 2007;46:1–9. doi: 10.1002/gcc.20384. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa S., Huibregtse J.M. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol Cell Biol. 2000;20:8244–8253. doi: 10.1128/mcb.20.21.8244-8253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Etienne-Manneville S. Polarity proteins in migration and invasion. Oncogene. 2008;27:6970–6980. doi: 10.1038/onc.2008.347. [DOI] [PubMed] [Google Scholar]
- 22.Steeg P.S. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 23.Nolan M.E., Aranda V., Lee S., Lakshmi B., Basu S., Allred D.C., Muthuswamy S.K. The polarity protein Par6 induces cell proliferation and is overexpressed in breast cancer. Cancer Res. 2008;68:8201–8209. doi: 10.1158/0008-5472.CAN-07-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scrib protein expression in human breast pathology. A: Validation of Scrib-antibody specificity. Immunohistochemistry with the primary antibody to Scrib was performed in A549 cells transfected with control or Scrib-targeting siRNA. B: Scrib protein expression was evaluated in a series of normal breast glands, ductal carcinoma in situ, and invasive cancers using a tissue microarray platform. C: Quantification of Scrib immunoreactivity (IHC intensity) in normal mammary parenchyma and breast diseases. The number of available areas per tissue category is indicated at the bottom of the bars. D: Representative images of Scrib expression pattern in normal breast glands, ductal carcinoma in situ (DCIS), HR-positive/Her2-negative (HR-POS/Her2-NEG) and HR-negative/Her2-positive (HR- NEG/Her2-POS) invasive cancers. Original magnification x80 (cores) and x400 (enlargments). *P < 0.05; **P < 0.01; ***P < 0.001
Oncomine Cancer Microarray database analysis of Scrib gene expression profiles. Public microarray datasets (Oncomine repository) were queried for expression of Scrib in carcinomas versus normal tissues (A) or in neoplasia with correlated molecular analysis, as k-Ras mutation status (B) or HPV-infection presence (C). Datasets are reported using the name of the involved organ and the name of the author as it appears in Oncomine repository. (ns, not significant; Ca, Carcinoma; Ad-Ca, Adenocarcinoma; SCC, Squamous Cell Carcinoma; SCLC, Small Cell Lung Cancer; HCC, Hepatocellular Carcinoma; AA, Anaplastic Astrocytoma; HOG, Human Oligodendroglioma; GBM, Glioblastoma; DA, Diffuse Astrocytoma; PCA, Prostate cancer; KRAS-WT, k-Ras wild-type; KRAS-Mut, Mutated k-Ras; HPV, Human Papilloma Virus).
A: Scrib expression was evaluated by IHC in cervical lesions and matched normal epithelium in cases of cervical dysplasia (CIN) or invasive carcinomas (SCC) positive for Human Papilloma Virus infection. B: Representative images of cervical lesions and normal epithelium. Original magnification x400. CIN, Cervical intraepithelial neoplasia; ISC, in situ cervical carcinoma; SCC, Squamous cell carcinoma.


