Abstract
Kaposi’s Sarcoma-associated herpesvirus encodes a homolog of the human cellular interleukin-6 that may play a formative role in many KSHV-related diseases. While the viral IL-6 can signal similarly to its human counterpart little is known about the role of vIL-6 during KSHV infection. Using homologous recombination and selection in eukaryotic cells, a KSHV isolate was purified that does not express vIL-6 as was a control recombinant that left vIL-6 intact. The two viruses establish and maintain latency to similar levels in BJAB B-cells, reactivate to similar levels in B-cells and Monkey kidney cells and have very similar KSHV gene expression patterns. BJAB cells expressing KSHV survive better than the parental BJAB cells in low serum and the vIL-6 deletion does not abrogate this growth advantage. Thus vIL-6 is not essential for establishment, maintenance, or reactivation from latency in cell culture and is not involved in the survival of infected BJAB B-cells in low serum.
Keywords: KSHV, Interleukin 6, IL-6, vIL-6, Kaposi’s Sarcoma
Introduction
Cytokine dysregulation has been proposed to play a role in Kaposi’s Sarcoma (KS), primary effusion lymphomas (PELs), and plasmablastic multicentric castleman’s disease (pMCD), all Kaposi’s Sarcoma-associated herpesvirus (KSHV)-related diseases. In particular, IL-6 activation is a prominent feature of all diseases and this dysregulation has been proposed to play a major role in genesis of these proliferative disorders. Intriguingly, KSHV encodes a homolog of human IL-6, called vIL-6, that may play a role in KSHV infection and replication, in immune evasion and in tumor induction and angiogenesis (Moore et al., 1996; Neipel et al., 1997; Nicholas et al., 1997). KSHV vIL-6 has nearly 25% amino acid identity with its human counterpart and is a necessary autocrine cytokine for primary effusion lymphoma cells (Jones et al., 1999). When an antibody that blocked signaling of vIL-6 was employed, low-density PEL cells did not divide to similar levels as control treated cells. Thus vIL-6 has been shown to play a role in at least one KSHV-related B-cell malignancy (Jones et al., 1999).
The KSHV vIL-6 activates the same downstream signaling pathways as human IL-6 but it can differ from human IL-6 in its receptor usage (Hideshima et al., 2000; Molden et al., 1997; Moore et al., 1996; Osborne et al., 1999). Both human and KSHV IL-6 activate signaling through the JAK/STAT pathway, specifically activating STAT3 transcriptional activity (Molden et al., 1997; Osborne et al., 1999). To activate signaling in a cell, human IL-6 requires gp130, a surface receptor common to many cytokines, and gp80, an IL-6 specific component. vIL-6 also uses the gp130–gp80 combination but does not have the same absolute requirement for the gp80 moiety (Boulanger et al., 2004; Li et al., 2001). This makes the viral IL-6 a more promiscuous activator of signaling since gp130 is present on many cell types while gp80 is limited to specific cells (Taga and Kishimoto, 1997). In addition, expression of vIL-6 in transgenic mice led to increased vascular endothelial growth factor (VEGF) secretion and new blood vessel formation (Aoki et al., 1999), indicating a potential role of vIL-6 in angiogenesis and the development of KSHV-related disorders.
There is differential expression of vIL-6 in different diseases (Cannon et al., 1999; Moore et al., 1996; Staskus et al., 1999). In KS tumors vIL-6 is expressed predominantly in the low percentage of cells undergoing lytic replication and there is little staining in the latently infected cells indicating that vIL-6 is likely expressed predominantly during lytic replication (Molden et al., 1997; Moore et al., 1996; Parravinci et al., 1997; Staskus et al., 1999). vIL-6 is expressed in a greater proportion of infected cells in MCD lesions but also tracks with the large percentage of cells undergoing lytic replication (Brousset et al., 2001; Staskus et al., 1999). In cultured PEL cells, vIL-6 behaves as an early lytic gene that is directly induced by the KSHV lytic switch protein, Rta (Song et al., 2003; Sun et al., 1999). In uninduced PEL cells, there are low levels of expression, likely due to the low percentage of cells undergoing lytic replication. Upon induction with phorbol esters or with sodium butyrate, vIL-6 expression is strongly induced with early gene kinetics and is not inhibited by Cidofovir, an inhibitor of herpesvirus replication (Lu et al., 2004; Nicholas et al., 1997). However, one study that stained a single PEL in vivo found that a large proportion of the infected cells expressed variable levels of vIL-6, indicating the possibility that in vivo, PELs express vIL-6 to low levels during latency (Staskus et al., 1999). Of relevance to this finding, upon stimulation of BCP-1 cells, a KSHV positive, EBV negative PEL line, vIL-6 expression is activated by type I interferons in the absence of other lytic gene expression (Chatterjee et al., 2002). However, this activation was not seen in all PEL lines (Pozharskaya et al., 2004). In addition, the presence of vIL-6 messenger RNA in KSHV virions (Bechtel et al., 2005) may provide immediate expression upon viral entry that could help create a cellular environment optimal for KSHV establishment of infection. Despite a plethora of data about the signaling activities and gene expression of vIL-6, little is known about its role during KSHV infection.
In this study, a vIL-6 deletion mutant was created by homologous recombination in BCBL-1 cells and purified in Vero African green monkey kidney cells. In addition, a recombinant was created with the selection markers inserted at the same site as the deletion but with the vIL-6 gene and promoter left intact to serve as a control for the insertion of the selectable markers at that site in the genome. Since B-cells are resistant to infection in culture, we utilized our previously characterized system to examine KSHV in BJAB cells (Chen and Lagunoff, 2005). ΔvIL-6 and the control virus established latency at similar levels. vIL-6 was also found to be non-essential for KSHV reactivation and lytic replication as both viruses reactivated and produced virus to similar levels in both BJAB cells and Vero cells. The deletion of vIL-6 did not disrupt KSHV lytic or latent gene expression, indicating that vIL-6 is not significantly involved KSHV gene regulation. Furthermore, we show that KSHV imparted a survival advantage to BJAB cells grown in low serum but there was no loss of growth advantage in the ΔvIL-6 virus infected cells. Thus, vIL-6 is non-essential for KSHV maintenance or replication in cell culture.
Results
Purification of a vIL-6 deletion mutant
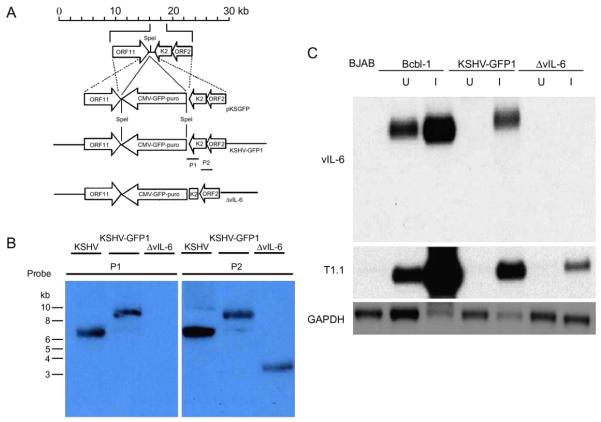
In order to determine the potential functions of viral interleukin 6 (vIL-6) in KSHV replication and pathogenesis, we created a control recombinant KSHV, KSHV-GFP1 (Chen and Lagunoff, 2005), and a vIL-6 deletion mutant (Fig. 1A). To create the vIL-6 deletion mutant, part of the open reading frame K2 gene, spanning between BC-1 positions 17089 and 17563 (Russo et al., 1996), was deleted and re-ligated with a poly-linker as described in Materials and Methods. Both KSHV-GFP1 recombinant virus and vIL-6 deletion mutants express a GFP-puromycin resistance fusion protein inserted at the same position in the KSHV genome. The GFP-puro marker was inserted into a SpeI site (BC-1 position nt 17089) between the polyadenylation signals of ORF11 and ORFK2 to avoid potential promoter interference effects. The control plasmid containing just the insertion or the ΔvIL-6 plasmid were electroporated into BCBL-1 cells and selected for puromycin resistance. The resulting BCBL-1 cell lines were induced with TPA and virus from the supernatant was used to infect Vero cells at a low multiplicity of infection to decrease the chance that multiple virus particles enter each cell. After selection, puromycin-resistant and GFP-expressing Vero cell lines were established. The resulting green Vero cells were then induced with Adeno-50 recombinant virus and sodium butyrate and the supernatant was used to infect a new batch of Vero cells at a low multiplicity of infection as previously described (Chen and Lagunoff, 2005; Vieira and O’Hearn, 2004). After at least five rounds of infection, selection, and purification, Vero cells harboring control recombinant virus, KSHV-GFP1, or vIL-6 deletion mutant, ΔvIL-6, were established. To demonstrate that vIL-6 was deleted from the viral genome, virion DNA from induced Vero cells containing the control or ΔvIL-6 virus was isolated, digested, and subjected to Southern blot analysis. As seen in Fig. 1B, probe P1 recognizing the vIL-6 region deleted recognizes a slightly slower-migrating species in the KSHV-GFP1 lane, indicative of the insertion of GFP-puro marker, but not in the ΔvIL-6 lane, indicative of the deletion of vIL-6. A slightly faster-migrating species is recognized by probe P2 that recognizes the remaining region of vIL-6 and full-length ORF2 in the ΔvIL-6 lane due to the insertion of an additional HindIII site in the poly-linker. The ΔvIL-6 virus is essentially pure while there is a miniscule amount of wild type virus seen with the control virus (Fig. 1B). To confirm the purity of the ΔvIL-6, recombinant PCR with vIL-6 primers was used. After more than 25 cycles of PCR, no wild type band could be detected in DNA from ΔvIL-6 virus while it was easily detected in wild type and the control recombinant virus. As vIL-6 is a cytokine that can act in a paracrine fashion, it is critical for the ΔvIL-6 virus to be pure.
Fig. 1.
Creation of the ΔvIL-6 recombinant virus. (A) Strategy for generation of the ΔvIL-6 recombinant virus. The structure of the first 30 kb of the KSHV genome is shown at the top. Viral genes are indicated as open boxes. Most of the vIL-6 gene was deleted as described in the Materials and methods. Genomic structure of the resulting reconstituted KSHV-GFP1 and ΔvIL-6 is shown. (B) Southern blot analysis of virion DNA isolated from the supernatants of induced BCBL-1 cells and Vero cells infected with KSHV-GFP1 or ΔvIL-6. The probes used are shown in panel A. Probe P1 corresponds to the vIL-6 region deleted (BC-1 positions 17089 to 17563). Probe P2 corresponds to the remaining genomic region of vIL-6 and full length of ORF2. (C) Northern blot analysis of uninduced and induced KSHV-GFP1 Vero cells and ΔvIL-6 Vero cells. BCBL-1 cells and Vero cells infected with KSHV-GFP1 or ΔvIL-6 recombinant viruses were induced with Adeno-50 virus and sodium butyrate. Forty-eight hours post induction, mRNA was extracted and analyzed. The Northern blot was probed with radiolabeled sequences containing viral IL-6, PAN RNA and GAPDH, respectively. U: uninduced. I: induced with Adeno-50 virus and sodium butyrate.
To demonstrate that the ΔvIL-6 virus does not express even the small portion of vIL-6 that was left in the genome, northern blot analysis was used to examine expression of vIL-6. Polyadenylated RNA was isolated from the established Vero cells and was subjected to Northern blot analysis. As seen in Fig. 1C, vIL-6 was easily detected in the Vero cells harboring control recombinant virus and BCBL-1 cells following lytic induction, but not in the Vero cells harboring vIL-6 deletion mutant. PAN RNA is strongly induced in the control and the ΔvIL-6 cells indicating that it is the lack of vIL-6 sequences not the loss of inducibility in selected cells leading to the lack of vIL-6 mRNA (Fig. 1C). The absence of T1.1 transcript in uninduced BJAB cells infected with KSHV-GFP1 or ΔvIL-6 recombinant virus, but not in uninduced BCBL-1 cells, correlates with the low spontaneous lytic reactivation rate in BJAB cells, which has been described previously (Chen and Lagunoff, 2005).
vIL-6 is not essential for establishment and maintenance of KSHV latency in BJAB cells
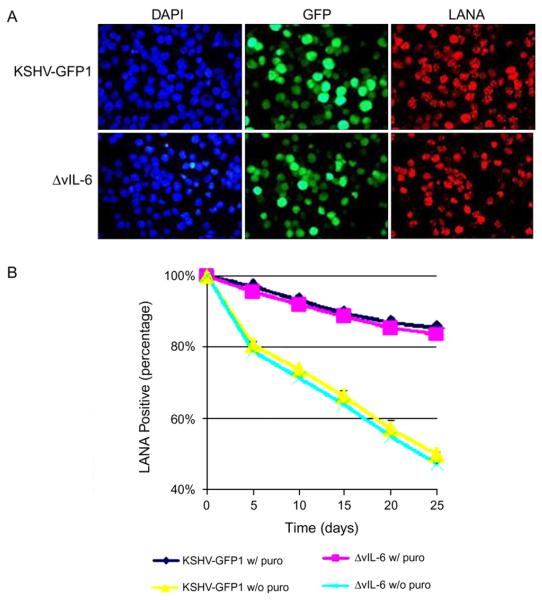
vIL-6 was shown to be an autocrine growth factor of lymphoma cells infected with KSHV (Jones et al., 1999). Therefore we wanted to examine the phenotype of the vIL-6 deletion in B-cells. However, to date B-cells in culture have been resistant to KSHV infection (Bechtel et al., 2003; Blackbourn et al., 2000; Renne et al., 1998). In a previous study, we showed that once KSHV DNA was introduced into B-cells, the virus could establish and maintain latency (Chen and Lagunoff, 2005). To determine the role of vIL-6 in KSHV replication, episomal KSHV DNA from the control recombinant or ΔvIL-6 was introduced into BJAB cells as previously described (Chen and Lagunoff, 2005). Forty-eight hours after electroporation, GFP expression was observed. After 2 weeks under selection, nearly 100% of the BJAB cells expressed GFP and reacted with the LANA antibody, showing the typical speckled pattern (Fig. 2A), which indicates that both KSHV-GFP1 and ΔvIL-6 recombinant virus established latency in an equivalent time period and a similar number of cells grew out of selection. In addition, similar numbers of LANA speckles were seen in BJAB cells containing KSHV-GFP1 or ΔvIL-6 recombinant viruses indicating that they contained similar numbers of episomal copies (L. Chen and M. Lagunoff, data not shown). Thus vIL-6 is not essential for the establishment of KSHV latency in BJAB cells.
Fig. 2.
vIL-6 is not essential for the establishment and maintenance of KSHV latency. (A) Photomicrographs of BJAB cells infected with KSHV-GFP1 or ΔvIL-6, showing 4′,6′-diamidino-2-phenylindole (DAPI) (left panel), GFP fluorescence (middle panel), and LANA red fluorescence (right panel). (B) The KSHV latent episome was gradually lost in BJAB cells. The percentage of LANA positive cells at each time-point is shown. Each value represents the average of three separate experiments at each time-point. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Similar to TIME cells and other cell types, the KSHV latent episome is lost in a significant percentage of BJAB cells over the course of one month (Chen and Lagunoff, 2005; Grundhoff and Ganem, 2004; Lagunoff et al., 2002). As seen in Fig. 2B, after 1 month in culture in the presence of puromycin selection, approximately 85% of the cells are still GFP positive and react with LANA antibody. After 1 month in culture without puromycin selection, approximately 50% of the cells remain GFP positive and express LANA. Over the time course of 1 month, the control recombinant virus and the vIL-6 deletion mutant are lost at a similar rate. Thus vIL-6 is not essential for the maintenance of KSHV latent episomes in BJAB cells. Interestingly, both control recombinant virus and vIL-6 deletion mutant are maintained in a small percentage of cells over the course of 5 months in the absence of puromycin selection (L. Chen and M. Lagunoff, data not shown).
To demonstrate that the vIL-6 deletion mutant indeed established and maintained latency in BJAB cells and did not integrate into the host genome, cell samples infected with control virus and ΔvIL-6 were subjected to Gardella gel analysis as previously described (Chen and Lagunoff, 2005). As seen in Fig. 3, in uninduced BJAB cells harboring control recombinant virus and BJAB cells harboring ΔvIL-6, the predominant species that reacts with KSHV sequences migrates similarly to episomal virus from BCBL-1 cells and much slower than KSHV virion DNA. Thus both control recombinant virus and ΔvIL-6 are maintained in an episomal form in BJAB cells. Due to a low percentage of spontaneously lytic reactivation, significant amounts of linear KSHV DNA are detected in uninduced BCBL-1 cells. There is no detectable linear or circular KSHV DNA in uninfected BJAB cells (Fig. 3). This has been repeated at different occasions with cells of different passage numbers and similar results were observed.
Fig. 3.
KSHV is episomal in ΔvIL-6 cells. BCBL-1, KSHV-GFP1-BJAB, and ΔvIL-6-BJAB cells were induced with Adeno-50 (Ad50) and 2 mM sodium butyrate (NaB). Forty-eight hours after induction, uninduced and induced cells were examined by Gardella gel electrophoresis to detect KSHV episomes. KSHV virion was used as a control. 32P-labeled KSHV genomic region spanning ORFK2 and ORF2 was used as a probe. U: uninduced. I: induced with Adeno-50 virus and sodium butyrate.
vIL-6 is not essential for KSHV lytic reactivation from latency in BJAB cells
The Control recombinant virus was previously shown to be able to reactivate in BJAB cells (Chen and Lagunoff, 2005). To determine if vIL-6 is involved in KSHV lytic reactivation from latency in BJAB cells, BJAB cells harboring control recombinant virus and BJAB cells harboring ΔvIL-6 were induced and subjected to Gardella gel analysis. Similar amounts of linear KSHV DNA are detected in induced BJAB cells harboring control recombinant virus and in BJAB cells harboring ΔvIL-6 (Fig. 3). The low amount of linear KSHV DNA detected corresponds to the low percentage of lytic reactivation in BJAB cells. To accurately measure and compare the percentage of lytic reactivation in BJAB cells, BJAB cells harboring control recombinant virus or ΔvIL-6 were induced with Adeno-50 virus and sodium butyrate and subjected to flow cytometric analysis as previously described (Lagunoff et al., 2001). Recombinant KSHV containing BJAB cells were induced for 24 h and the number of cells expressing ORF 59, a marker of lytic replication, was determined by flow cytometry. As seen in Fig. 4, we performed the experiment with 3 separate cell lines created on 3 different occasions for the control virus, KSHV-GFP1 and for ΔvIL-6. The average rate of lytic induction for the KSHV-GFP1 containing cell lines was 1.36%, while in the ΔvIL-6 BJAB cells, it was 1.98% indicating no significant difference in reactivation (Fig. 4).
Fig. 4.
Evaluation of Lytic reactivation of KSHV-GFP1 and ΔvIL-6 recombinant KSHV in BJAB cells. BJAB cells containing KSHV-GFP1 or ΔvIL-6 were induced with Adeno-50 recombinant virus and 3 mM sodium butyrate. Cells were fixed and stained for flow cytometry 24 h after induction. The top panel lists the percentage of live cells expressing ORF 59. The bottom panel shows an example of the primary flow cytometry data. The x axis is set for GFP detection, while the y axis is set for ORF59 antibody conjugated to phycoerythrin. The gate shown was based on negative and positive controls, and applied to all samples.
To further demonstrate that the ΔvIL-6 virus can be induced to undergo lytic replication cycle, Vero cells harboring control recombinant virus or ΔvIL-6 were induced with Adeno-50 recombinant virus and sodium butyrate. Forty-eight hours post induction, immunofluorescence assay was performed with the ORF59 antibody. As seen in Fig. 5, no significant difference in lytic reactivation rates was observed between control recombinant virus and ΔvIL-6. The percentage of ORF59 positive cells, indicative of lytic KSHV replication, was 10.8% and 8.5%, respectively. To demonstrate that ΔvIL-6 can complete the entire lytic cycle and produce infectious virus, infection assays were done with induced supernatants. Seventy-two hours after induction of KSHV-GFP1 and ΔvIL-6 Vero cells, the supernatant of each sample was harvested and was used to infect a fresh batch of Vero cells. Viral infection rates were determined by counting the number of cells expressing GFP, indicative of recombinant virus infection. No GFP expression is detected when virus harvested from uninduced KSHV-GFP1 Vero cells or ΔvIL-6 Vero cells is used as the inoculum. Virus harvested from supernatants of KSHV-GFP1 Vero cells induced with Adeno-50 virus and sodium butyrate yields on average 3200 GFP-positive cells per 25 × 106 cells induced, while virus isolated from supernatants of ΔvIL-6 Vero cells induced with same reagents yields on average 2840 GFP-expressing cells per 25 × 106 cells induced (numbers are the average of three separate experiments).
Fig. 5.
Photomicrographs of uninduced and induced KSHV-GFP1 Vero cells and ΔvIL-6 Vero cells. Top panels shows nuclear staining (4′,6′-diamidino-2-phenylindole or DAPI), and lower panels show ORF59 fluorescence in red. Vero cells (leftmost panels) were included as negative control. Quantification is described in the text. U: uninduced. I: induced with Adeno-50 virus and sodium butyrate. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Deletion of vIL-6 does not disrupt KSHV gene expression
To determine if the deletion of vIL-6 disrupts overall viral gene expression, a viral DNA array was established to measure the expression level of representative latent, immediate-early, early, and late genes. The quality of the spotting of the PCR based array was first confirmed with total KSHV DNA. Polyadenylated RNA was isolated from uninduced and induced BCBL-1 cells and BJAB cells harboring control recombinant virus or ΔvIL-6. Polyadenylated RNA was then reverse transcribed, labeled with 32P, and subjected to viral DNA array assay as described in Materials and methods. As seen in Fig. 6, both control recombinant virus and ΔvIL-6 shared similar latent, immediate-early, early, and late gene expression patterns with their parental BCBL-1 virus. The expression level of major latent transcripts including LANA, viral FLIP, and viral cyclin is not strongly increased upon induction while the Kaposin transcript is greatly increased (Fig. 6A). Lytic gene expression is greatly increased following TPA or Adeno-50 virus treatment of all cell types (Figs. 6B–D). In comparison to that in BCBL-1 cells, the lower level of immediate-early, early, and late viral gene expression in BJAB cells harboring KSHV-GFP1 and ΔvIL-6 is consistent with the lower lytic spontaneous and induced reactivation rate in induced BJAB cells infected with control recombinant virus or ΔvIL-6. Thus, the deletion of vIL-6 does not disrupt viral gene expression during latent or lytic replication cycles. Data shown here (Fig. 6) are representative of three experiments repeated on different occasions.
Fig. 6.
vIL-6 deletion does not disrupt KSHV gene expression patterns. KSHV membrane arrays were hybridized with oligo(dT) primed radiolabeled cDNA synthesized from BCBL-1 cells, KSHV-GFP1 BJAB cells, and ΔvIL-6 BJAB cells at 48 hpi in the absence or presence of induction. U: uninduced. I: induced with Adeno-50 virus and sodium butyrate. Major latent genes (A), immediate-early genes (B), early genes (C) and late genes (D) were shown. Data shown here are representative of three experiments repeated on different occasions.
Both control recombinant virus and wild type KSHV isolated from induced BCBL-1 cells share similar ORF11 and ORF2 gene expression patterns, which indicates that the insertion of GFP-puromycin resistance marker into the SpeI site (BC-1 position nt 17089) did not interfere with flanking viral promoter activities. This is expected as the GFP-puromycin resistance marker was inserted between the polyadenylation signals of ORF11 and ORFK2. However, the deletion of vIL-6 leads to the higher expression of ORF11 expression in uninduced ΔvIL-6 infected cells (L. Chen and M. Lagunoff, data not shown), which indicates that the vIL-6 region deleted (BC-1 positions 17089 to 17563) may be involved in the regulation of ORF11 expression in the lytic cascade. The exact function of ORF11 is still unknown; however, our data suggest that the deregulation of ORF11 expression is not critical for either latent or lytic KSHV replication.
The presence of vIL-6 does not account for the survival advantage of KSHV infected BJAB cells in low serum
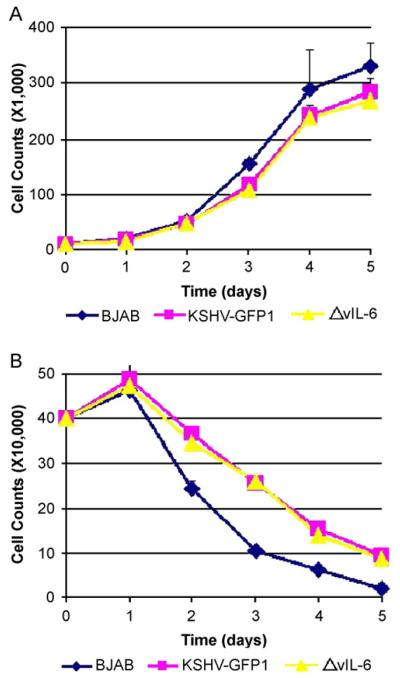
The inhibition of vIL-6 signaling by dominant-negative STAT3 expression induced apoptosis in primary effusion lymphoma cells (Aoki et al., 2003). To determine the role of vIL-6 in BJAB cell growth in the context of KSHV infection, live cells were measured by trypan-blue staining. Methyl thiazolyl tetrazolium (MTT) assay was also performed as described in Materials and methods. As seen in Fig. 7A, under normal (10%) serum conditions, BJAB cells infected with KSHV-GFP1 and BJAB cells infected with ΔvIL-6 grow at a similar rate. In the absence of puromycin selection, both KSHV-GFP1 and ΔvIL-6 infected BJAB cell lines grow at a slightly slower rate than their parental BJAB cells. No growth advantage was observed when vIL-6 was over-expressed in BJAB cells. The above data were also confirmed by MTT assays.
Fig. 7.
The presence of vIL-6 does not provide growth advantage to KSHV infected BJAB cells. Cell counts from Recombinant Infected BJAB cells growing under normal (10%) serum conditions (A) or under serum starvation (1%) conditions (B). The average number of live cells at each time-point is shown. Each value represents the average of three separate experiments at each time-point. The data were also confirmed by Methyl thiazolyl tetrazolium (MTT) assay as described in the text.
KSHV-GFP1 infected BJAB cells are more resistant to serum starvation induced apoptosis than their parental BJAB cells (Fig. 7B). However, the ΔvIL-6 infected BJAB cells maintained the same growth advantage as the control virus indicating that presence of vIL-6 does not impart the growth advantage to infected BJAB cells. Similar resistance to low serum induced apoptosis is also observed under 0.1%, 0.5%, and 2% serum conditions. Thus, a KSHV gene (or genes) other than vIL-6 is responsible for the resistance of serum starvation induced apoptosis in BJAB cells. Further investigation is ongoing to understand which viral gene is responsible for the resistance and the underlying mechanism.
Discussion
The data presented here provide the first analysis of a KSHV gene knockout created by homologous recombination in eukaryotic cells and purified by infection of eukaryotic cells. The function of many KSHV genes has been elucidated by over-expression of the viral gene in cultured cells or in mice in the absence of other KSHV gene expression. However, this method takes the gene out of the context of total KSHV gene expression and the tight regulation of gene expression exerted by herpesviruses. The classic method to determine the role of herpesviral genes during infection is to delete a single gene for the genome and analyze phenotypic changes during infection with the deletion recombinant as compared to wild type virus. While the generation of recombinant KSHV isolates has proved to be difficult, a couple of systems have recently emerged. A bacterial artificial chromosome containing the entire KSHV genome has successfully been used to make recombinant KSHV isolates (Krishnan et al., 2005; Luna et al., 2004; Xu et al., 2005; Ye et al., 2004; Zhou et al., 2002), though these viruses all contain the approximately 10 kb insertion of the BAC sequences. Recently, the Vieira lab (Vieira and O’Hearn, 2004) and we (Chen and Lagunoff, 2005) have showed that KSHV mutants could be generated by homologous recombination in PEL cell lines and subsequently be purified in Green monkey kidney (Vero) cells. While generation of recombinant viruses is possible, analysis of the recombinant viruses has proved difficult due to the lack of efficient infection systems. In particular B-cells in culture are resistant to infection with KSHV (Bechtel et al., 2003). To analyze the effects of KSHV infection on B-cells in culture, we overcame the block in infection by introducing the entire genome of a recombinant KSHV directly into BJAB cells by electroporation and selecting for cells containing KSHV containing a selectable marker. Thus we have circumvented the lack of infection in culture and can examine recombinant viruses in B-cells in a controlled fashion (Chen and Lagunoff, 2005). We used this system to provide a controlled B-cell system to determine the role of vIL-6 in the context of KSHV infection.
Many roles for vIL-6 have been proposed during KSHV infection and pathogenesis. Since the vIL-6 mRNA is present in the virion, it was proposed that it might play a role in creating an environment that promotes KSHV establishment of infection (Bechtel et al., 2005). It has also been proposed that vIL-6 signaling may play a role in viral replication or viral gene regulation. The studies presented here demonstrate that if vIL-6 plays any role in KSHV establishment of latency, reactivation, or lytic replication, it is a relatively minor one or limited to specific cell types. In addition our studies show that vIL-6 does not play a role in the regulation of KSHV transcription.
To examine the potential role of vIL-6 in cell proliferation, we grew BJAB cells infected with the control recombinant or ΔvIL-6 in low serum conditions. KSHV infection of BJAB cells provides a significant and reproducible increase in cell number at multiple time points. We used a number of different KSHV-BJAB isolates established on different occasions and reproducibly see a 2-fold increase in cell number when the cells are grown in low serum conditions. We found an identical growth curve with the BJAB cells containing the ΔvIL-6 virus indicating that vIL-6 is not essential for the KSHV induced protection of BJAB cell from apoptosis induced by serum starvation. One caveat to this set of experiments is that BJAB cells are already immortal and do not rely on the presence of KSHV for growth as PEL cells do. However, to date there is no system for analyzing these effects in primary B-cells. Our attempts to eliminate vIL-6 in PEL cells using RNAi lead to a greater than 50% decrease in vIL-6 (Chen and Lagunoff, unpublished). However, for a low abundance cytokine, even low-level expression may allow full signaling and will likely not be indicative of the function of the gene. Thus the deletion virus approach is critical. It is very intriguing that KSHV provides some growth advantage to BJAB cells in restrictive conditions. Further studies are underway to address this growth advantage in low serum provided by KSHV.
In the generation of PELs, the infected B-cells have become reliant on vIL-6 signaling. Our studies show that this reliance is not due to the role of vIL-6 in KSHV latency or replication but rather likely play a direct role in the B-cells immortal growth. The data presented here show that our BJAB cell system allows for the analysis of KSHV recombinants and that it allows the determination of the essential nature of a KSHV gene in culture as well as its effects on viral gene regulation. However, this cell culture system does not allow the functional identification of KSHV genes that play a role in immune evasion or directing angiogenesis. The information that can be gleaned from cell culture models that exist is an important first step of elucidating the role of viral genes in the context of KSHV. Further studies to determine the role of vIL-6 in altering the host cell are underway.
Materials and methods
Cells and media
BCBL-1 cells were grown as previously described (Renne et al., 1996). BJAB cells are KSHV- and EBV-negative B-cell lymphoma cells. BCBL-1 cells and BJAB cells were carried in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum, penicillin, streptomycin, glutamine, and β-mercaptoethanol. TIME cells are hTERT-immortalized dermal microvascular endothelial cells (Lagunoff et al., 2002). TIME cells were maintained in an EBM-2 medium bullet kit with supplements (Cambrex). African green monkey kidney (Vero) cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin, streptomycin, and glutamine.
Construction of recombinant KSHV
The constructs and creation of the GFP-puro control recombinant virus were described earlier (Chen and Lagunoff). For the construction of the vIL-6 deletion mutant (ΔvIL-6), pKS11K2 was digested with EcoRI and PstI to drop out 303 bases of the vIL-6 gene, and the resulting fragment was religated with an EcoRI–BamHI–HindIII–XhoI–PstI polylinker, creating pKS11ΔK2. pKSGFP and pKS11ΔK2 were then electroporated into BCBL-1 cells. Twenty-four hours post-transfection, BCBL-1 cells were selected with puromycin. After selection for 1 month, all cells expressed GFP and were puromycin resistant. Recombinant virus was harvested from these cells as previously described (Lagunoff et al., 2002). African green monkey kidney (Vero) cells were then infected with recombinant KSHV. Forty-eight hours post-KSHV infection, medium was replaced with fresh medium containing 10 μg of puromycin per milliliter. The culture medium was replaced with fresh puromycin-containing medium every day to remove dead cells until green colonies were visible. The purification of recombinant KSHV virus was done similarly to a previous description (Vieira and O’Hearn, 2004). Briefly, supernatant from KSHV-Vero cells induced with a defective Adenovirus expressing the KSHV ORF 50 gene (a kind gift of Don Ganem) and 3 mM sodium butyrate were used to infect new Vero cells at a low multiplicity of infection (MOI). The MOI was determined empirically by determining the dilution of virus stock that yielded infection of less than 1% of the cells as determined by visualization of GFP. Single GFP-expressing colonies were isolated with a cloning cylinder. Each colony was tested for the purity of recombinant virus by PCR and/or Southern analysis. Vero cells carrying a higher percentage of recombinant KSHV were induced, and supernatant was used to infect new Vero cells. Single green colonies were isolated and screened. This procedure was repeated at least five times, and KSHV-Vero and ΔvIL-6-Vero cell lines were established and used in the subsequent study.
Viruses and induction
KSHV was harvested as previously described (Lagunoff et al., 2002). For infections, the viral pellet was resuspended in the indicated medium and used to inoculate corresponding cell cultures for 2 h. The cell monolayer was then washed once and overlaid with fresh complete medium. For Vero cell inductions, the cell monolayer was infected with Adenovirus recombinant expressing KSHV ORF50 (Adeno-50) at a density of approximately 4000 particles per cell for 2 h. The Adenovirus was pre-incubated with 1 μg/ml polylysine (Sigma) in medium for 100 min at room temperature (Liang and Ganem, 2003). After infection, the cell monolayer was washed three times and overlaid with fresh complete medium containing 3 mM sodium butyrate. For BJAB cell inductions, cells were pelleted and the cell pellet was resuspended with 0.5 ml of polylysine-treated Adeno-50 virus inoculum. Cells were infected with Adeno-50 virus at a density of 4000 to 5000 particles per cell for 2 h. After infection, the cells were washed three times and resuspended in fresh complete medium containing 2 mM sodium butyrate. BCBL-1 cells were induced with phorbol 12-myristate-13-acetate (PMA) at a final concentration of 20 ng/ml.
BJAB lytic reactivation assay
To quantify the induction rate using Adeno-50 with sodium butyrate, three different KSHV-GFP1-BJAB and ΔvIL-6-BJAB isolates were performed and the rate of reactivation was quantified by flow cytometry analysis for ORF 59 as described before (Lagunoff et al., 2001). KSHV-GFP1-BJAB and ΔvIL-6-BJAB cells (2 × 107) growing to log phase were induced with Adeno-50 virus (4000 particles/cell) and 3 mM sodium butyrate. Twenty-four hours after induction, cells were washed in phosphate-buffered saline (PBS); fixed in 1% paraformaldehyde; washed in PBS with 1% bovine serum albumin, 0.02% saponin, and 0.1% sodium azide; incubated with mouse monoclonal antibody to ORF59 in the PBS-saponin buffer; washed; incubated with secondary goat anti-mouse phycoerythrin antibody; washed; and run on an influx flow cytometer. Gating on live cells, one laser filter was set to measure GFP while the other was set for phycoerythrin. BJAB and BCBL-1 cells were used as negative and positive controls respectively.
Episomal DNA preparation and electroporation into BJAB cells
KSHV episomes were isolated from KSHV-Vero and ΔvIL-6-Vero cells (5 × 106) by an alkaline lysis procedure as previously described (Chen and Lagunoff, 2005; Simpson and Huxley, 1996). KSHV episomal DNA was then transfected into 2×107 BJAB cells via electroporation (Bio-Rad Gene Pulser II; 210 V, 960 μF). Transfected cells were resuspended in 20 ml of culture medium. At 2 days post-transfection, medium was replaced with fresh medium containing 10 μg of puromycin per milliliter. Half of the culture medium was replaced with fresh puromycin-containing medium every 5 days.
Immunofluorescence assay
Immunofluorescence assays were done as previously described (Chen and Lagunoff, 2005; Lagunoff et al., 2002). The primary antibodies used were an anti-LANA peptide rabbit polyclonal antiserum (a kind gift from A. Polson and D. Ganem) and mouse monoclonal antibodies that recognize ORF 59 (Advanced Biotechnologies Inc.). The primary antibodies were diluted 1:1000, and secondary antibodies – anti-rabbit Alexa fluor 488 and anti-mouse Alexa fluor 594 (Molecular Probes) – were diluted 1:1000.
Gardella gel electrophoresis
The Gardella gel assay was performed as previously described (Gardella et al., 1984). Uninduced and induced BJAB cells latently infected with KSHV or vIL-6 deletion mutant were washed twice in phosphate-buffered saline. The cell pellets (1 × 106 cells) were resuspended in loading buffer containing 5% Ficoll and 40 μg/ml RNase A in Tris–borate-EDTA and then loaded onto the gels. The gels were run at 40 V for 4 h and then at 120 V for an additional 22 h in Tris–borate–EDTA at 4 °C. The gels were then dried, rehydrated in 0.5 M NaOH–0.15 M NaCl buffer, and directly probed with 32P-radiolabeled DNA corresponding to KSHV sequences.
Southern and Northern blot analyses
Virion DNA was purified from BCBL-1, KSHV-BJAB, and ΔvIL-6-BJAB cells as previously described (Chen and Lagunoff, 2005). Isolated virion DNA was digested with HindIII and subjected to Southern blot analysis as previously describe (Chen and Lagunoff, 2005; Lagunoff and Ganem, 1997). The blot was probed with 32P-radiolabeled probe P1 and P2, respectively. Total RNA was extracted from 2.5×107 uninduced and induced BCBL-1, KSHV-BJAB, or ΔvIL-6-BJAB cells with RNA-Bee RNA isolation reagents (Tel-Test, Friendswood, TX). mRNA was then extracted with an Oligotex direct mRNA kit, following the manufacturer’s instructions (QIAGEN). For Northern blot hybridization, 2 μg of mRNA from each sample was separated in a 1% agarose gel containing 18% formaldehyde and transferred to a nylon membrane, which was then hybridized with 32P-radiolabeled KSHV-specific probes.
DNA array construction and probe hybridization
KSHV DNA array was constructed and hybridized similarly to a previous description (Martinez-Guzman et al., 2003). Briefly, primers were designed to amplify 200–500 bp regions of cDNA sequence of known KSHV ORFs. Primers were chosen to amplify a sequence from the human cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene that was used as a positive control. The PCR products were confirmed by agarose gel electrophoresis and were then ethanol precipitated and resuspended in water at 100 ng/μl. Approximately 20 ng of DNA was spotted in quadruplicate on Hybond-N membrane (Amersham, Piscataway, N.J.) by using a replication system (V and P Scientific, Inc. San Diego, CA). The DNA on the arrays was denatured for 5 min (0.5 M NaOH, 1.5 M NaCl), neutralized for 5 min (0.5 M Tris, 1.5 M NaCl), and UV cross-linked to the membrane (Stratalinker). The quality of viral DNA array elements was evaluated by the hybridization of 32P-labeled KSHV-specific probes. 32P-labeled KSHV-specific probes were generated from purified virion DNA using the Amersham Rediprime™ II random prime labeling system (GE Healthcare). To determine the viral gene expression level, RNA–Bee RNA isolation reagents (Tel-Test, Friendswood, TX). Polyadenylated RNA was then extracted with an Oligotex direct mRNA kit, following the manufacturer’s instructions (QIAGEN). 500 ng polyadenylated RNA was used in each reverse transcription (RT) reaction. The labeled cDNA probe synthesis was prepared by using a Strip-EZ RT kit (Ambion, Austin, TX) with [α-32P]dCTP (Perkin-Elmer). The labeled probe was denatured at 100 °C. The prehybridization, hybridization, and wash steps were carried out as previously described (Lagunoff and Ganem, 1997).
Methyl thiazolyl tetrazolium (MTT) assay
Methyl thiazolyl tetrazolium (MTT) assay was done according to manufacture’s manual (Chemicon). Briefly, cells were seeded into 96-well plates at a concentration of 1 × 104 cells per ml. At 0, 1, 2, 3, 4, and 5 days, cell proliferation was evaluated using the MTT assay, in which 0.01 ml of MTT was added to each well and then incubated at 37 °C for 4 h. At the end of the assay, 0.1 ml color development solution was added to each well to dissolve the blue formazan reaction product. The absorbance was measured at 570 nm using a spectrophotometer. The background absorbance produced by wells containing medium only was subtracted from all wells. The results are presented as the growth of each culture at each time point relative to the initial number of cells that is presented on day 0.
Acknowledgments
The authors would like to thank Don Ganem for a kind gift of the Adenovirus expressing ORF50 and the anti-LANA antibody, and the Rabinovitch laboratory for the help with flow cytometry experiments and analysis. ML is supported by a grant (R01 CA097934-01A1) from the National Cancer Institute, a research scholar grant from the American Cancer Society, the PEW scholars program in biological sciences sponsored by the PEW Charitable Trust, and a pilot grant from the Puget Sound Partners in Global Health.
References
- Aoki Y, Jaffe ES, Chang Y, Jones K, Teruya-Feldstein J, Moore PS, Tosato G. Angiogenesis and hematopoiesis induced by Kaposi’s sarcoma-associated herpesvirus-encoded interleukin-6. Blood. 1999;93(12):4034–4043. [PubMed] [Google Scholar]
- Aoki Y, Feldman GM, Tosato G. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood. 2003;101(4):1535–1542. doi: 10.1182/blood-2002-07-2130. [DOI] [PubMed] [Google Scholar]
- Bechtel JT, Liang Y, Hvidding J, Ganem D. Host range of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J. Virol. 2003;77(11):6474–6481. doi: 10.1128/JVI.77.11.6474-6481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtel J, Grundhoff A, Ganem D. RNAs in the virion of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2005;79(16):10138–10146. doi: 10.1128/JVI.79.16.10138-10146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackbourn DJ, Lennette E, Klencke B, Moses A, Chandran B, Weinstein M, Glogau RG, Witte MH, Way DL, Kutzkey T, Herndier B, Levy JA. The restricted cellular host range of human herpesvirus 8. AIDS. 2000;14(9):1123–1133. doi: 10.1097/00002030-200006160-00009. [DOI] [PubMed] [Google Scholar]
- Boulanger MJ, Chow DC, Brevnova E, Martick M, Sandford G, Nicholas J, Garcia KC. Molecular mechanisms for viral mimicry of a human cytokine: activation of gp130 by HHV-8 interleukin-6. J. Mol. Biol. 2004;335(2):641–654. doi: 10.1016/j.jmb.2003.10.070. [DOI] [PubMed] [Google Scholar]
- Brousset P, Cesarman E, Meggetto F, Lamant L, Delsol G. Colocalization of the viral interleukin-6 with latent nuclear antigen-1 of human herpesvirus-8 in endothelial spindle cells of Kaposi’s sarcoma and lymphoid cells of multicentric Castleman’s disease. Hum. Pathol. 2001;32(1):95–100. doi: 10.1053/hupa.2001.21131. [DOI] [PubMed] [Google Scholar]
- Cannon JS, Nicholas J, Orenstein JM, Mann RB, Murray PG, Browning PJ, DiGiuseppe JA, Cesarman E, Hayward GS, Ambinder RF. Heterogeneity of viral IL-6 expression in HHV-8-associated diseases. J. Infect. Dis. 1999;180(3):824–828. doi: 10.1086/314956. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Osborne J, Bestetti G, Chang Y, Moore PS. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science. 2002;298(5597):1432–1435. doi: 10.1126/science.1074883. [DOI] [PubMed] [Google Scholar]
- Chen L, Lagunoff M. Establishment and maintenance of Kaposi’s sarcoma-associated herpesvirus latency in B cells. J. Virol. 2005;79(22):14383–14391. doi: 10.1128/JVI.79.22.14383-14391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardella T, Medveczky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J. Virol. 1984;50(1):248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Ganem D. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Invest. 2004;113(1):124–136. doi: 10.1172/JCI200417803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima T, Chauhan D, Teoh G, Raje N, Treon SP, Tai YT, Shima Y, Anderson KC. Characterization of signaling cascades triggered by human interleukin-6 versus Kaposi’s sarcoma-associated herpes virus-encoded viral interleukin 6. Clin. Cancer Res. 2000;6(3):1180–1189. [PubMed] [Google Scholar]
- Jones KD, Aoki Y, Chang Y, Moore PS, Yarchoan R, Tosato G. Involvement of interleukin-10 (IL-10) and viral IL-6 in the spontaneous growth of Kaposi’s sarcoma herpesvirus-associated infected primary effusion lymphoma cells. Blood. 1999;94(8):2871–2879. [PubMed] [Google Scholar]
- Krishnan HH, Sharma-Walia N, Zeng L, Gao SJ, Chandran B. Envelope glycoprotein gB of Kaposi’s sarcoma-associated herpesvirus is essential for egress from infected cells. J. Virol. 2005;79(17):10952–10967. doi: 10.1128/JVI.79.17.10952-10967.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi’s sarcoma-associated herpesvirus. Virology. 1997;236(1):147–154. doi: 10.1006/viro.1997.8713. [DOI] [PubMed] [Google Scholar]
- Lagunoff M, Lukac DM, Ganem D. Immunoreceptor tyrosine-based activation motif-dependent signaling by Kaposi’s sarcoma-associated herpesvirus K1 protein: effects on lytic viral replication. J. Virol. 2001;75(13):5891–5898. doi: 10.1128/JVI.75.13.5891-5898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunoff M, Bechtel J, Venetsanakos E, Roy AM, Abbey N, Herndier B, McMahon M, Ganem D. De novo infection and serial transmission of Kaposi’s sarcoma-associated herpesvirus in cultured endothelial cells. J. Virol. 2002;76(5):2440–2448. doi: 10.1128/jvi.76.5.2440-2448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang H, Nicholas J. Detection of direct binding of human herpesvirus 8-encoded interleukin-6 (vIL-6) to both gp130 and IL-6 receptor (IL-6R) and identification of amino acid residues of vIL-6 important for IL-6R-dependent and-independent signaling. J. Virol. 2001;75(7):3325–3334. doi: 10.1128/JVI.75.7.3325-3334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ganem D. Lytic but not latent infection by Kaposi’s sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc. Natl. Acad. Sci. U. S. A. 2003;100(14):8490–8495. doi: 10.1073/pnas.1432843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Suen J, Frias C, Pfeiffer R, Tsai MH, Chuang E, Zeichner SL. Dissection of the Kaposi’s sarcoma-associated herpesvirus gene expression program by using the viral DNA replication inhibitor cidofovir. J. Virol. 2004;78(24):13637–13652. doi: 10.1128/JVI.78.24.13637-13652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RE, Zhou F, Baghian A, Chouljenko V, Forghani B, Gao SJ, Kousoulas KG. Kaposi’s sarcoma-associated herpesvirus glycoprotein K8.1 is dispensable for virus entry. J. Virol. 2004;78(12):6389–6398. doi: 10.1128/JVI.78.12.6389-6398.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Guzman D, Rickabaugh T, Wu TT, Brown H, Cole S, Song MJ, Tong L, Sun R. Transcription program of murine gammaherpesvirus 68. J. Virol. 2003;77(19):10488–10503. doi: 10.1128/JVI.77.19.10488-10503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molden J, Chang Y, You Y, Moore PS, Goldsmith MA. A Kaposi’s sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J. Biol. Chem. 1997;272(31):19625–19631. doi: 10.1074/jbc.272.31.19625. [DOI] [PubMed] [Google Scholar]
- Moore PS, Boshoff C, Weiss RA, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274(5293):1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- Neipel F, Albrecht JC, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 1997;71(6):4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas J, Ruvolo VR, Burns WH, Sandford G, Wan X, Ciufo D, Hendrickson SB, Guo HG, Hayward GS, Reitz MS. Kaposi’s sarcoma-associated human herpesvirus-8 encodes homologues of macro-phage inflammatory protein-1 and interleukin-6. Nat. Med. 1997;3(3):287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- Osborne J, Moore PS, Chang Y. KSHV-encoded viral IL-6 activates multiple human IL-6 signaling pathways. Hum. Immunol. 1999;60(10):921–927. doi: 10.1016/s0198-8859(99)00083-x. [DOI] [PubMed] [Google Scholar]
- Parravinci C, Corbellino M, Paulli M, Magrini U, Lazzarino M, Moore PS, Chang Y. Expression of a virus-derived cytokine, KSHV vIL-6, in HIV-seronegative Castleman’s disease. Am. J. Pathol. 1997;151(6):1517–1522. [PMC free article] [PubMed] [Google Scholar]
- Pozharskaya VP, Weakland LL, Offermann MK. Inhibition of infectious human herpesvirus 8 production by gamma interferon and alpha interferon in BCBL-1 cells. J. Gen. Virol. 2004;85(Pt. 10):2779–2787. doi: 10.1099/vir.0.80214-0. [DOI] [PubMed] [Google Scholar]
- Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 1996;2(3):342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J. Virol. 1998;72(6):5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc. Natl. Acad. Sci. U. S. A. 1996;93(25):14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson K, Huxley C. A shuttle system for transfer of YACs between yeast and mammalian cells. Nucleic Acids Res. 1996;24(23):4693–4699. doi: 10.1093/nar/24.23.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MJ, Deng H, Sun R. Comparative study of regulation of RTA-responsive genes in Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 2003;77(17):9451–9462. doi: 10.1128/JVI.77.17.9451-9462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskus KA, Sun R, Miller G, Racz P, Jaslowski A, Metroka C, Brett-Smith H, Haase AT. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. J. Virol. 1999;73(5):4181–4187. doi: 10.1128/jvi.73.5.4181-4187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi’s sarcoma-associated herpesvirus gene expression. J. Virol. 1999;73(3):2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu. Rev. Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- Vieira J, O’Hearn PM. Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology. 2004;325(2):225–240. doi: 10.1016/j.virol.2004.03.049. [DOI] [PubMed] [Google Scholar]
- Xu Y, AuCoin DP, Huete AR, Cei SA, Hanson LJ, Pari GS. A Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 2005;79(6):3479–3487. doi: 10.1128/JVI.79.6.3479-3487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye FC, Zhou FC, Yoo SM, Xie JP, Browning PJ, Gao SJ. Disruption of Kaposi’s sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J. Virol. 2004;78(20):11121–11129. doi: 10.1128/JVI.78.20.11121-11129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Zhang YJ, Deng JH, Wang XP, Pan HY, Hettler E, Gao SJ. Efficient infection by a recombinant Kaposi’s sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 2002;76(12):6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]