Abstract
Patr-AL is an expressed, non-polymorphic MHC class I gene carried by ∼50% of chimpanzee MHC haplotypes. Comparing Patr-AL+ and Patr-AL- haplotypes showed Patr-AL defines a unique 125kb genomic block flanked by blocks containing classical Patr-A and pseudogene Patr-H. Orthologous to Patr-AL are polymorphic orangutan Popy-A and the 5′ part of human pseudogene HLA-Y, carried by ∼10% of HLA haplotypes. Thus the AL gene alternatively evolved in these closely related species to become classical, non-classical and non-functional. Although differing by 30 amino acid substitutions in the peptide-binding α1 and α2 domains, Patr-AL and HLA-A*0201 bind overlapping repertoires of peptides; the overlap being comparable to that between the A*0201 and A*0207 subtypes differing by one substitution. Patr-AL thus has the A02 supertypic peptide-binding specificity. Patr-AL and HLA-A*0201 have similar three-dimensional structures, binding peptides in similar conformation. Although comparable in size and shape, the B and F specificity pockets of Patr-AL and HLA-A*0201 differ in both their constituent residues and contacts with peptide anchors. Uniquely shared by Patr-AL, HLA-A*0201, and other members of the A02 supertype are the absence of serine at position 9 in the B pocket and the presence of tyrosine at position 116 in the F pocket. Distinguishing Patr-AL from HLA-A*02 is an unusually electropositive upper face on the α2 helix. Stimulating PBMC from Patr-AL- chimpanzees with B cells expressing Patr-AL, produced potent alloreactive CD8 T cells with specificity for Patr-AL and no crossreactivity toward other MHC class I, including HLA-A*02. PBMC from Patr-AL+ chimpanzees are tolerant of Patr-AL.
Introduction
Major histocompatibility complex (MHC) class I molecules bind intracellular peptides and transport them to the cell surface (1). There they interact with the receptors of natural killer (NK) cells (2) and CD8 T cells (3), lymphocytes of innate and adaptive immunity that defend against intracellular infection and cancer. Because of the strong and varying selection pressures from viruses and other pathogens, the family of genes encoding MHC class I molecules is fast-evolving and varies between species (4, 5). Common features of the MHC class I gene family are highly polymorphic genes encoding classical class I molecules, conserved genes encoding non-classical class I molecules, and a variety of class I pseudogenes and gene fragments (6, 7). Polymorphism provides diversity and change in the peptide-binding repertoire of MHC class I molecules, whereas conservation allows consistently valuable functions to be kept and shared by all members of a species.
Although chimpanzee and human genomes share >95% sequence similarity (8, 9), chimpanzees are less susceptible to many human diseases, including malaria, hepatitis B virus (HBV), and cancer, as well as HIV-1 (10, 11). In this context, any difference between the immune system genes of the two species becomes a potential candidate for contributing to disease resistance or susceptibility. Humans have six functional MHC class I genes of which HLA-A, B and C are highly polymorphic and HLA-E, F and G are conserved (12, 13). Chimpanzees have orthologs (Patr-A, B, C, E, F and G) of all these genes (14, 15), and their protein products have similar functional properties to their human counterparts (16, 17, 18, 19, 20, 21, 22).
Distinguishing the chimpanzee MHC is a seventh MHC class I gene, Patr-AL, with no obvious human counterpart (23, 24). In comparison to the other expressed class I genes, Patr-AL is most similar in sequence to Patr-A and HLA-A (hence the name A-like), from which it is estimated to have diverged >20 mya, long before the separation of chimpanzee and human ancestors 6-10 mya (23). Whereas the other human and chimpanzee MHC class I genes are present on all MHC haplotypes, Patr-AL is present only on ∼50% of chimpanzee MHC haplotypes (23); an even distribution suggestive of a balancing selection that maintains MHC haplotypes with and without Patr-AL. Such selection is a general feature of MHC variation (25). As a consequence of this distribution, a majority of chimpanzees have Patr-AL, but, importantly, a significant minority does not. Indeed, the chimpanzee MHC haplotype sequenced by Anzai et al (15) has Patr-A, B, C, E, F, and G, but lacks Patr-AL. Accordingly, one objective of our investigation was to define the location and environment of the Patr-AL gene in the chimpanzee MHC, thus defining the genes and genomic region that humans have lost.
In previous analysis we showed that Patr-AL exhibits modest polymorphism and in this regard resembles the non-classical HLA-E, F, and G genes (23). From a functional perspective, however, Patr-AL can be considered as a gene having two balanced alleles with dramatic functional difference: one makes a functional protein, the other (gene absence) does not. Although related to MHC-A, Patr-AL differs from Patr-A and HLA-A by >40 amino-acid substitutions, including >30 in the α1 and α2 domains that form the peptide-binding site. Thus a second objective for this investigation was to determine if Patr-AL has peptide-binding function and with what specificity and structural nuance. Out third and final objective, was to determine if Patr-AL has the potential to function as a histocompatibility antigen and be recognized by T-cell receptors.
Materials and Methods
DNA sequencing and analysis
The CHORI-251 BAC library (Children's Hospital of Oakland Research Institute, Oakland, CA) was screened with a Patr-AL cDNA probe. Positive clones were end-sequenced to determine their relative location in the chimpanzee MHC and clone 639P10 was selected based on its coverage of the entire region containing Patr-AL. A shotgun library with insert sizes of 1 to 2 kb was made using the TOPO Shotgun kit (Invitrogen, Carlsbad, CA). Two thousand clones were sequenced by Sanger sequencing. Finishing was performed at the Stanford Genome Technology Center (Palo Alto, CA) using pyrosequencing on the 454 platform, as previously described (26). The sequence of the region was extended by primer-walking that generated an additional 8 kb of sequence from the overlapping BAC clone 243L17. Sequences were assembled using Staden 1.6.0 (27). The complete sequence has been deposited with GenBank, accession number HM629932. In addition, 5′ flanking sequences of Gogo-A*0401, Gogo-A*0501 and intron 3 of Gogo-A*0501 were amplified, cloned, and sequenced from the gorilla Radi; similarly, the 5′ flanking sequence of the human HLA-Y pseudogene was obtained from the B-cell line WON-M. The sequences have been deposited in GenBank, accession numbers HM629928 (Gogo-A*0501 5′ flanking sequence), HM629929 (Gogo-A*0401 5′ flanking sequence), HM629930 (HLA-Y 5′ flanking sequence), and HM629931 (intron 3 of Gogo-A*0501). The primers A210_SENSE (5′ GTGGCATGATCACCATGCACTGC) and A_X2_ANTI (5′ GAGCGCGATCCGCAGGC), annealing with the 5′ flanking region and exon 2, respectively, were used for amplification of the 5′ region and the primers G5_X3_SENSE (5′ GTGGAGTGGCTCCGCAGATA 3′) and G5_X4_ANTI (5′ CCTCATGGTCAGAGACAGCTTGG 3′) annealing to sites in exons 3 and 4 were used for the amplification of intron 3.
Large-scale sequence alignment was performed and visualized using the VISTA program (www-gsd.lbl.gov/vista/). Local alignments were performed using MAFFT (28). Phylogenetic analysis for the T, W, and K pseudogenes was performed using MEGA3 (29). Analysis of divergence time for the T, W, and K pseudogenes was performed using the MCMCtree program implemented in the Phylogenetic Analysis by Maximum Likelihood package (PAML, (30)) calibrated by fossil-based speciation time estimates for Macaca mulata (23-33 mya), Gorilla gorilla (10.5-12 mya), and Pan troglodytes (7-9 mya) (31, 32).
We assumed that the ancestor of PatrAL and HLA-Y was not fixed at the time of the human/chimpanzee divergence, because neither is fixed in the modern species and the deletions in the Patr-AL and HLA-Y haplotypes share the same breakpoints. Simulations tested if both PatrAL+ and PatrAL- haplotypes could be retained under neutrality (i.e. in the absence of selection to keep both of them in the population) from the time of the human/chimpanzee divergence until present. The simulations recorded the allele-frequency change per generation and stopped when one haplotype was lost. Forward-time population simulation was performed using simuPOP (33), assuming a generation time of 15 years, Ne=30,000 (34), random mating, and starting haplotype frequencies of 50%. The simulations were conservative; because reduction in population size, generation time, or unequal starting frequencies would increase the probability of losing one haplotype, as would selection for one haplotype.
Phylogenetic analysis of MHC-A, MHC-H and MHC-A related gene sequences MHC-A, MHC-H and MHC-A related gene sequences were aligned using MAFFT (28) and manual correction of the resulting alignments. The aligned sequences were then investigated for the presence of recombinant segments using a combination of domain-by-domain phylogenetic analyses and recombination detection methods, as implemented in the recombination detection program, RDP (35). Neighbour-Joining (NJ) analyses were conducted with MEGA4 (29) using the Tamura-Nei method with 500 replicates; windows including one to eight segments (introns and/or exons) were used. To confirm the results of the recombination analysis, phylogenetic analyses were conducting on ten datasets representing the full MHC class I gene sequence (with some overlap) with three methods: Maximum-Likelihood (ML), NJ and Maximum-Parsimony (MP). NJ analyses were performed as indicated above. PAUP*4.0b10 (36), with the tree bisection-reconnection branch swapping algorithm, was used for MP analyses with 500 replicates and a heuristic search. ML analyses were performed with RAxML7 (37) under the GTR+CAT model with 500 replicates (rapid bootstrapping).
For the full-gene analysis, the final set of sequences was obtained by excluding the recombinant sequences, the pseudogenes, as well as exon 4, encoding the α3 domain, and the non-coding regions in the 5′ and 3′ of the gene. To complement this analysis and extend the set of sequences that could be included, we analyzed a smaller gene segment beginning 300bp upstream of the ATG start codon and ending in exon 2. Phylogenetic analyses were conducted with three methods, as indicated above.
Ancestral sequence reconstructions for the peptide-binding domain were performed with the CODEML program of the PAML package (30) using the marginal reconstruction approach and the M0 model. The tree topologies used for these reconstructions were obtained using the ML approach described above.
Characterization of Patr-AL and HLA-A peptide pools
Soluble Patr-AL, HLA-A*0201, and HLA-A*0207 were secreted by transfected 721.221 cells (hereafter shortened to 221 cells) grown for 60 days in a Unisyn CP2500 bioreactor (Biovest International, Minneapolis, MN) as described (38). Soluble class I protein was affinity-purified and the peptides acid-eluted. Peptide pools were subjected to Edman sequencing on a model 492A pulsed liquid phase protein sequencer (Perkin-Elmer, Waltham, MA), with cysteine underivatized, to determine the peptide motif. Alternatively, pools were fractionated by reverse-phase HPLC, and fractions analyzed using a Q-Star QTOF mass spectrometer (PerSeptive Sciex, Foster City, CA). Sequence assignment was performed using MASCOT (Matrix Science, London, UK).
Combinatorial prediction of the overlap between the Patr-AL and HLA-A*02 peptide pools was performed assuming average 40% overlap between the Patr-AL pool and individual HLA-A*02 allotype pools, and 60% overlap between HLA-A*02 allotype pools. The binomial expansion describing the percentage of peptides unique to the Patr-AL pool converges to 1-0.4/0.6 = 33.3%, indicating 66.6% overlap between the Patr-AL pool and the collective HLA-A*02 pool.
Comparison of peptide-binding profiles
From the sequences of nonamer binding peptides, either deposited for 16 HLA-A allotypes in the SYFPETHI database (39) or obtained in this study for Patr-AL, A*0201 and A*0207, we compiled 19 allotype-specific datasets. Only peptides originating from uninfected cells and allotypes having 5 or more peptides defined were included in the analysis. For each dataset, peptide scoring matrices (representing the peptide-binding profile) were generated using the PROPHECY program of the EMBOSS package (40) [Gribskov scoring scheme, gap open and extension penalties of 500]. The matrices were built using the complete nonamer sequences, thereby giving an unbiased estimation of the residue preference at each of the nine positions. The 19 matrices were then used to score the peptides of the 19 peptide datasets, using the PROPHET program of the EMBOSS package (40). Each resulting matrix:peptide score gives a relative measure of the ability of that particular peptide to bind to the MHC allotype represented by that particular matrix. We reasoned that by comparing means of the scores obtained for each of the matrix:peptide combinations we would obtain an indication of how the peptide-binding properties of the MHC allotypes are related to each other. To compare how the 19 different scoring matrices responded to each individual peptide set, we tabulated the absolute difference (|D|) between every pair of mean scores that was obtained using that peptide set. The resulting 19 tables were condensed to a single pairwise distance table by calculating the mean |D| for each peptide:matrix combination. Using this pairwise table as input, a neighbor-joining tree was generated with the NEIGHBOR program of the PHYLIP package (41).
Expression, crystallization, and structure determination
Soluble Patr-AL was produced in E. coli and re-folded with β2-microglobulin and peptide ALDKATVLL (Anaspec, Fremont, CA), as described (42). Complexes were affinity-purified by Ni-NTA chromatography (Qiagen, Valencia, CA) using histidine-tagged heavy chain, S200 gel filtration (Amersham, Piscataway, NJ), and MonoQ ion exchange (Amersham). Protein was concentrated to 10 mg/ml and crystals grown by sitting-drop vapor diffusion at 22°C, with well solution containing 22% PEG-12000, 100 mM Tris pH 8.5, 200 mM ammonium sulfate. X-ray diffraction data were collected at the Stanford Synchotron Radiation Laboratory and processed using the HKL-3000 suite (43). The structure was determined by molecular replacement using the CCP4 package (44). HLA-A*1101 (PDB ID 1QVO) was used as the phasing model. Figures were created with the PyMOL Molecular Graphics System, Version 1.3, (Schrödinger, LLC, Portland, OR).
Structure Analysis and Comparisons
For comparison of peptide conformation, residues 1-180 of Patr-AL were aligned using PyMOL to the following structures (Protein Data Bank ID indicated in brackets): HLA-A*0201-LLFGYPVYV (1HHK), HLA-A*0201 FLWGPRALV (1QEW), HLA-A*0201 ALWGFFPVL (1B0G), HLA-A*0201 GILGFVFTL (1HHI), HLA-A*0201 TLTSCNTSV (1HHG), HLA-B*4401 (1SYV), HLA-B*4403 (1SYS), HLA-A*0101 (1W72), HLA-A*0201 (1HHJ), HLA-B*5301 (1A1O), HLA-C*0401 (1QQD), HLA-B*2705 (1A83), HLA-B*3501 (2CIK), HLA-E*0101 (1MHE), HLA-B*1501 (1XR9), HLA-Cw*0301 (1EFX), HLA-G*0101 (2D31), HLA-B*0801 (1M05), HLA-A*1101 (2HN7), HLA-A*1101 (1X7Q), HLA-A*2402 (2BCK). Root mean-square distances between Cα carbons of peptides in different structures were measured in PyMOL. For comparison of the Patr-AL isoelectric point with those of other MHC class I, human HLA allotype sequences were extracted from from IMGT HLA (13) (http://www.ebi.ac.uk/imgt/hla/). Non-human MHC class I protein sequences were obtained from GenBank: Chicken, NP_001026509; rainbow trout, BAF37937; Rhesus macaque, CAL30052; nurse shark, AAL59860; mouse, AAB18955; goose, CAJ43115; rat, XP_001075560; dog, AAR27883; wallaby, ABC17813; cattle, CAA63476; cat, NP_001041626; pig, NP_001090900; horse, NP_001075975. Isoelectric points were estimated using the EMBL calculator (http://www.embl-heidelberg.de/cgi/pi-wrapper.pl).
CTL lines
Chimpanzee peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by Ficoll gradient separation. Alloreactive CTL lines were generated by stimulating chimpanzee PBMC, with autologous feeder cells and γ-irradiated 221 cells expressing Patr-AL, in culture for 14 days. CTL produced during the MLR were used as effector cells in standard 51Cr release cell killing assays against a target panel of 221 cell transfectants, each expressing a different MHC class I allotype.
Results
Patr-AL marks a unique genomic block absent from human MHC haplotypes
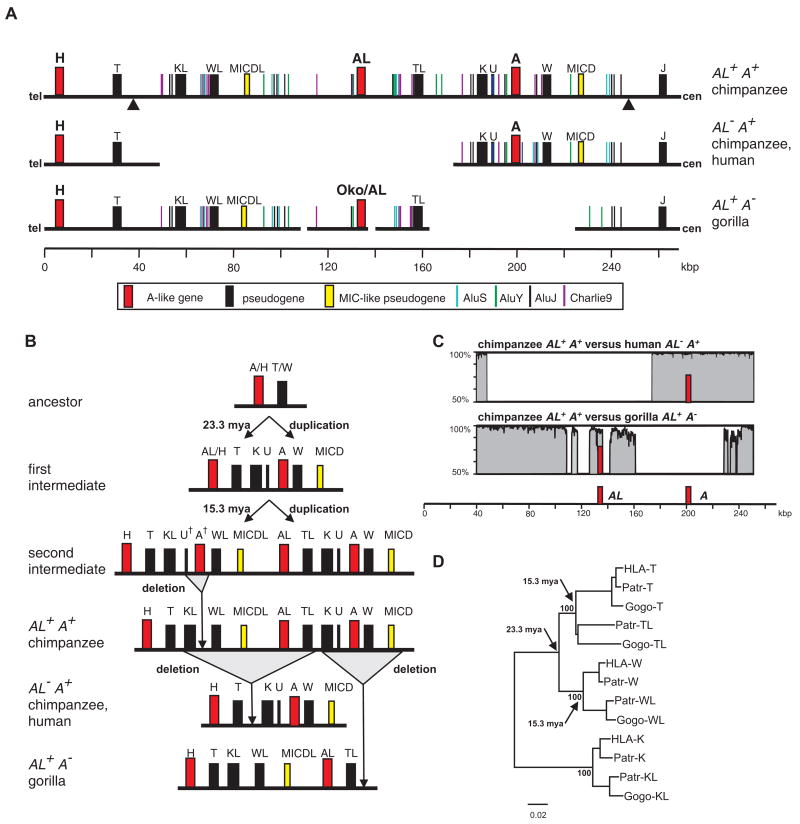
A Patr-AL cDNA probe was used to screen a BAC library made from genomic DNA of a Patr-AL+ chimpanzee. By sequencing two overlapping BAC clones we defined a 214 kb region that contained Patr-AL. In this region the telomeric 22 kb segment and the centromeric 67 kb segment corresponded to contiguous sequence in the Patr-AL- haplotype described by Anzai et al (15). In contrast, the central 125 kb region, which contains the Patr-AL gene, had no counterpart in the Patr-AL- haplotype. Eight sequenced HLA haplotypes (45, 46, 47) also lack this 125 kb genomic block and have the same breakpoints with the Patr-AL+ haplotype as the Patr-AL- haplotype (Fig. 1). The absence of MHC-AL from these human and chimpanzee MHC haplotypes almost certainly derives from one deletion event in an ancestral MHC-AL+ haplotype, which occurred prior to separation of human and chimpanzee ancestors >6 mya (31).
Figure 1. The MHC-AL+ haplotype arose through two MHC class I block duplications and one deletion, and gave rise to other hominid MHC haplotypes through deletion of either the MHC-A or MHC-AL blocks.
(A) Compares the genomic organization of the chimpanzee AL+A+ haplotype to human (GenBank ID BA000025) and chimpanzee AL- A+ haplotypes (GenBank ID BA000041), and to the gorilla AL+A- haplotype (GenBank IDs CU104658, CU104664). Genes, pseudogenes, gene fragments, and informative repetitive elements are shown. (B) Model for the evolution through duplication and deletion of MHC haplotypes containing the H, AL, and A genes. U† and A† are hypothetical paralogs that have yet to be indentified in any hominoid species or on any sequenced MHC haplotype. (C) Shows the sequence similarity between the chimpanzee AL+A+ haplotype and both the human AL- A+ (upper panel) and gorilla AL+A- (lower panel) haplotypes. A sliding window of 100 bp was analyzed. Positions of the AL and A genes are indicated. (D) Shows a neighbor-joining phylogenetic tree of the genomic segments containing the pseudogenes related to HLA-T, HLA-W, and HLA-K that was used to estimate the time of the two key duplication events. Arrows show the nodes used for maximum likelihood time estimates of the duplications.
From forward simulations under neutral evolution, we estimate that the mean time required for either loss or fixation of Patr-AL in the chimpanzee population would have been 83,300 generations (1.24 million years), a time period much shorter than that elapsing since separation of human and chimpanzee ancestors >6 mya (31). Consistent with this observation, only 15 of the 10,000 simulations retained both PatrAL+ and PatrAL- haplotypes throughout the 400,000 generations that followed separation of the human and chimpanzee lines to the present day. The simulations indicate that the probability of retaining both PatrAL+ and PatrAL- haplotypes on the chimpanzee line under neutral evolution was less than 1% (p=0.015). In conclusion, the observed presence of PatrAL+ and PatrAL- haplotypes in modern chimpanzee populations is most likely due to persistent balancing selection for both haplotype forms.
The Patr-AL genomic block is flanked by blocks containing Patr-A and Patr-H
Patr-A, the ortholog of HLA-A, lies in the centromeric 67 kb segment of the 214kb region we sequenced, at a distance of 70 kb from Patr-AL (Fig. 1A). Flanking Patr-A are several MHC class I pseudogenes and gene fragments (Patr-K and Patr-U upstream, Patr-W and MICD downstream) in identical configuration to that observed in the Patr-AL- and HLA haplotypes (Fig. 1A). Patr-AL is also flanked by pseudogenes and gene fragments, which we term Patr-K-like (Patr-KL), Patr-W-like (Patr-WL), MICD-like (MICDL), and Patr-T-like (Patr-TL), according to their similarities with human pseudogenes (48). Importantly, the organization of flanking pseudogenes and gene fragments differs between Patr-AL and Patr-A/HLA-A. Firstly, the W pseudogene is downstream from Patr-A and HLA-A, whereas WL is upstream of Patr-AL. Secondly, the TL pseudogene downstream from Patr-AL has no counterpart in the genomic block containing Patr-A. These differences show that the blocks containing Patr-A and Patr-AL are not simply products from a single duplication of a common ancestral block. Upstream of the block containing Patr-AL, lies the block containing the Patr-H pseudogene and downstream of the block containing Patr-A is the Patr-J pseudogene. Like Patr-AL, the H pseudogene (49) is related to MHC-A, as is the J pseudogene, but to lesser extent (50). That the TL pseudogene downstream of Patr-AL corresponds to the T pseudogene downstream of Patr-H and HLA-H, raised the possibility that the blocks containing Patr-H and Patr-AL were the duplicated products of a common ancestor (Fig. 1B).
Analysis of a gorilla MHC haplotype (GenBank accession numbers CU104658 and CU104664), showed it aligns with the chimpanzee and human haplotypes and has the blocks containing MHC-H and MHC-J flanking a block containing AL (Fig. 1A and 1C). Analysis of repetitive elements, as well as MHC class I pseudogenes and gene fragments, demonstrated that the block containing MHC-A is absent from the gorilla haplotype while a block syntenic to the Patr-AL block is present (Fig. 1A and 1C). Syntenic to the Patr-AL gene itself is a gene corresponding to a previously characterized cDNA, called Gogo-Oko, which has a divergent recombinant structure that puts it apart from other cDNA sequences named in the Gogo-A series (51, 52). From domain-by-domain phylogenetic analysis we now see that Gogo-Oko has segments in common with AL, H and the A2 lineage of A (Fig. 2B). That only the 5′ end of Gogo-Oko remains orthologous to Patr-AL shows the extent to which this putative gorilla equivalent of Patr-AL has been replaced by segments of H and A. That several gorilla Gogo-A are more related to HLA-A and Patr-A than Gogo-Oko (51) raises the possibility that that some gorilla MHC haplotypes have retained the block containing the A locus.
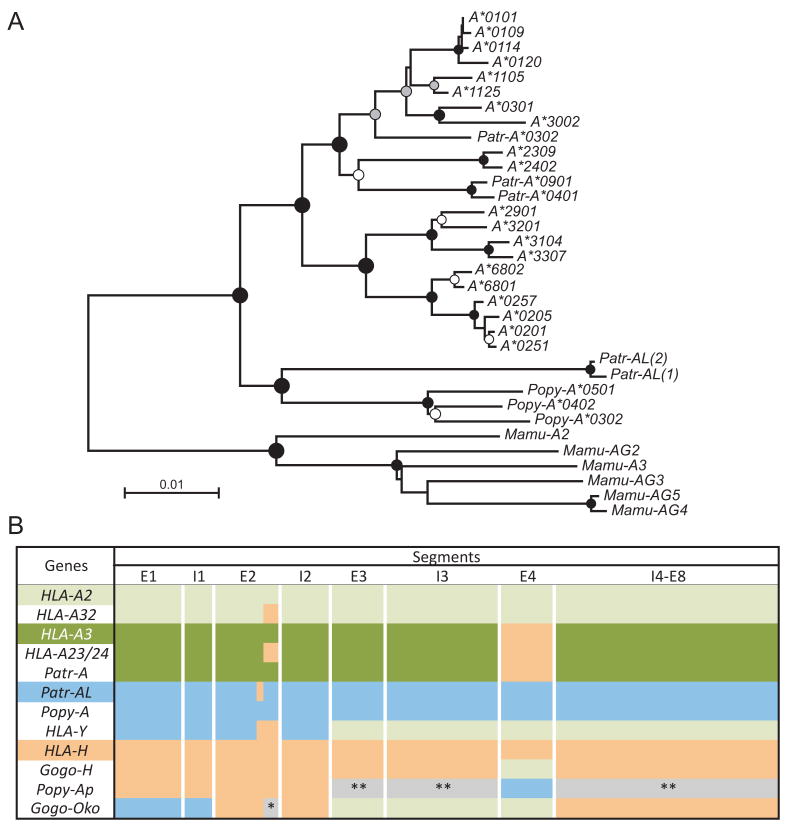
Figure 2. Patr-AL is orthologous to orangutan Popy-A and to 3′ segments of human HLA-Y and gorilla Gogo-Oko.
(A) Phylogenetic reconstruction was performed using neighbor-joining, maximum-likelihood, and maximum-parsimony methods. Shown is the neighbour-joining tree, with midpoint rooting and black, grey and white circles indicating ≥95, 80 and 60% bootstrap support, respectively, with all three methods. For the analysis exon 4, encoding the α3 domain, and the non-coding regions in the 5′ and 3′ of the gene were excluded. (B) Schematic showing the phylogenetic relationships between the MHC-A, MHC-H and MHC-AL and related MHC class I gene sequences. Equivalent segments between or within species share the same colors. These conclusions are based on the results from domain-by-domain phylogenetic analyses, and recombination detection methods (see Methods). HLA-A2 and HLA-A3 refer to the two major lineages of HLA-A alleles. *, MHC-A/AL group. **, outgroup to both MHC-A and -AL. Mamu, Macaca mulatta; Popy, Pongo pygmaeus; Patr, Pan troglodytes; Gogo, Gorilla gorilla.
Evolution of duplicated blocks and haplotypes containing H, AL and A genes
In higher primates the MHC class I gene families have expanded and diversified through duplications, followed by deletions, of a building block of ∼50 kb containing one MIC gene and one MHC class I gene (53). Evidence for these processes is seen in Fig. 1A. Our results are consistent with an evolutionary model in which the modern genomic blocks containing H, AL, and A evolved from a common ancestor by two successive duplications (Fig. 1B). The initial duplication produced one block containing the common ancestor of AL and H, and a second block containing the ancestor of A and a fourth unidentified gene (A† in Fig. 1B). The second duplication produced a haplotype with four blocks: three ancestral to the modern blocks containing A, AL and H, whereas the fourth block lost the unidentified A† class I gene and its associated U† pseudogene, as part of the deletion that gave rise to the structure of the AL+ haplotype, with its unique set of genes and gene fragments upstream of MHC-AL (Fig. 1B). From this haplotype, the AL- haplotype was formed by deletion of the 125 kb block containing AL, whereas the A- haplotype arose by deletion of the ∼80 kb block containing A. Phylogenetic analysis of the hominoid T, W, and K pseudogenes is consistent with the model (Fig. 1D) and allowed us to estimate the time of the duplications using a Bayesian approach (30). The first duplication occurred 23.3 mya (95% confidence interval 15.9-30.3 mya), consistent with the earlier estimate of 26 mya for divergence of A and AL (23); the second occurred 15.3 mya (95% confidence interval 11.6-23.2 mya) (Fig. 1D).
Species-specific evolution of AL gene diversity and function
To search for counterparts of Patr-AL in other hominoid species we performed phylogenetic analyses of MHC class I gene sequences (Fig. 2). Fig. 2A shows the tree obtained with the complete full-length gene sequences. The sequences were also divided into smaller segments that were separately subjected to phylogenetic analysis and a summary of the relationships observed is given in Fig. 2B by colored shading of the segments.
Most closely related to Patr-AL is orangutan Popy-A (Fig. 2A), which is orthologous to Patr-AL throughout the gene, with the exception of a ∼10bp segment in exon 2 (encoding residues 65-67 of the α1 domain) where Patr-AL is more related to H (Fig. 2B), a likely consequence of gene conversion. Unlike Patr-AL, Popy-A is a highly polymorphic gene, and for this reason was previously considered to be orthologous to HLA-A and Patr-A (54). Since the time of the last common ancestor of chimpanzee and orangutan, the AL gene evolved differently in the two species. In orangutan it became a polymorphic classical MHC class I gene, whereas in chimpanzee it became a conserved non-classical class I gene.
Although complete Patr-AL orthologs were not identified in gorilla and human, both species have MHC class I genes with segments related to AL. Exon 1 and intron 1 of gorilla Gogo-Oko and exons 1, 2 and introns 1, 2 of a human pseudogene, HLA-Y (45, 55, 56) are orthologous to Patr-AL. In contrast the remaining exons and introns of HLA-Y appear orthologous to the A locus. Of two ancient lineages of HLA-A alleles (54, 57), HLA-Y is closer to the A2 family than the A3 family (Fig. 2B). As is the case for Patr-AL, HLA-Y is not fixed (45, 55, 56), and neither is it represented in the eight sequenced HLA haplotypes (45). Although the precise genomic location of HLA-Y remains unknown, linkage disequilibrium between HLA-Y and a subset of HLA-A demonstrates its presence in the MHC. HLA-Y has been detected on all haplotypes that have HLA-A*2901, *3001, *33, *3401, or *6802 and on most haplotypes that have HLA-A*0203, *0205, and *31 (55, 56, 58, 59). The HLA-A alleles associated with HLA-Y are more frequent in non-Caucasian populations and we estimate that ∼20% of the human population carry HLA-Y. Whereas AL sequences contribute to expressed functional genes in chimpanzee, orangutan and gorilla, in humans the AL sequences appear only present in the non-functional form of HLA-Y.
The peptide-binding specificity and repertoire of Patr-AL is like that of HLA-A*02
Patr-AL was affinity-purified from the supernatant of 221 cells secreting soluble Patr-AL (38). Edman-sequencing showed that the peptides bound to Patr-AL were predominantly nonameric peptides constrained by three anchor positions: P2 preference for leucine, smaller aliphatic residues and glutamine; P3 preference for aspartate at P3, and C-terminal preference for aliphatic residues. This motif resembles that common to several HLA-A*02 subtypes (Fig. 3A) and differs from the peptide-binding motifs of other HLA-A and Patr-A (SYFPEITHI database, (39)) (Fig. S1).
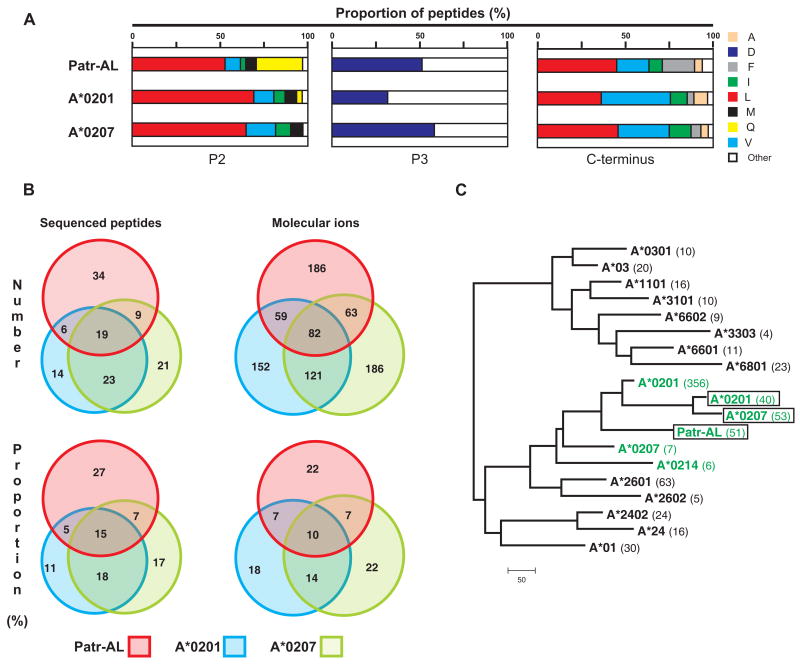
Figure 3. Extensive overlap between the repertoires of peptides bound by Patr-AL and HLA-A*02.
(A) Distribution of amino acid residues at the three anchor positions of the bound peptide: position 2 (P2), position 3 (P3), and the C-terminus. Proportions were calculated from the amino-acid sequences of 126 binding peptides for Patr-AL, HLA-A*0201 and HLA-A*0207. (B) Overlap of the peptide repertoires bound by Patr-AL, HLA-A*0201 and HLA-A*0207. The peptides binding to each MHC class I are defined by the differently colored circles: Patr-AL, red; A*0201, blue; and A*0207, green. The four overlapping regions between the circles defines the peptides bound by all three MHC class I and by the three combinations of two of them. On the left under ‘Sequenced peptides’ is shown the analysis for the peptides for which the amino-acid sequences are known. On the right under ‘Molecular ions’ is shown a second, independent study in which peptides were defined by the weight of their molecular ions. (C) Neighbor-joining phylogenetic tree to compare the peptide-binding specificities of Patr-AL, A*0201, and A*0207 as defined in our analysis (green and boxed), with those previously defined for A*0201, A*0207, A*0214 (green) and sixteen other HLA-A allotypes obtained from the SYFPEITHI database (39). The number of peptides in each dataset is shown in parentheses following the allotype name.
The pool of Patr-AL binding peptides was fractionated by reverse-phase HPLC and individual peptides were sequenced by tandem mass spectrometry. Because initial analysis identified ALDKATVLL, a known HLA-A*0207 binding peptide (60), we systematically compared the peptides eluted from Patr-AL with those obtained from HLA-A*0207, and also HLA-A*0201, the prototypical HLA-A*02. Sequencing 126 abundant peptides (Fig. S2) uncovered extensive overlap between the peptides bound by Patr-AL, HLA-A*0207 and HLA-A*0201 (Fig. 3B, left). This was pursued by examining 849 molecular ions, each randomly selected from one of the peptide profiles and then assessed for its presence in the other two profiles (Fig. 3B, right).
In magnitude, the overlap between peptides bound by Patr-AL and either A*0201 or A*0207 was comparable to that between the two A*02 subtypes (Fig. 3B, right): 52% of Patr-AL-bound peptides also bound A*0201 or A*0207; 49% of A*0207-bound peptides also bound A*0201. We estimate that 66% of the peptides bound by Patr-AL also bind to an HLA-A*02 subtype, and that 40% of the peptides bound by the entire family of A*02 subtypes can bind to Patr-AL. Conversely, review of the literature and the content of the SYFPEITHI database for MHC ligands and peptide motifs (39) showed that none of the peptides bound by Patr-AL has been found to bind to any MHC class I allotype other than HLA-A*02. Thus the peptide-binding function of chimpanzee Patr-AL is like that expected of a novel HLA-A*02 subtype. Such similarity was totally unexpected, because Patr-AL differs from A*0201 by 30 amino acid substitutions in the peptide-binding α1 and α2 domains (Fig. S3) (23), 13 of which are predicted to contact peptide (61). By contrast substitution of tyrosine for cysteine at position 99 is all that distinguishes A*0207 from A*0201.
Although the above analysis demonstrated considerable overlap between the repertoires of peptides bound by Patr-AL and HLA-A*02 and failed to find any commonality with other HLA class I, we developed an independent method to give unbiased comparison of the peptide-binding specificity of Patr-AL with a broader range of HLA-A allotypes embracing both the A2 and A3 ancient families of HLA-A alleles. We created a peptide-binding scoring matrix for each of the 16 HLA-A allotypes for which sufficient information was available in the SYFPEITHI database (39). The peptides eluted in this study from the HLA-A*0201 and A*0207 allotypes were also used to generate independent scoring matrices. Each matrix was then used to score each set of eluted peptides and pairwise comparisons of the scores used to generate a distance-tree. In this tree the Patr-AL peptide-binding specificity is clearly seen to cluster within the group of HLA-A*02 subtypes and to be apart from the other HLA-A (Fig. 3C).
Although the Patr-AL and A*02 specificities are clearly differentiated from those of other HLA-A, the peptides defined in our analysis group A*0201 and A*0207 more closely together, than is seen from analysis of the A*0201 and A*0207 binding peptides defined in other studies (Fig. 3C). This likely reflects differences in the methods used to assay and define the binding peptides. All our data came from sequence analysis of peptides eluted from HLA class I secreted from 221 cells, whereas the sequences in SYFPETHI derive from a variety of cellular and molecular methods (62, 63). For example, a study based on the binding of synthetic peptides found that the peptide-binding repertoire of A*0207 was largely limited to a subset of that bound by A*0201 (60), whereas in our analysis 30 of the 72 sequenced peptides eluted from A*0207 were not among the 49 peptides eluted from A*0201 (Fig. 3B). It is also likely that the small number of nonamer sequences in SYFPETHI representing some allotypes led to imprecision in the values for their scoring matrices.
Patr-AL and HLA-A*02 bind peptides with similar conformation
The complex of Patr-AL bound to the ALDKATVLL peptide was crystallized and a three-dimensional structure determined at 2.7Å resolution (Table I). Patr-AL has a typical MHC class I structure (Fig. 4A), in which the Cα traces of the heavy chain and β2-microglobulin superimpose with their counterparts in other HLA class I structures. Notably, the root mean square deviation (RMSD) between Cα carbons of the Patr-AL and A*0201 chains was 0.557 Å.
Table I.
Summary of crystallographic analysis.
| Data collection and processing | |
| Space group | P4322 |
| Cell dimensions | |
| a, b, c (Å) | 116.08, 116.08, 82.41 |
| α, β, γ (°) | 90, 90, 90 |
| Rmerge | 0.134 (0.694) |
| Mean I/σI | 18.2 (3.5) |
| Total number of observations | 205551 (28445) |
| Total number of unique observations | 16014 (2294) |
| Completeness | 99.9% (100%) |
| Redundancy | 12.8 (12.4) |
| Refinement | |
| Resolution limits (Å) | 50-2.70 |
| Reflection σ cutoff | I > 0 σ |
| Reflections (total / test) | 15812 / 771 |
| R / Rfree factors | 0.226 / 0.282 |
| Number of atoms | 3197 |
| RMSD bond length (Å) | 0.008 |
| RMSD bond angle (°) | 1.38 |
| Average B factor | |
| MHC heavy chain (A, 2256 atoms) | 49.27 |
| Light chain β2m (B, 833 atoms) | 47.47 |
| Peptide (C, 66 atoms) | 43.22 |
| Water (42 atoms) | 44.12 |
| PROCHECK statistics | |
| Residues in most favored regions | 86.6% |
| Residues in additional allowed regions | 12.8% |
| Residues in generously allowed regions | 0.6% |
| Residues in disallowed regions | 0.0% |
Figure 4. Despite their distinctive molecular surfaces, Patr-AL and HLA-A*0201 bind peptides in similar conformation.
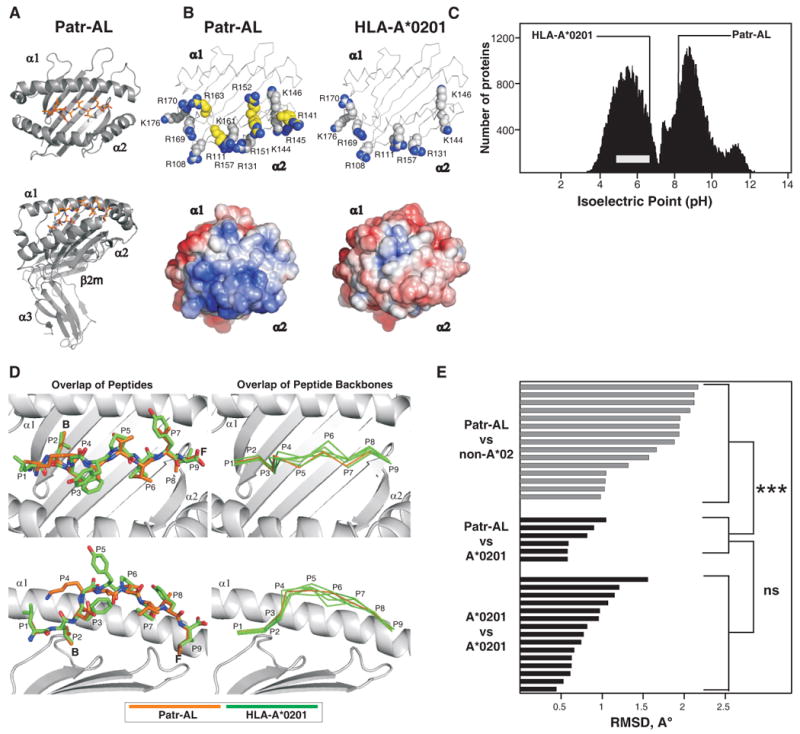
(A) Ribbon diagrams of the crystallographic structure of the complex of peptide ALDKATVLL (colored red) bound to Patr-AL. The upper diagram is a top view showing the peptide bound by the α1and α2 domains. The lower diagram is a side view showing all four extracellular Patr-AL domains.(B) Compares the distribution of electropositive residues on the top face of Patr-AL (PDB ID pending) and HLA-A*0201(PDB ID 1HHK). The upper diagrams show the position of positively-charged residues; those unique to Patr-AL colored yellow. In the lower diagrams, Poisson-Boltzmann electrostatic potentials of Patr-AL and HLA-A*0201 are projected onto solvent-exposed surfaces and colored from red (-5.0) to blue (+5.0). (C) Compares the isoelectric point of Patr-AL to those of other cellular proteins. The bimodal black-shaded distribution represents all cellular proteins (88), the grey bar shows the range of isoelectric point for MHC class I other than Patr-AL. The isoelectric points of Patr-AL and HLA-A*02 are indicated by the lines. (D) Visual comparison of the conformation of peptide bound by Patr-AL and HLA-A*020. The left panels show least-squares superimposition of Patr-AL and HLA-A*0201 peptide-binding domains, which results in close alignment of peptide Cα backbones and similar side chain orientations. The positions of peptide residues and pockets B and F are indicated. The right panels show alignment of the Patr-AL bound peptide backbone with its counterparts in six structures of different peptides bound to HLA-A*0201 (PDB IDs 1HHK, 1QEW, 1B0G, 1HHI, 1HHG, 1HHJ). (E) Quantitative comparison of the conformation of peptide bound to Patr-AL and HLA class I. RMSDs of the Cα backbone for Patr-AL bound peptides fall within the range observed for HLA-A*0201 peptides, and are significantly lower than for peptides bound to non-A*02 HLA class I. structures. The details of the allotypes and peptides compared are in supplementary Fig. S5.
Despite the common structure and peptide-binding specificity, Patr-AL is distinguished from HLA-A*02 and all other forms of MHC class I by the unusually electropositive solvent-accessible surface of its α2 helix. The α2 helix of Patr-AL has one lysine (position 161) and five arginine (positions 141, 145, 151, 152, and 163) residues in addition to the three lysines (positions 144, 146, and 176) and six arginines (positions 108, 111, 131, 157, 169, and 170) present in A*0201 (Fig. 4B). This preponderance of positive charge is such that Patr-AL is the only known MHC class I isoform with a basic isoelectric point (pI = 8.5) (Fig. 4C) and it is poorly resolved by the conditions usually used to distinguish HLA-A and B variants (64). For other MHC class I, this surface of the α2 helix binds to the Vα domain of T cell receptors (65).
The conformation of bound ALDKATVLL peptide and its interactions with the binding groove are well resolved in the Patr-AL structure (Fig. S4). The N-terminus is deeply buried in the groove, and the side chain of the P2 anchor points into the B pocket between the α1 helix and the β-sheet floor. The peptide backbone then arches up to overcome an obstruction in the peptide-binding groove caused by the bulky His70 and Tyr99 side-chains. The arch peaks at residue P4 and then slopes down into the groove, allowing the P9 anchor to engage the F pocket. This conformation is very similar to those observed for six different peptides bound to HLA-A*0201 (Fig. 4D) (PDB IDs 1QEW, 1B0G, 1HHG, 1HHI, 1HHJ, 1HHK (66, 67)).
We quantified the conformational differences between the peptides bound to Patr-AL and other class I by calculating the pair-wise RMSDs of the peptide Cα backbones. Comparison of Patr-AL with HLA-A*0201 gave RMSDs of 0.58-1.05 Å, well within the range defined by the six A*0201-binding peptides (0.44-1.56 Å). In contrast, comparison of Patr-AL with 15 other HLA class I isoforms gave RMSDs of 1.03-2.17 Å (Fig. 4E). The striking conformational similarity of peptides bound to Patr-AL and HLA-A*02 cannot be attributed solely to anchor residue preferences, because the distantly related HLA-B*0801, E*0101 and G*0101 isoforms share Patr-AL's preference for aliphatic anchors at P2 and P9, yet their bound peptides deviate by 2.07 Å, 1.88 Å, and 1.95 Å from Patr-AL's, respectively.
The specificity-determining pockets of Patr-AL and HLA-A*0201 have similar architecture despite containing non-conservative substitutions
Because the B and F specificity-determining pockets play a major role in determining which peptides bind to MHC class I (68), we compared their architecture in Patr-AL and HLA-A*0201 (Fig. 5). As a negative control we also examined HLA-B*0801, which has a non-overlapping peptide-binding repertoire with Patr-AL and A*0201.
Figure 5. Patr-AL and HLA-A*0201 (1HHK) but not HLA-B*0801 (1M05) share binding site architecture despite non-conservative changes in specificity-determining pockets B and F.
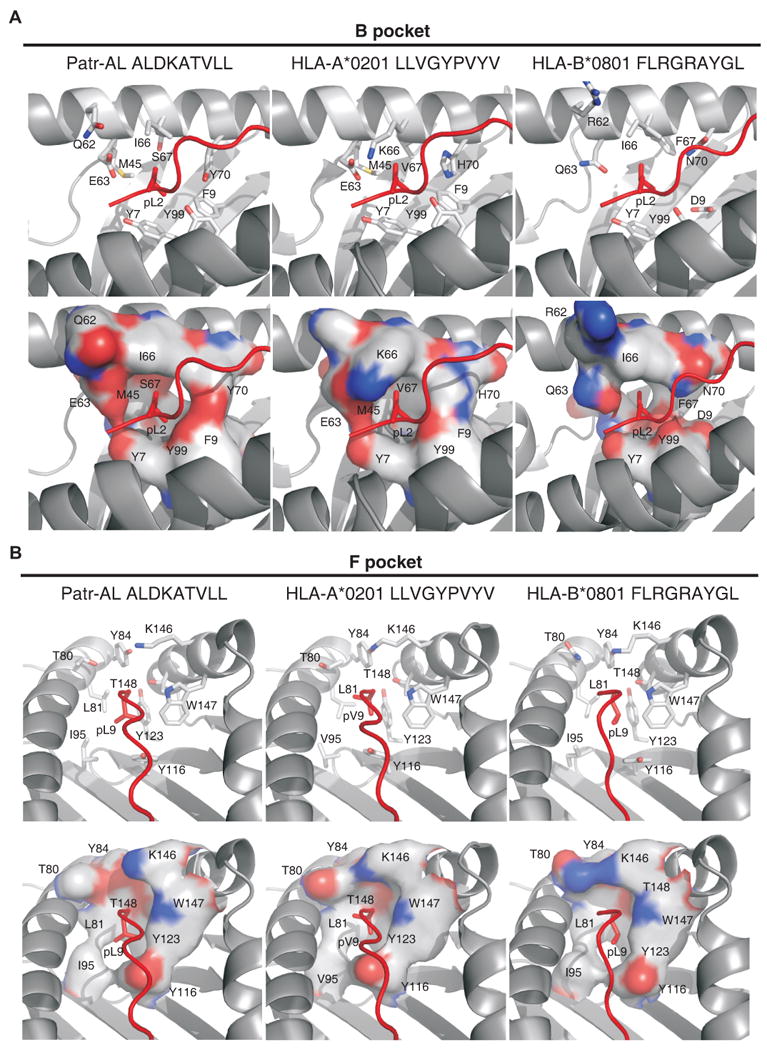
The architecture of (A) the B pocket and (B) the F pocket in the structures of Patr-AL, HLA-A*0201 (1HHK) and HLA-B*0801 (1M05) in complexes with peptide are compared. Peptide is shown in red. Top panels, stick representations of side chains forming the pockets are shown in gray; bottom panels, molecular surfaces of the pockets are shown. Residues and approximate positions of the B and F pockets are indicated.
The B pockets of Patr-AL (Fig 5A left) and A*0201 (Fig 5A center) are both deep and hydrophobic. Despite their similarities in size and shape, the two pockets differ by non-conservative substitutions at positions 66, 67 and 70. Substitution of lysine 66 in A*0201 for isoleucine in A*0201 appears functionally neutral, because the lysine side chain contributes four aliphatic carbons to the wall of the pocket while the charged ε-amino group remains solvent-accessible at the top of the groove. Serine 67 at the bottom of the Patr-AL B pocket is similar in size to valine 67 in A*0201, but its hydroxyl group is available for hydrogen bonding which could explain the preference of Patr-AL for glutamine at P2 (Fig. 3A, left). Histidine 70 in A*0201 obstructs the groove's floor, like tyrosine 70 in Patr-AL. The hydrophobic edge of the indole ring of histidine 70 faces the B pocket and preserves its hydrophobic nature. Contrasting with these similarities, the B pocket of B*0801 differs in size, shape and composition from its Patr-AL and A*0201 B counterparts (Fig 5A, right). Bulky phenylalanine 67 makes the pocket shallower, and substitution of aspartate for phenylalanine at position 9 allows tyrosine 99 to adopt a different rotamer, which disrupts the pocket wall causing the bound peptide to sink deeper into the groove.
With the exception of position 95, all the residues lining the F pocket are conserved in Patr-AL, HLA-A*0201, and HLA-B*0801 (Fig. 5B). At the bottom of the F pocket A*0201 and B*0801 have valine 95, whereas Patr-AL has the larger isoleucine, which is accommodated by a different rotamer of leucine 81. Consequently, the F pocket of Patr-AL is wider and shallower, consistent with its increased capacity to bind peptides with C-terminal phenylalanine (Fig. 3A). From this analysis we see that the specificity determining pockets of A*02 and Patr-AL accomplish the same functional effect, but in different ways, using different amino acid residues and different molecular contacts.
Evolution of the peptide-binding specificity shared by Patr-AL and HLA-A*02
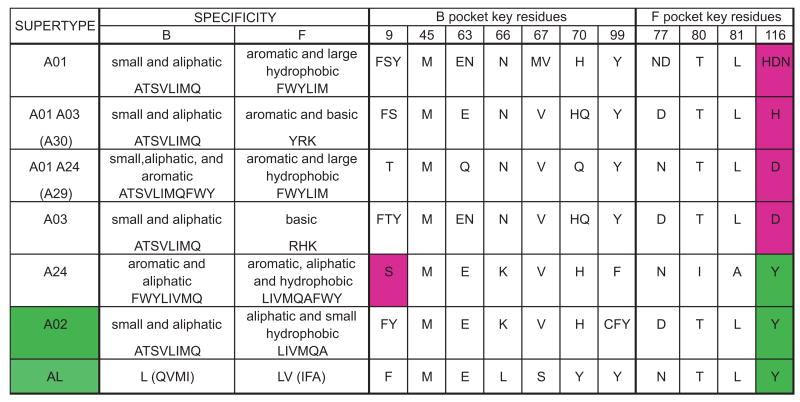
MHC class I allotypes can be clustered according to their peptide-binding specificity, as assessed by the combination of peptide anchor residues preferentially bound by the B and F pockets of the peptide-binding site; such clusters being referred to as supertypes. HLA-A allotypes have been grouped into six supertypes (A01, A02, A03, A24, A01-A03, and A01-A24) (68). The A02 supertype principally consists of A*02, and the related A*69 and some A*68 subtypes. From examining the patterns of substitution in the B and F pockets, we discovered that A02 peptide specificity correlates simply with the amino acid residues at position 9 in the B pocket and 116 in the F pocket (Fig. 6). Only allotypes of the A02 and A24 supertypes can accommodate aliphatic residues in the F-pocket, a feature correlating with presence of tyrosine at position 116. Distinguishing A24 from A02 is the capacity to bind aromatic residues in the B-pocket, which is dependent on serine at position 9. Thus, the A02 supertype is uniquely defined by the lack of serine at position 9 and the presence of tyrosine at position 116. Consistent with this definition, Patr-AL has phenylalanine at position 9 and tyrosine at position 116, as do two of three Popy-A allotypes (Fig. 6). In contrast, none of the 30 Patr-A allotypes has this motif, as is also true for five bonobo Papa-A allotypes, four Gogo-A allotypes and Gogo-Oko, and two gibbon Hyla-A allotypes (69). This correlation is only relevant in the context of the A-related genes, because many HLA-B allotypes and some HLA-C allotypes have the combination of tyrosine 116 without serine 9 (including HLA-B*0801, Fig. 5) but do not have peptide-binding specificities similar to HLA-A*02. Thus other residues that distinguish HLA-A from HLA-B and C (70), make important contributions to the A02 supertype. However, in the context of A-related genes the residues at positions 9 and 116 provide simple evolutionary switches that can introduce or take away the A02 supertype specificity.
Figure 6. The A02 supertype can be simply described by the residues at positions 9 and 116 of the peptide binding domain.
Shown are the supertypes defined by Sidney et al (68), the characteristics of the residues accommodated by the B and F pockets and the amino acids found at the key residues of these pockets for all allotypes. All allotypes assigned to a supertype were aligned and the amino acids present at the key residue positions were determined; they are shown using the single letter code. Only A02 and A24 can accommodate aliphatic residues in the F pocket, a feature correlating with the presence of tyrosine at position 116 of the peptide-binding domain (highlighted green in the figure). They differ in the residues that can be accommodated in the B pocket, with A24 able to accommodate aromatic residues, a distinction that correlates with the presence of serine at position 9 of the peptide-binding domain (highlighted purple).
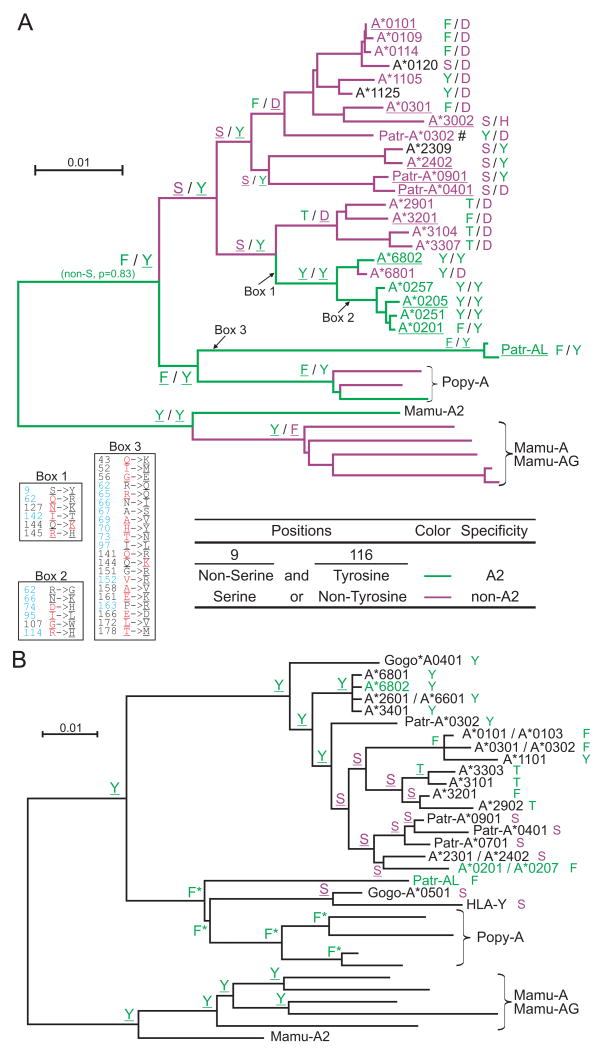
To track how the A02 specificity has evolved, we performed ancestral sequence reconstructions at positions 9 and 116. Our goal was to assess if the shared peptide binding specificity of A*02 and Patr-AL had been maintained since the time of their common ancestor or if it has periodically been lost and regained. The phylogenetic tree in Fig. 7A examines both positions 9 and 116, whereas that in Fig. 7B concentrates on position 9. The common ancestor of Patr-AL and HLA-A is predicted to have had phenylalanine 9 and tyrosine 116, the combination retained by Patr-AL. Thus we predict this ancestor had the A*02/AL peptide binding specificity. In contrast, the last common ancestor of all forms of HLA-A had the combination of serine 9 and tyrosine 116. Thus, we predict that A*02/AL binding specificity was lost during the early evolution of the HLA-A locus and then regained in the common ancestor of the A*02 and A*68 allotypes. Subsequently it was lost by the majority of A*68 allotypes but retained by A*02. Both this analysis and the structural differences in the B and F pockets indicate that the common peptide-binding specificity of Patr-AL and A*02 has a dynamic evolution in which it is periodically lost and regained through substitution at positions 9 and 116.
Figure 7. Evolution of the A2 peptide-binding specificity at the HLA-A gene.
Combination of residues at positions 9 and 116 of the MHC-A and -AL peptide binding domain correlates with the presence or absence of the A2 peptide specificity (summarized in Fig. 6). Panel A shows the results of ancestral sequence reconstruction for both positions on the phylogenetic tree shown in Fig. 2. To complement this analysis, and extend the set of sequences that could be included in the analysis, we generated a phylogenetic tree on a smaller gene segment beginning 300bp upstream of the ATG start codon and ending in exon 2 (supplemental Fig. S6) and used this tree for ancestral reconstruction of position 9 (panel B). (A) The identities at positions 9 and 116 from the ancestral sequence reconstruction are shown for thirteen nodes. The branches of the tree and the names of the sequences are colored according to peptide specificity (green for A02 and purple for non-A02): for the branches, peptide specificity is based on the residues predicted at positions 9 and 116. For contemporary allotypes, the specificities were obtained from Sidney et al. (68) (underlining indicates ‘observed’ specificities). For each sequence the residues at positions 9 and 116 are shown to the right. The amino acid changes that occurred along three branches are given in boxes: positions colored in blue represent peptide-binding sites (89), residues colored in red along the HLA-A2 branches represent residues found in Patr-AL, while residues colored in red along the Patr-AL branch represent HLA-A*0201 residues. Underlined residues have p>0.95. #, specificity is based on Patr-A*0301. (B) Identity of position 9 from the ancestral sequence reconstruction is shown for all nodes. For each sequence the residue at position 9 is shown to the right. Serine is colored purple, all others are green. Names of MHC class I with A02 peptide specificity are green. Underlined residues have p>0.95. *, non-serine residue (p=1.0).
Patr-AL is recognized by T-cell receptors and influences the T cell repertoire
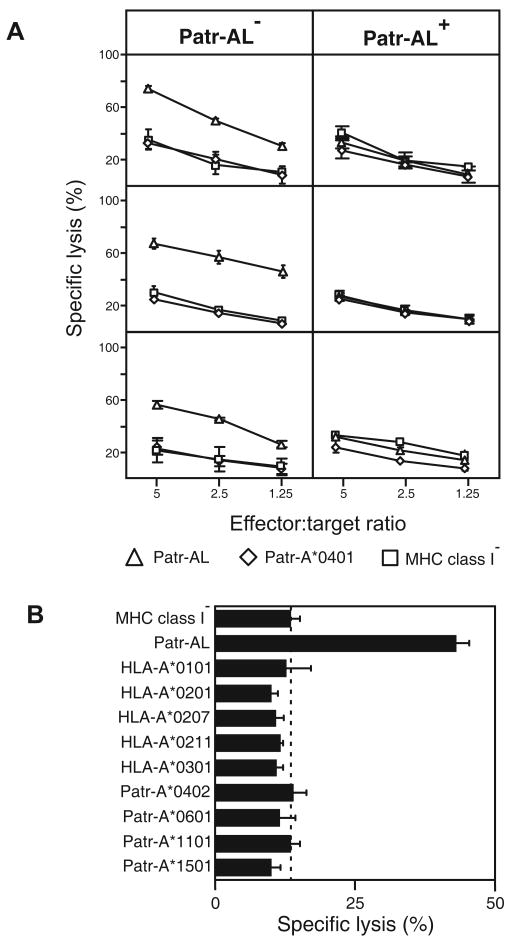
Because Patr-AL is not fixed in the chimpanzee genome (23) individual chimpanzees can either have or lack Patr-AL. To see if Patr-AL functions as a T cell alloantigen, peripheral blood mononuclear cells (PBMC) from three Patr-AL- chimpanzees were stimulated with class I-deficient 221 cells transfected with Patr-AL. Vigorous cellular proliferation yielded CD8+ CTL lines that killed 221 cells transfected with Patr-AL, but not untransfected cells or cells transfected with Patr-A (Fig. 8A, left panels). As a control, PBMC from three Patr-AL+ chimpanzees were similarly cultured with Patr-AL-expressing 221 cells. This stimulation gave less proliferation and the T cells produced did not kill Patr-AL-expressing 221 cells (Fig. 8A, right panels). This result shows that Patr-AL is recognized functionally by T cell receptors and induces self tolerance in Patr-AL+ individuals. It also implies that Patr-AL is expressed in the thymus where it participates in negative selection of the T-cell repertoire.
Figure 8. Patr-AL is an alloantigen that stimulates highly specific CD8 T cells.
(A) Stimulating PBMC from Patr-AL- chimpanzees with Patr-AL expressing transfected 221 cells led to the generation of Patr-AL specific CD8+ cytotoxic T cell lines (CTL). As shown for CTL produced from PBMC of three individual Patr-AL- chimpanzees (left panels), these T cells killed 221 target cells expressing Patr-AL but not 221 cells expressing either Patr-A*0401 or untransfected 221 cells. In contrast, when PBMC from three Patr-AL+ chimpanzees were stimulated with 221 cells expressing Patr-AL, no CTL reactive with Patr-AL were produced (right panels). Patr-AL is thus seen to be an alloantigen for Patr-AL- individuals and to generate T cell tolerance in Patr-AL+ individuals. (B) Shown are the results of cytotoxicity assays performed with Patr-AL specific CTL from one individual chimpanzee and transfected 221 target cells expressing a range of human and chimpanzee MHC-A allotypes. The CTL are exquisitely specific for Patr-AL, exhibiting no significant cross-reactivity with any of the MHC-A variants tested. Assays shown were performed with an effector:target cell ratio of 2.5. Data in A and B are shown as mean ± SD for triplicates and represent at least two independent experiments.
The CTL raised against Patr-AL were tested for their capacity to kill 221 transfected cells expressing a variety of Patr-A and HLA-A allotypes, including HLA-A*0201. The CTL were exquisitely specific for Patr-AL, showing no reactivity with 221 cells expressing any other form of MHC class I. This was not so surprising a result given the extensive sequence divergence of Patr-AL in the upper faces of the α1 and α2 helices that interact with TCR (23), particularly the uniquely electropositive face of the Patr-AL α2 helix predicted to interact with the TCRα chain (71).
Discussion
Coding-region sequences group HLA-A alleles into six families, roughly corresponding to the broad serological types, derived from two ancient lineages. The A3 lineage comprises the A9, A80, and A1/A3/A11 families, the A2 lineage comprises the A2, A10, and A19 families (54, 57, 72). Patr-A, the chimpanzee ortholog of HLA-A, has only A1/3/11 family alleles of the A3 lineage (73). This restriction reduces Patr-A diversity compared to HLA-A, whereas Patr-B and C, are more diverse than human HLA-B and C. Because chimpanzee genomes are overall more diverse than human genomes (8, 74, 75), this unusual reversal for MHC-A suggested that pathogen-mediated selection has favored preservation of A2 lineage alleles on the human line and/or their extinction on the chimpanzee line (76).
On examining non-coding sequences, notably intron 2 that separates exons 2 and 3 encoding the MHC class I peptide-binding site, de Groot et al (77, 78) detected lower diversity in Patr-A, B and C than in their human orthologs. This pointed to chimpanzees experiencing general reduction in MHC class I diversity during the selective sweep, which did not affect other gene systems. Another potential consequence of the sweep was fixation in chimpanzee of the deletion that recombined MICA with MICB to give the chimeric MICA/B gene (15, 79). The sweep was estimated to have occurred after separation of human and chimpanzee ancestors 6-9 mya, but before chimpanzee subspeciation ∼1.5 mya. de Groot et al speculated that the selective sweep was caused by a simian ancestor of HIV-1, which could explain why modern chimpanzees are more resistant to HIV-1 infection than humans (78). Circumstantial evidence suggests HIV-1 has adapted to HLA-A*02 during the current epidemic and that T cells responding to viral antigens presented by A*02 are ineffective (80, 81, 82). Even with the application of modern medicine this situation is expected to lead to reduced A*02 frequencies in human populations and the possibility of its extinction in some of them. de Groot et al proposed that comparable adaptation of pathogen to A2 lineage alleles led to their extinction in ancestral chimpanzee populations.
The peptide-binding specificities of HLA-A allotypes have been grouped into 6 supertypes: A01, A02, A03, A24, A01-A03, and A01-A24 (68). Extending the analysis to chimpanzee MHC class I identified examples of the A01, A03 and A24 supertypes but no equivalent to the A02 supertype (21, 83, 84, 85), an absence correlating with the loss of A2 lineage MHC-A alleles. Our investigation shows that chimpanzee Patr-AL, the product of a non-classical MHC class I gene situated near Patr-A in the MHC, not only has the A02 supertype but binds a peptide repertoire that overlaps extensively with that of HLA-A*02. Worldwide, HLA-A*02, the most frequent HLA-A allotype, comprises >250 subtypes, most differing from A*0201 by one amino acid substitution (http://www.ebi.ac.uk/imgt/hla/ (13)). Here we compared Patr-AL, with widespread A*0201 and A*0207. A*0207 is local to East Asian populations, where it achieves gene frequencies up to 23%, comparable to Caucasian A*0201 frequencies.
Although Patr-AL differs from A*0201 by 30 residues in the peptide-binding domains, including many that modulate HLA class I function (61, 70, 86, 87), the perturbation this has on the peptide-binding repertoire is little different from that achieved by the single substitution distinguishing A*0207 from A*0201. Key features shared by Patr-AL and A*02, and which distinguish A02 from other supertypes, are presence in the F pocket of tyrosine 116 and absence from the B pocket of serine 9. Patr-AL and HLA-A*02 have very similar three-dimensional structures and conformations of bound peptide. However, the anchoring interactions of peptide residues 2 and 9 with the B and F pockets, respectively, differ significantly in detail because of non-conservative substitutions in the residues lining these pockets in Patr-AL and HLA-A*0201.
We demonstrate that chimpanzee Patr-AL binds peptides like HLA-A*02 and is of the A02 supertype. Neither Patr-AL nor HLA-A*02 is fixed and their gene frequencies are comparably high. HLA-A*02 and Patr-AL are both alloantigens that interact with the αβ receptors of CD8 T cells and will induce T-cell tolerance when expressed as self MHC class I. Thus Patr-AL has the potential to contribute to chimpanzee immunity by presenting peptide antigens to CD8 T cells. Although Patr-AL binds many of the same peptides as HLA-A*02, Patr-AL specific T cells do not recognize HLA-A*02 and other MHC class I allotypes, which we attribute to the numerous non-conservative substitutions that distinguish Patr-AL from other MHC class I in the upward face of the class I molecule that contacts T cell receptors. Further differentiating Patr-AL from HLA-A*02 and other MHC-A, is its much reduced polymorphism, lower levels of gene and cell-surface expression, and a more restricted tissue distribution (23). In aggregate these differences argue for chimpanzee Patr-AL and human HLA-A*02 having qualitatively different functions.
Although losing the ancient A2 lineage of MHC-A alleles, the chimpanzee has not lost the peptide-binding specificity of the A02 supertype. In considering the overall reduced diversity of chimpanzee MHC class I compared to human MHC class I, Patr-AL stands out as a factor that chimpanzees have and humans lack. Clearly Patr-AL survived the selective sweep postulated by de Groot et al (77, 78), and conceivable is that its function was beneficial and has been a target for positive selection, as is consistent with our demonstration that Patr-AL has been subject to balancing selection. In this scheme of things, absence of a human equivalent of Patr-AL could make humans more susceptible to HIV/AIDS.
HLA-A*02, Patr-AL and orangutan Popy-A last shared a common ancestor 14-30 mya. We predict this ancestor had phenylalanine 9 and tyrosine 116, the combination retained by Patr-AL and one of three Popy-A allotypes (Popy-A*03) (69). Thus it is likely that functional MHC class I of the A02 supertype was maintained throughout the evolution of Patr-AL on the chimpanzee line. That is not the case for the human line, because ancestral HLA-A had serine 9 and tyrosine 116, which lacks the A02 supertype. Acquisition of phenylanine 9 by A*02 was an event specific to human evolution. Thus the A02 supertype appears to have been eliminated at some point during human evolution and then regained much later. This provides potential precedent in human history for a pathogen-mediated selective sweep that eliminated MHC-A allotypes of the A02 supertype. Since then the A02 supertype evolved anew and was driven by selection to high frequency in the modern human population. Future epidemiological studies should determine if the current epidemic of HIV/AIDS is acting to reverse that trend and reduce the frequency of HLA-A*02 and the A02 supertype.
Supplementary Material
Acknowledgments
The authors would like to thank the Yerkes Regional Primate Center for the samples of chimpanzee peripheral blood.
Footnotes
This work was supported by the following: National Institutes of Health grant AI031168 (to P.P.), and RO1-AI4840 (to K.C.G.), the Howard Hughes Medical Institute (K.C.G.), the Smith Stanford Graduate Fellowship and the Howard Hughes Medical Institute Pre-Doctoral Fellowship (to M.G.). We also acknowledge the support of the Yerkes Center Base Grant RR000165.
References
- 1.Peaper DR, Cresswell P. Regulation of MHC class I assembly and peptide binding. Annu Rev Cell Dev Biol. 2008;24:343–368. doi: 10.1146/annurev.cellbio.24.110707.175347. [DOI] [PubMed] [Google Scholar]
- 2.Cheent K, Khakoo SI. Natural killer cells: integrating diversity with function. Immunology. 2009;126:449–457. doi: 10.1111/j.1365-2567.2009.03045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenkins MK, Chu HH, McLachlan JB, Moon JJ. On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu Rev Immunol. 2010;28:275–294. doi: 10.1146/annurev-immunol-030409-101253. [DOI] [PubMed] [Google Scholar]
- 4.Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev. 2001;183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x. [DOI] [PubMed] [Google Scholar]
- 5.Shiina T, Ota M, Shimizu S, Katsuyama Y, Hashimoto N, Takasu M, Anzai T, Kulski JK, Kikkawa E, Naruse T, Kimura N, Yanagiya K, Watanabe A, Hosomichi K, Kohara S, Iwamoto C, Umehara Y, Meyer A, Wanner V, Sano K, Macquin C, Ikeo K, Tokunaga K, Gojobori T, Inoko H, Bahram S. Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics. 2006;173:1555–1570. doi: 10.1534/genetics.106.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelley J, Walter L, Trowsdale J. Comparative genomics of major histocompatibility complexes. Immunogenetics. 2005;56:683–695. doi: 10.1007/s00251-004-0717-7. [DOI] [PubMed] [Google Scholar]
- 7.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 8.Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 9.Kehrer-Sawatzki H, Cooper DN. Understanding the recent evolution of the human genome: insights from human-chimpanzee genome comparisons. Hum Mutat. 2007;28:99–130. doi: 10.1002/humu.20420. [DOI] [PubMed] [Google Scholar]
- 10.Bontrop RE, Watkins DI. MHC polymorphism: AIDS susceptibility in non-human primates. Trends Immunol. 2005;26:227–233. doi: 10.1016/j.it.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Muchmore EA. Chimpanzee models for human disease and immunobiology. Immunol Rev. 2001;183:86–93. doi: 10.1034/j.1600-065x.2001.1830107.x. [DOI] [PubMed] [Google Scholar]
- 12.Koller BH, Geraghty DE, DeMars R, Duvick L, Rich SS, Orr HT. Chromosomal organization of the human major histocompatibility complex class I gene family. J Exp Med. 1989;169:469–480. doi: 10.1084/jem.169.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams EJ, Parham P. Genomic analysis of common chimpanzee major histocompatibility complex class I genes. Immunogenetics. 2001;53:200–208. doi: 10.1007/s002510100318. [DOI] [PubMed] [Google Scholar]
- 15.Anzai T, Shiina T, Kimura N, Yanagiya K, Kohara S, Shigenari A, Yamagata T, Kulski JK, Naruse TK, Fujimori Y, Fukuzumi Y, Yamazaki M, Tashiro H, Iwamoto C, Umehara Y, Imanishi T, Meyer A, Ikeo K, Gojobori T, Bahram S, Inoko H. Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc Natl Acad Sci U S A. 2003;100:7708–7713. doi: 10.1073/pnas.1230533100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowen DG, Walker CM. Mutational escape from CD8+ T cell immunity: HCV evolution, from chimpanzees to man. J Exp Med. 2005;201:1709–1714. doi: 10.1084/jem.20050808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper S, Erickson AL, Adams EJ, Kansopon J, Weiner AJ, Chien DY, Houghton M, Parham P, Walker CM. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439–449. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 18.de Groot NG, Heijmans CM, Zoet YM, de Ru AH, Verreck FA, van Veelen PA, Drijfhout JW, Doxiadis GG, Remarque EJ, Doxiadis, van Rood JJ, Koning F, Bontrop RE. AIDS-protective HLA-B*27/B*57 and chimpanzee MHC class I molecules target analogous conserved areas of HIV-1/SIVcpz. Proc Natl Acad Sci U S A. 2010;107:15175–15180. doi: 10.1073/pnas.1009136107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG, Canavez F, Cooper SL, Valiante NM, Lanier LL, Parham P. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12:687–698. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- 20.Kowalski H, Erickson AL, Cooper S, Domena JD, Parham P, Walker CM. Patr-A and B, the orthologues of HLA-A and B, present hepatitis C virus epitopes to CD8+ cytotoxic T cells from two chronically infected chimpanzees. J Exp Med. 1996;183:1761–1775. doi: 10.1084/jem.183.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinney DM, Erickson AL, Walker CM, Thimme R, Chisari FV, Sidney J, Sette A. Identification of five different Patr class I molecules that bind HLA supertype peptides and definition of their peptide binding motifs. J Immunol. 2000;165:4414–4422. doi: 10.4049/jimmunol.165.8.4414. [DOI] [PubMed] [Google Scholar]
- 22.Mizukoshi E, Nascimbeni M, Blaustein JB, Mihalik K, Rice CM, Liang TJ, Feinstone SM, Rehermann B. Molecular and immunological significance of chimpanzee major histocompatibility complex haplotypes for hepatitis C virus immune response and vaccination studies. J Virol. 2002;76:6093–6103. doi: 10.1128/JVI.76.12.6093-6103.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams EJ, Cooper S, Parham P. A novel, nonclassical MHC class I molecule specific to the common chimpanzee. J Immunol. 2001;167:3858–3869. doi: 10.4049/jimmunol.167.7.3858. [DOI] [PubMed] [Google Scholar]
- 24.Geller R, Adams EJ, Guethlein LA, Little AM, Madrigal JA, Parham P. Linkage of Patr-AL to Patr-A and- B in the major histocompatibility complex of the common chimpanzee (Pan troglodytes) Immunogenetics. 2002;54:212–215. doi: 10.1007/s00251-002-0452-x. [DOI] [PubMed] [Google Scholar]
- 25.Solberg OD, Mack SJ, Lancaster AK, Single RM, Tsai Y, Sanchez-Mazas A, Thomson G. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum Immunol. 2008;69:443–464. doi: 10.1016/j.humimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 28.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 31.Benton MJ, Donoghue PC. Paleontological evidence to date the tree of life. Mol Biol Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- 32.Gibbons A. Paleoanthropology. Fossil teeth from Ethiopia support early, African origin for apes. Science. 2007;317:1016–1017. doi: 10.1126/science.317.5841.1016a. [DOI] [PubMed] [Google Scholar]
- 33.Peng B, Kimmel M. simuPOP: a forward-time population genetics simulation environment. Bioinformatics. 2005;21:3686–3687. doi: 10.1093/bioinformatics/bti584. [DOI] [PubMed] [Google Scholar]
- 34.Won YJ, Hey J. Divergence population genetics of chimpanzees. Mol Biol Evol. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- 35.Martin DP, Williamson C, Posada D. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- 36.Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4.0. Sinauer; Sunderland, MA: 2001. [Google Scholar]
- 37.Stamatakis A. RAxML-VI-HPC maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 38.Hickman HD, Luis AD, Buchli R, Few SR, Sathiamurthy M, VanGundy RS, Giberson CF, Hildebrand WH. Toward a definition of self: proteomic evaluation of the class I peptide repertoire. J Immunol. 2004;172:2944–2952. doi: 10.4049/jimmunol.172.5.2944. [DOI] [PubMed] [Google Scholar]
- 39.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 40.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 41.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington; Seattle: 2005. Distributed by the author. [Google Scholar]
- 42.Garboczi DN, Hung DT, Wiley DC. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci U S A. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minor W, Cymbrowki M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 44.Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, Almeida J, Forbes S, Gilbert JG, Halls K, Harrow JL, Hart E, Howe K, Jackson DK, Palmer S, Roberts AN, Sims S, Stewart CA, Traherne JA, Trevanion S, Wilming L, Rogers J, de Jong PJ, Elliott JF, Sawcer S, Todd JA, Trowsdale J, Beck S. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, Forbes S, Halls K, Howson JM, Humphray SJ, Hunt S, Mungall AJ, Osoegawa K, Palmer S, Roberts AN, Rogers J, Sims S, Wang Y, Wilming LG, Elliott JF, de Jong PJ, Sawcer S, Todd JA, Trowsdale J, Beck S. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14:1176–1187. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Traherne JA, Horton R, Roberts AN, Miretti MM, Hurles ME, Stewart CA, Ashurst JL, Atrazhev AM, Coggill P, Palmer S, Almeida J, Sims S, Wilming LG, Rogers J, de Jong PJ, Carrington M, Elliott JF, Sawcer S, Todd JA, Trowsdale J, Beck S. Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet. 2006;2:e9. doi: 10.1371/journal.pgen.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernandez-Vina M, Geraghty DE, Holdsworth R, Hurley CK, Lau M, Lee KW, Mach B, Maiers M, Mayr WR, Muller CR, Parham P, Petersdorf EW, Sasazuki T, Strominger JL, Svejgaard A, Terasaki PI, Tiercy JM, Trowsdale J. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zemmour J, Koller BH, Ennis PD, Geraghty DE, Lawlor DA, Orr HT, Parham P. HLA-AR, an inactivated antigen-presenting locus related to HLA-A. Implications for the evolution of the MHC. J Immunol. 1990;144:3619–3629. [PubMed] [Google Scholar]
- 50.Messer G, Zemmour J, Orr HT, Parham P, Weiss EH, Girdlestone J. HLA-J, a second inactivated class I HLA gene related to HLA-G and HLA-A. Implications for the evolution of the HLA-A-related genes. J Immunol. 1992;148:4043–4053. [PubMed] [Google Scholar]
- 51.Lawlor DA, Warren E, Taylor P, Parham P. Gorilla class I major histocompatibility complex alleles: comparison to human and chimpanzee class I. J Exp Med. 1991;174:1491–1509. doi: 10.1084/jem.174.6.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watkins DI, Chen ZW, Garber TL, Hughes AL, Letvin NL. Segmental exchange between MHC class I genes in a higher primate: recombination in the gorilla between the ancestor of a human non-functional gene and an A locus gene. Immunogenetics. 1991;34:185–191. doi: 10.1007/BF00205822. [DOI] [PubMed] [Google Scholar]
- 53.Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H. Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol Rev. 2002;190:95–122. doi: 10.1034/j.1600-065x.2002.19008.x. [DOI] [PubMed] [Google Scholar]
- 54.Lawlor DA, Warren E, Ward FE, Parham P. Comparison of class I MHC alleles in humans and apes. Immunol Rev. 1990;113:147–185. doi: 10.1111/j.1600-065x.1990.tb00040.x. [DOI] [PubMed] [Google Scholar]
- 55.Coquillard G, Lau M, Kletzel M, Rodriguez-Marino SG. Identification of two pseudogenes with sequence homology to human and gorilla MHC class IA genes: ancestral haplotype in the Filipino population. Hum Immunol. 2004;65:665–673. doi: 10.1016/j.humimm.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 56.Williams F, Curran MD, Middleton D. Characterisation of a novel HLA-A pseudogene, HLA-BEL, with significant sequence identity with a gorilla MHC class I gene. Tissue Antigens. 1999;54:360–369. doi: 10.1034/j.1399-0039.1999.540405.x. [DOI] [PubMed] [Google Scholar]
- 57.Domena JD, Hildebrand WH, Bias WB, Parham P. A sixth family of HLA-A alleles defined by HLA-A*8001. Tissue Antigens. 1993;42:156–159. doi: 10.1111/j.1399-0039.1993.tb02186.x. [DOI] [PubMed] [Google Scholar]
- 58.Swelsen WT, Voorter CE, van den Berg-Loonen EM. Sequence-based typing of the HLA-A10/A19 group and confirmation of a pseudogene coamplified with A*3401. Hum Immunol. 2005;66:535–542. doi: 10.1016/j.humimm.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe Y, Tokunaga K, Geraghty DE, Tadokoro K, Juji T. Large-scale comparative mapping of the MHC class I region of predominant haplotypes in Japanese. Immunogenetics. 1997;46:135–141. doi: 10.1007/s002510050252. [DOI] [PubMed] [Google Scholar]
- 60.Sidney J, del Guercio MF, Southwood S, Hermanson G, Maewal A, Appella E, Sette A. The HLA-A*0207 peptide binding repertoire is limited to a subset of the A*0201 repertoire. Hum Immunol. 1997;58:12–20. doi: 10.1016/s0198-8859(97)00206-1. [DOI] [PubMed] [Google Scholar]
- 61.Saper MA, Bjrkman PJ, Wiley DC. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J Mol Biol. 1991;219:277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- 62.Fleischhauer K, Tanzarella S, Russo V, Sensi ML, van der Bruggen P, Bordignon C, Traversari C. Functional heterogeneity of HLA-A*02 subtypes revealed by presentation of a MAGE-3-encoded peptide to cytotoxic T cell clones. J Immunol. 1997;159:2513–2521. [PubMed] [Google Scholar]
- 63.Sudo T, Kamikawaji N, Kimura A, Date Y, Savoie CJ, Nakashima H, Furuichi E, Kuhara S, Sasazuki T. Differences in MHC class I self peptide repertoires among HLA-A2 subtypes. J Immunol. 1995;155:4749–4756. [PubMed] [Google Scholar]
- 64.Yang SY, Morishima Y, Collins NH, Alton T, Pollack MS, Yunis EJ, Dupont B. Comparison of one-dimensional IEF patterns for serologically detectable HLA-A and B allotypes. Immunogenetics. 1984;19:217–231. doi: 10.1007/BF00364765. [DOI] [PubMed] [Google Scholar]
- 65.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol. 2002;38:1007–1021. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]
- 66.Madden DR, Garboczi DN, Wiley DC. The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell. 1993;75:693–708. doi: 10.1016/0092-8674(93)90490-h. [DOI] [PubMed] [Google Scholar]
- 67.Zhao R, Loftus DJ, Appella E, Collins EJ. Structural evidence of T cell xeno-reactivity in the absence of molecular mimicry. J Exp Med. 1999;189:359–370. doi: 10.1084/jem.189.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson J, Mistry K, McWilliam H, Lopez R, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic Acids Res. 2010;38:D863–869. doi: 10.1093/nar/gkp879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parham P, Lomen CE, Lawlor DA, Ways JP, Holmes N, Coppin HL, Salter RD, Wan AM, Ennis PD. Nature of polymorphism in HLA-A, -B, and -C molecules. Proc Natl Acad Sci U S A. 1988;85:4005–4009. doi: 10.1073/pnas.85.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 72.Kato K, Trapani JA, Allopenna J, Dupont B, Yang SY. Molecular analysis of the serologically defined HLA-Aw19 antigens. A genetically distinct family of HLA-A antigens comprising A29, A31, A32, and Aw33, but probably not A30. J Immunol. 1989;143:3371–3378. [PubMed] [Google Scholar]
- 73.McAdam SN, Boyson JE, Liu X, Garber TL, Hughes AL, Bontrop RE, Watkins DI. Chimpanzee MHC class I A locus alleles are related to only one of the six families of human A locus alleles. J Immunol. 1995;154:6421–6429. [PubMed] [Google Scholar]
- 74.Fischer A, Wiebe V, Paabo S, Przeworski M. Evidence for a complex demographic history of chimpanzees. Mol Biol Evol. 2004;21:799–808. doi: 10.1093/molbev/msh083. [DOI] [PubMed] [Google Scholar]
- 75.Kaessmann H, Wiebe V, Weiss G, Paabo S. Great ape DNA sequences reveal a reduced diversity and an expansion in humans. Nat Genet. 2001;27:155–156. doi: 10.1038/84773. [DOI] [PubMed] [Google Scholar]
- 76.Adams EJ, Cooper S, Thomson G, Parham P. Common chimpanzees have greater diversity than humans at two of the three highly polymorphic MHC class I genes. Immunogenetics. 2000;51:410–424. doi: 10.1007/s002510050639. [DOI] [PubMed] [Google Scholar]
- 77.de Groot NG, Heijmans CM, de Groot N, Otting N, de Vos-Rouweller AJ, Remarque EJ, Bonhomme M, Doxiadis GG, Crouau-Roy B, Bontrop RE. Pinpointing a selective sweep to the chimpanzee MHC class I region by comparative genomics. Mol Ecol. 2008;17:2074–2088. doi: 10.1111/j.1365-294X.2008.03716.x. [DOI] [PubMed] [Google Scholar]
- 78.de Groot NG, Otting N, Doxiadis GG, Balla-Jhagjhoorsingh SS, Heeney JL, van Rood JJ, Gagneux P, Bontrop RE. Evidence for an ancient selective sweep in the MHC class I gene repertoire of chimpanzees. Proc Natl Acad Sci U S A. 2002;99:11748–11753. doi: 10.1073/pnas.182420799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Groot NG, Garcia CA, Verschoor EJ, Doxiadis GG, Marsh SG, Otting N, Bontrop RE. Reduced MIC gene repertoire variation in West African chimpanzees as compared to humans. Mol Biol Evol. 2005;22:1375–1385. doi: 10.1093/molbev/msi127. [DOI] [PubMed] [Google Scholar]
- 80.Altfeld M, Allen TM, Kalife ET, Frahm N, Addo MM, Mothe BR, Rathod A, Reyor LL, Harlow J, Yu XG, Perkins B, Robinson LK, Sidney J, Alter G, Lichterfeld M, Sette A, Rosenberg ES, Goulder PJ, Brander C, Walker BD. The majority of currently circulating human immunodeficiency virus type 1 clade B viruses fail to prime cytotoxic T-lymphocyte responses against an otherwise immunodominant HLA-A2-restricted epitope: implications for vaccine design. J Virol. 2005;79:5000–5005. doi: 10.1128/JVI.79.8.5000-5005.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ferrari G, Neal W, Ottinger J, Jones AM, Edwards BH, Goepfert P, Betts MR, Koup RA, Buchbinder S, McElrath MJ, Tartaglia J, Weinhold KJ. Absence of immunodominant anti-Gag p17 (SL9) responses among Gag CTL-positive, HIV-uninfected vaccine recipients expressing the HLA-A*0201 allele. J Immunol. 2004;173:2126–2133. doi: 10.4049/jimmunol.173.3.2126. [DOI] [PubMed] [Google Scholar]
- 82.Goulder PJ, Altfeld MA, Rosenberg ES, Nguyen T, Tang Y, Eldridge RL, Addo MM, He S, Mukherjee JS, Phillips MN, Bunce M, Kalams SA, Sekaly RP, Walker BD, Brander C. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J Exp Med. 2001;193:181–194. doi: 10.1084/jem.193.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sidney J, Asabe S, Peters B, Purton KA, Chung J, Pencille TJ, Purcell R, Walker CM, Chisari FV, Sette A. Detailed characterization of the peptide binding specificity of five common Patr class I MHC molecules. Immunogenetics. 2006;58:559–570. doi: 10.1007/s00251-006-0131-4. [DOI] [PubMed] [Google Scholar]
- 85.Sidney J, Peters B, Moore C, Pencille TJ, Ngo S, Masterman KA, Asabe S, Pinilla C, Chisari FV, Sette A. Characterization of the peptide-binding specificity of the chimpanzee class I alleles A 0301 and A 0401 using a combinatorial peptide library. Immunogenetics. 2007;59:745–751. doi: 10.1007/s00251-007-0243-5. [DOI] [PubMed] [Google Scholar]
- 86.Bjorkman PJ, Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- 87.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 88.Wu S, Wan P, Li J, Li D, Zhu Y, He F. Multi-modality of pI distribution in whole proteome. Proteomics. 2006;6:449–455. doi: 10.1002/pmic.200500221. [DOI] [PubMed] [Google Scholar]
- 89.Chelvanayagam G. A roadmap for HLA-A, HLA-B, and HLA-C peptide binding specificities. Immunogenetics. 1996;45:15–26. doi: 10.1007/s002510050162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.