Abstract
Interactions between HLA class I and killer cell immunoglobulin-like receptors (KIR) diversify human NK cell responses. Dominant KIR ligands are the C1 and C2 epitopes of MHC-C, a young locus restricted to humans and great apes. C1 and C1-specific KIR evolved first, being present in orangutan and functionally like their human counterparts. Orangutans lack C2 and C2-specific KIR, but have a unique C1+C2 specific KIR that binds equally to C1 and C2. Such a receptor was likely the mechanism by which C2-KIR interaction evolved from C1-KIR while avoiding a non-functional intermediate: either orphan receptor or ligand. Orangutan inhibitory MHC-C reactive KIR pair with activating receptors of identical avidity and specificity, contrasting with the selective attenuation of human activating KIR. The orangutan C1-specific KIR reacts or cross-reacts with all four polymorphic epitopes (C1, C2, Bw4, and A3/11) recognized by human KIR, revealing their structural commonality. Saturation mutagenesis at specificity-determining position 44, demonstrates that KIR are inherently restricted to binding just these four epitopes, either individually or in combination. This restriction frees the majority of HLA-A and –B variants to be dedicated T-cell receptor ligands, not subject to conflicting pressures from the NK cell and T cell arms of the immune response.
Introduction
Characterization of alloreactive NK cell specificities led to the identification of inhibitory receptors for polymorphic HLA class I determinants (1, 2). Because of their functional similarity to the Ly49 receptors of mouse NK cells, it was commonly anticipated that the human NK cell receptors would also be Ly49 molecules (3). However, the subsequent molecular analyses demonstrated that structurally unrelated proteins, the killer cell immunoglobulin-like receptors (KIR) were the functional analogues of mouse Ly49 (4–7). This unambiguous example of convergent evolution provided a first indication of the rapid evolution of NK cell receptors and the strong, but variable, selection that acts upon NK cell function in the early phases of both the immune response and placental reproduction (8, 9).
Like HLA class I, the human KIR gene family exhibits extraordinary genetic and functional diversity, but one built upon gene content variation as well as allelic polymorphism (10, 11). Combinations of KIR and HLA class I factors distinguish individuals and populations, and correlate with a broad range of clinical conditions that includes infection, autoimmunity, transplantation, and pregnancy syndromes (12, 13). Many of these associations involve differences between the group A and B KIR haplotypes, the former containing mainly inhibitory KIR genes, the latter being enriched for activating KIR genes (14).
Human KIR recognize all HLA-C variants, but only some HLA-A and –B variants (15). Interactions between three inhibitory KIR (KIR2DL1, KIR2DL2/3, and KIR3DL1) and their respective ligands, the C1 and C2 epitopes of HLA-C and the Bw4 epitope of HLA-A and –B, influence NK cell education during development and NK cell function during an immune response (16–19). Structurally similar activating receptors pair with these inhibitory receptors, but their functions remain speculative, because of the weak (KIR2DS1) or undetectable (KIR2DS2 and KIR3DS1) binding to ligand caused by acquired mutation (20–23). A fourth inhibitory receptor (KIR3DL2), which recognizes an epitope shared by HLA-A3 and HLA-A11 (24–26), does not contribute to NK cell education (16, 19), but is paired with a structurally divergent activating receptor (KIR2DS4) that recognizes HLA-A11, as well as a subset of HLA-C allotypes (27).
To gain distance and perspective on these enigmatic properties of the human KIR we have studied the KIR genes in other mammalian species (28). In several non-primate species KIR is either absent, inactivated or represented by a single gene (29). Although the distantly related KIR3DX gene is diversified in cattle (30), expansion of the KIR3DL gene is restricted to the simian primates (monkeys, apes and human) (31), the prosimian primates having but a single non-functional KIR3DL gene (32). Whereas New World monkeys (the platyrrhines) have their own distinctive forms of KIR3DL (33), Old World monkeys, apes and human (the catarrhines) share a common set of four KIR3DL lineages defined by phylogenetic analysis (34, 35). Lineage I, comprises KIR2D with D0 and D2 domains, such as KIR2DL4 that binds MHC-G (36, 37); lineage II are KIR3DL that bind MHC-A and –B epitopes (38, 39); lineage III comprises KIR3D and KIR2D with D1 and D2 domains, including receptors specific for the C1 epitope carried by MHC-B and –C and the C2 epitope carried by MHC-C (2, 40–44); and lineage V KIR is represented by KIR3DL3, for which no ligand specificity has been determined (45). The expansion of KIR lineages correlates with presence of their cognate MHC class I KIR ligands. Thus lineage II KIR expanded in Old World monkeys (32, 33), species where MHC-A and –B are first detected. Similarly, lineage III KIR expanded in orangutan (34), the species where MHC-C is first detected (46), and continued in chimpanzee and human with accompanying reduction in the numbers of lineage II KIR (47, 48).
Whereas HLA-C is fixed in the human MHC and carries both C1 and C2 epitopes, orangutan Popy-C is not fixed, being present on ~50% of MHC haplotypes, and carrying only the C1 epitope (46). On the basis of these properties, we hypothesized that orangutan lineage III KIR and MHC-C represent an evolutionary intermediate of the human system, one having C1 and C1-specific KIR but neither C2 nor C2-specific KIR (49). In proving this point, the functional study described here discovered a novel type of C1+C2 receptor that preceded the C2 half of the system, and likely facilitated its evolution. The orangutan activating KIR provided valuable insight regarding the state of their human counterparts, and the relative plasticity of the orangutan KIR prompted further experiments in mutagenesis that revealed an inherent limitation to the recognition of HLA class I by KIRs.
Materials and Methods
KIR-Fc fusion proteins
The methods were based on those of Winter et al (50), as described by Moesta et al (44). Exons encoding the Ig-like domains and stem of orangutan and human lineage III KIR (2DL1*001 and 2DL3*001) were amplified by PCR and fused in-frame with the region encoding the Fc portion of the human IgG1 H chain. Single point mutants of KIR2DLA were made using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Wild type and mutant KIR-Ig constructs were cloned into the pACgp67A transfer vector and cotransfected with linearized baculovirus (BD Biosciences) into Sf9 cells using Cellfectin according to the manufacturer’s directions (Invitrogen). Two additional rounds of amplification were performed to obtain high-titer virus. Hi5 insect cells were infected with the high-titer virus for 72 hours and then centrifuged at 2,500g for 15 min. The supernatant was collected, filtered to remove residual particulate material, and neutralized with 10× HEPES buffered saline (pH 7.2). The supernatant was then incubated overnight at 4°C with protein A-Sepharose beads (Invitrogen). The KIR-Fc fusion protein was eluted from the protein A-Sepharose beads with sequential aliquots of 800µl 0.1 M glycine (pH 2.7) and neutralized using 200µl 1 M Tris (pH 9.0). Sephadex G-25M columns were used to desalt the protein into DPBS. At this stage KIR-Fc protein concentration was determined using the Bradford assay (BioRad). Proper folding of KIR-Fc was assessed by the binding to conformation-dependent antibodies: NKVFS1 (mouse anti-KIR; Serotech) and goat anti-human IgG (Immunotech). To do this KIR-Fc was first bound to beads coated with anti-human IgG (Bangs Laboratories) and subsequent staining with the PE-labeled conformation-dependent antibodies was measured by flow cytometry.
Binding of KIR-Fc to beads coated with HLA class I
Binding of KIR-Fc fusion proteins to a panel of 29 HLA-A, 50 HLA-B and 16 HLA-C allotypes was assessed using LABScreen single-antigen bead sets (One Lambda). These beads, originally developed for characterizing the specificity of antibodies for HLA class I (51, 52), have proved incisive in defining the MHC class I specificity of KIR (27, 43, 44). KIR-Fc fusion proteins and W6/32, a mouse monoclonal antibody which recognizes all HLA class I isoforms with similar avidity (53–55), were incubated at 100µg/ml with LABScreen microbeads for 1 hour at 4°C with shaking. Beads were washed four times and then incubated with either anti-human Fc-PE for KIR-Fc or anti-mouse IgG-PE for W6/32. The fluorescent signal intensity of the attached secondary antibody and the unique fluorescent level for each HLA bead were read on a Luminex 100 instrument. Reported are the mean fluorescence levels of at least 200 events for KIR-Fc, expressed as a ratio to W6/32 binding, which served as a control for variation in the amount of HLA class I bound to each bead (44).
NKL and KIR transductants
NKL, a cell line derived from a human leukemia that resembles activated NK cells and expresses no endogenous cell-surface KIR (56), was maintained as described (57). Transduced NKL cells expressing a KIR of choice were generated as previously described (44). Full-length coding regions for wild-type and mutant KIR were amplified by PCR and cloned into pBMN retroviral vector. Point mutations were made using the QuikChange mutagenesis kit (Stratagene). Recombinant retrovirus was generated by transfection into Phi-NX cells using standard protocols and used to infect/transduce NKL cells. KIR-expressing cells were sorted for equivalent cell-surface expression levels using a FACStar cell sorter (BD Biosciences). The pBMN vector and Phi-NX cells were a gift from Garry Nolan, Stanford University, Stanford, CA.
721.221 and HLA transfectants
721.221 (221 for short), a MHC class I deficient EBV-transformed human B cell line (58) was maintained as described (39). Popy-C*0301 was isolated from cDNA of the orangutan, Allen, using primers 5’-CTCGAGGCCGCCACCATGGCCGTCATGGCGCCCGTAACCCTCCTCC-3’ and 5’-GCGGCCGCTCAGGCTTTACAAGCAATGAGAGACT-3’. The mutant Popy C*0301-80K, in which asparagine at position 80 was replaced by lysine, was made using the QuickChange mutagenesis kit (Stratagene). Transfectants of 221 cells expressing Popy C*0301 and C*0301-80K were generated as previously described, in which critical leader peptide residues were mutated to prevent HLA-E binding to CD94:NKG2A(44).
Cellular cytotoxicity assay
Killing of transfected and untransfected 221 cells by NKL transductants was assayed as previously described(44). NKL effector cells were mixed with 51Cr-loaded 221 target cells for 4h at 37°C at ratios of 40:1, 20:1, and 10:1. Supernatants were harvested and the levels of 51Cr quantified using a Wallac β-scintillation counter. Specific lysis was calculated as the specific release minus the spontaneous release divided by the total release less the spontaneous release. Because the killing of 221 cells by NKL cells is close to the maximum possible, we could not assess the activating potential of orangutan activating lineage III KIR using this system.
Results
Structural comparison of human and orangutan KIR identified two candidate inhibitory MHC-C receptors: Popy2DLA and Popy2DLB (49). Like the human C1 receptor KIR2DL3, Popy2DLA has lysine at position 44, the specificity-determining residue (59, 60), suggesting that Popy2DLA too has C1 specificity. By contrast, Popy2DLB has glutamate 44, a non-conservative substitution absent from human KIR, suggesting a novel MHC class I specificity (Fig. 1). To test these hypotheses, we determined ligands and signaling function for Popy2DLA and 2DLB. Throughout this investigation human KIR2DL1*001 and KIR2DL3*001 were used as controls.
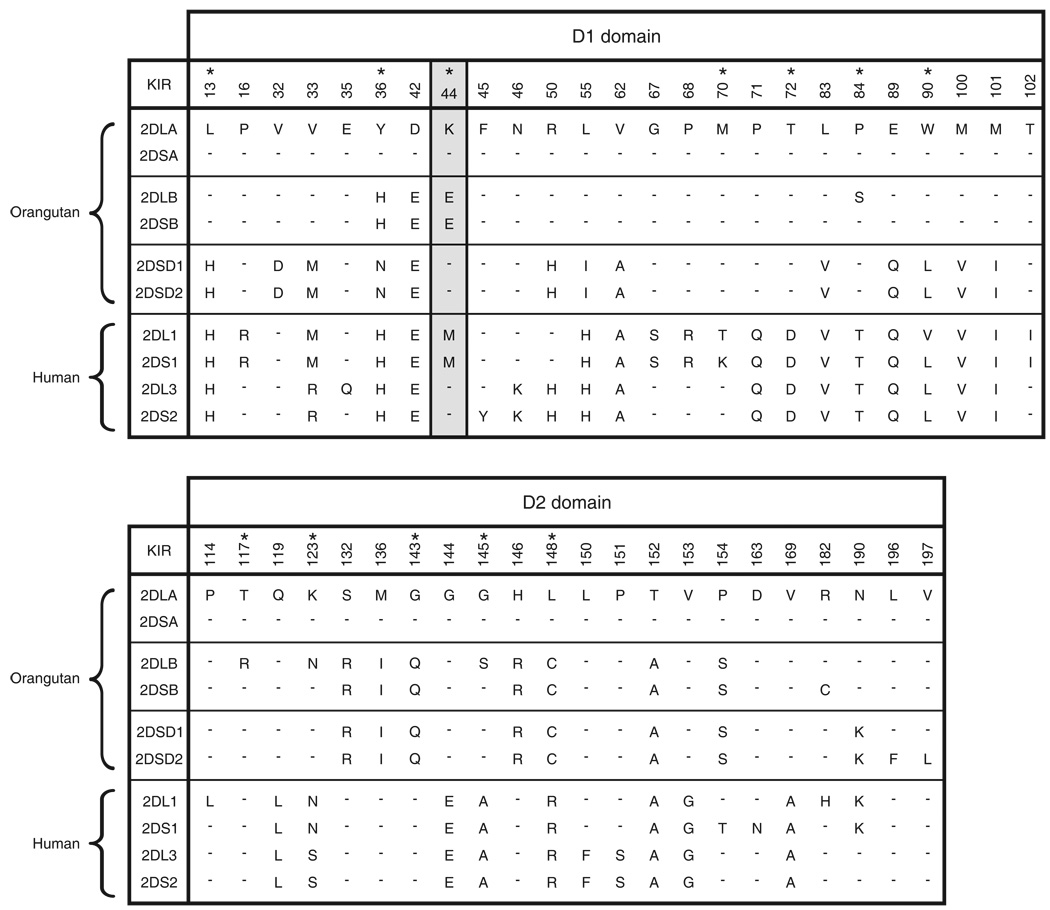
Figure 1. Sequence variation among orangutan and human lineage III KIR.
The alignment shows the positions of amino-acid substitution in the D1 and D2 ligand-binding domains of human and orangutan KIR. Dashes indicate identities with the reference sequence, orangutan Popy2DLA. The orangutan sequences come from our initial characterization of PopyKIR (49). We have renamed the activating KIR to reflect better their functional pairing with particular inhibitory KIR. Thus, 2DSA was previously called 2DSD (AF470364), 2DSB was 2DSC (AF470362), 2DSD1 was 2DSB (AF470361), and 2DSD2 was 2DSA (AF470360). 2DSD1 and 2DSD2 are so named because they likely represent alleles at a single locus. Asterisks indicate positions where sequence diversification is the consequence of positive natural selection. Position 44 (shaded grey) determines HLA-C specificity of human KIR2DL: lysine confers C1 specificity, methionine confers C2 specificity (60). Glutamate 44 is absent from human KIR, but present in orangutan KIR.
Orangutan Popy2DLA is an inhibitory receptor recognizing C1 like human KIR2DL3
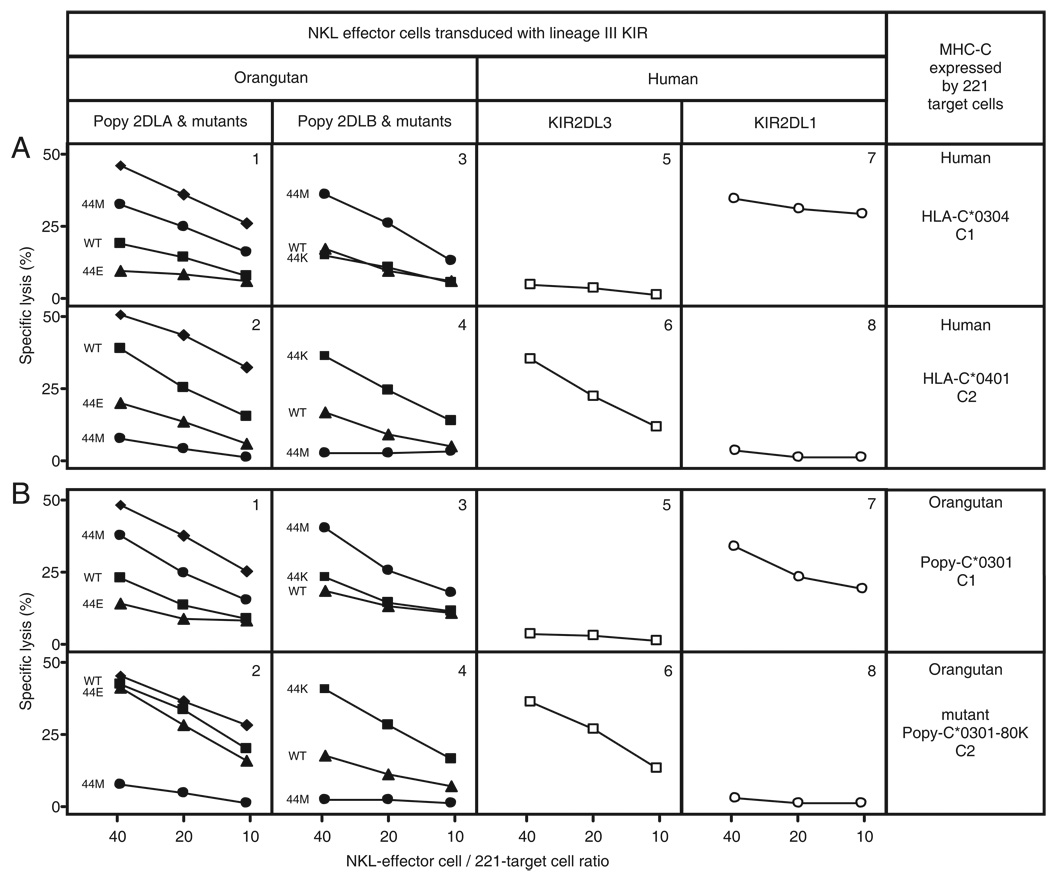
Functional interactions of KIR with MHC-C bearing either the C1 or C2 epitope were examined in cytotoxicity assays where NKL effector cells expressed a single KIR and 221 target cells expressed a single MHC class I (Figure 2). NKL cells expressing Popy2DLA were inhibited by 221 cells expressing C1+C*0304 (Fig. 2A1), but killed class I-deficient 221 cells and 221 cells expressing C2+C*0401 (Fig. 2A2). This engagement of C1 was like that of human KIR2DL3 (Fig. 2A5, 6) and different from C2-recognition by KIR2DL1 (Fig. 2A7, 8). The dependence on lysine 44 of C1-recognition by Popy2DLA was investigated using a Popy2DLA mutant having methionine 44, the specificity-determining residue of KIR2DL1. NKL cells expressing this Popy2DLA-44M mutant were inhibited by 221 cells expressing the C2 epitope (Fig. 2A2), but not by 221 cells expressing the C1 epitope (Fig. 2A1). This reaction pattern is like that of C2-specific KIR2DL1 (Fig. 2A7, 8). Popy2DLA was thus demonstrated to be a functional inhibitory receptor that recognizes C1 in a lysine 44 dependent manner.
Figure 2. Human and orangutan have functionally compatible MHC-C ligands and inhibitory MHC-C reactive KIR.
Shown are results of cytotoxicity assays between NKL effector cells expressing various KIR2DL and 221 target cells expressing either C1 or C2. In panel A, the target cells express human HLA-C: either C*0304 (C1) or C*0401 (C2); in panel B the target cells express orangutan Popy-C: either C*0301 (C1) or mutant C*0301-80K (C2). The NKL effector cells express wild type and mutant forms of Popy2DLA and Popy2DLB, having lysine 44 (▪), methionine 44 (●) or glutamate 44 (▲) (panels 1–4). In panels 1 and 2, control killing of untransfected 221 cells is also shown (♦). Control NKL effector cells expressed human C1-specific KIR2DL3*001 (□) and C2-specific KIR2DL1*001 (○) (panels 5–8). All NKL cells expressing KIR with lysine 44 were inhibited from killing C1 transfected 221 cells. NKL cells expressing KIR with methionine 44 were inhibited from killing C2 transfected 221 cells. NKL cells expressing KIR with glutamate 44 were inhibited from killing C1 and C2 transfected 221 cells. An exception is 2DLA-44E that lysed targets expressing orangutan C2, showing that variation at position 80 in MHC-C is not the only factor influencing interaction with KIR2DL. All experiments were performed in triplicate with a minimum of three independent experiments for each combination of target and effector. Shown are the data from a representative experiment.
Popy2DLB is an inhibitory receptor that recognizes both C1 and C2 epitopes
NKL cells expressing Popy2DLB killed untransfected 221 cells, but were inhibited by 221 cells expressing either C1+C*0304 (Fig. 2A3) or C2+C*0401 (Fig. 2A4). To investigate this dual C1+C2 reactivity, we studied Popy2DLB mutants in which glutamate 44 was replaced by either lysine (Popy2DLB-44K) or methionine (Popy2DLB-44M). Both mutations abrogated the dual reactivity, demonstrating its dependence upon glutamate 44. The Popy2DLB-44K mutant recognized C1 (Fig. 2A3) but not C2 (Fig. 2A4); the Popy2DLB-44M mutant recognized C2 (Fig. 2A4) but not C1 (Fig. 2A3). To determine if glutamate 44 is sufficient to confer the dual C1+C2 reactivity, we studied a mutant (Popy2DLA-44E) of Popy2DLA in which lysine 44 was replaced with glutamate. NKL cells expressing Popy2DLA-44E were inhibited by 221 cells expressing either C2 (Fig. 2A2) or C1 (Fig. 2A1), but killed untransfected 221 cells efficiently. These results show that glutamate 44 is sufficient to confer C1+C2 reactivity on Popy2DLA, despite the fact that Popy2DLA differs from Popy2DLB by an additional thirteen amino acid substitutions in the D1 and D2 domains forming the ligand-binding site (Fig. 1). In conclusion, glutamate 44 is both necessary and sufficient to confer recognition of C1+C2 by orangutan lineage III KIR.
Popy-C allotypes are recognized by orangutan and human KIR
Having established that Popy2DLA and Popy2DLB recognize well-characterized human C1 and C2 epitopes, we studied their capacity to recognize orangutan MHC-C (Popy-C), the physiologically relevant ligand. Because all Popy-C allotypes have asparagine 80 and are thus predicted to carry C1(46, 49), we constructed a mutant, Popy-C*0301-80K, in which asparagine 80 was replaced by lysine (the residue defining the C2 epitope of HLA-C). Transfected 221 cells expressing Popy-C*0301 or Popy-C*0301-80K were tested for their capacity to be killed by NKL cells expressing Popy2DLA, Popy2DLB, KIR2DL3 and KIR2DL1 (Fig. 2B).
NKL cells expressing Popy2DLA, Popy2DLB or KIR2DL3 were strongly inhibited by 221 cells expressing Popy-C*0301 (Fig. 2B1, 3 and 5); and similar properties were observed for the Popy-C*01 and Popy-C*02 allotypes (data not shown). In contrast, NKL cells expressing KIR2DL1 (Fig. 2B7) were not inhibited by target cells expressing Popy-C*0301. The natural Popy-C allotypes are thus seen to carry C1 epitopes that function as ligands for inhibitory orangutan and human KIR. Popy-C*0301 induced stronger inhibition on engaging KIR2DL3 (Fig. 2B5) than either Popy2DLA (Fig. 2B1) or Popy2DLB (Fig. 2B3), an effect that could arise from either stronger avidity or signal transduction of the human receptor.
That target cells expressing Popy-C*0301-80K were killed by NKL cells expressing KIR2DL3 (Fig. 2B6), but not by NKL cells expressing KIR2DL1 (Fig. 2B8), shows that the C1 epitope of Popy-C*0301 was replaced by a functional C2 epitope in this mutant. Further supporting this interpretation, 221 cells expressing Popy-C*0301-80K were killed by NKL cells expressing Popy2DLA (Fig. 2B2), but not by NKL cells expressing Popy2DLB (Fig. 2B4). Here Popy-C*0301-80K behaved similarly to C2+HLA-C*0401, being recognized by Popy2DLB but not by 2DLA. Consistent with these observations, C1+Popy-C*0301 was shown to be a good ligand for the Popy2DLA-44E (Fig. 2B1) and Popy2DLB-44K (Fig. 2B3) mutants, but a weak ligand for the Popy2DLA-44M (Fig. 2B1) and Popy2DLB-44M (Fig. 2B3) mutants. Conversely, C2+Popy-C*0301-80K proved a good ligand for Popy2DLA-44M (Fig. 2B2) and Popy2DLB-44M (Fig. 2B4), but a weak ligand for Popy2DLA-44E (Fig. 2B2) and Popy2DLB-44K (Fig. 2B4). This analysis demonstrates that natural Popy-C allotypes carry C1 epitopes that behave like human C1 epitopes in their interactions with inhibitory lineage III KIR. Our results also show that only one point mutation is necessary for the orangutan to acquire C2+Popy-C. And should it occur, the C2+Popy-C would immediately be functional as a ligand for Popy2DLB.
High resolution HLA class I specificity of human and orangutan inhibitory KIR
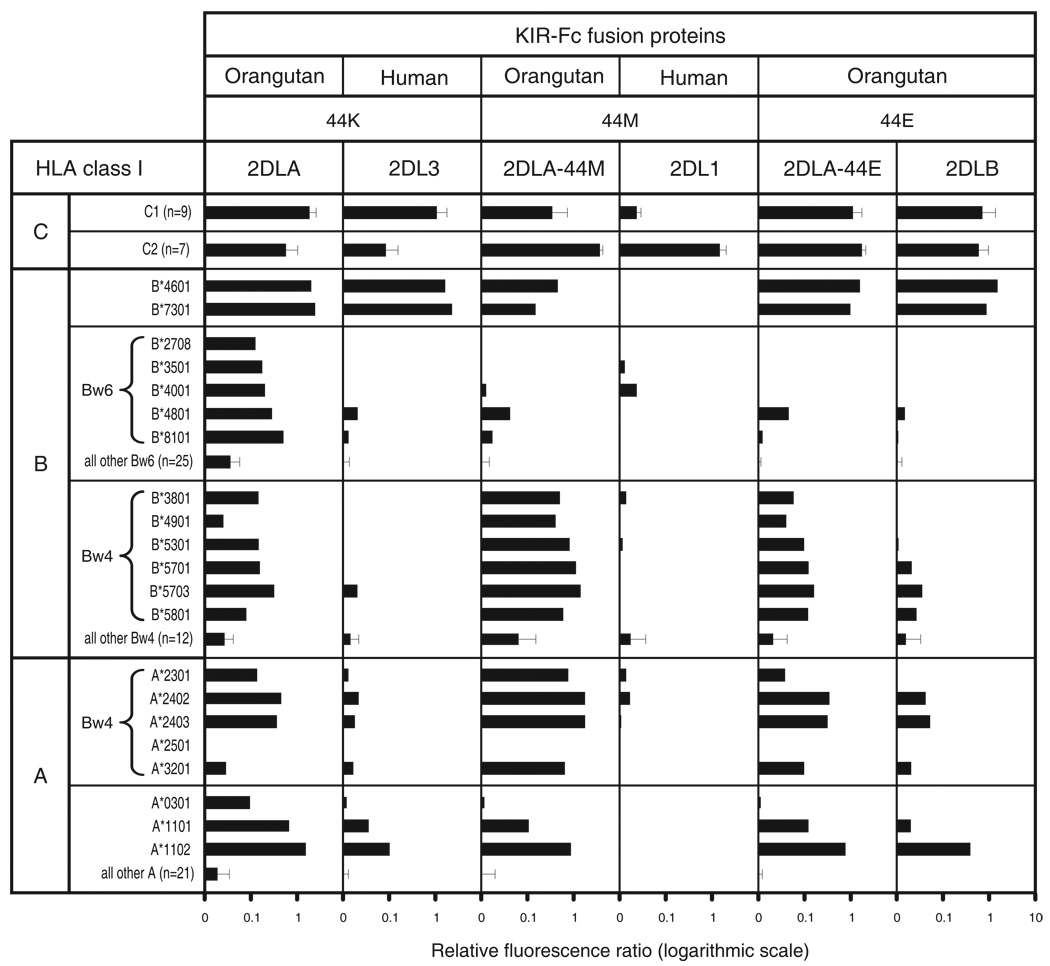
To complement and extend the results from cellular cytotoxicity assays (Fig. 2), we measured the binding of KIR-Fc fusion proteins to beads coated with one of 29 HLA-A, 50 HLA-B and 16 HLA-C allotypes. Figure 3 shows mean values for groups of functionally similar allotypes, while Figure 4 shows the binding to individual HLA-C allotypes.
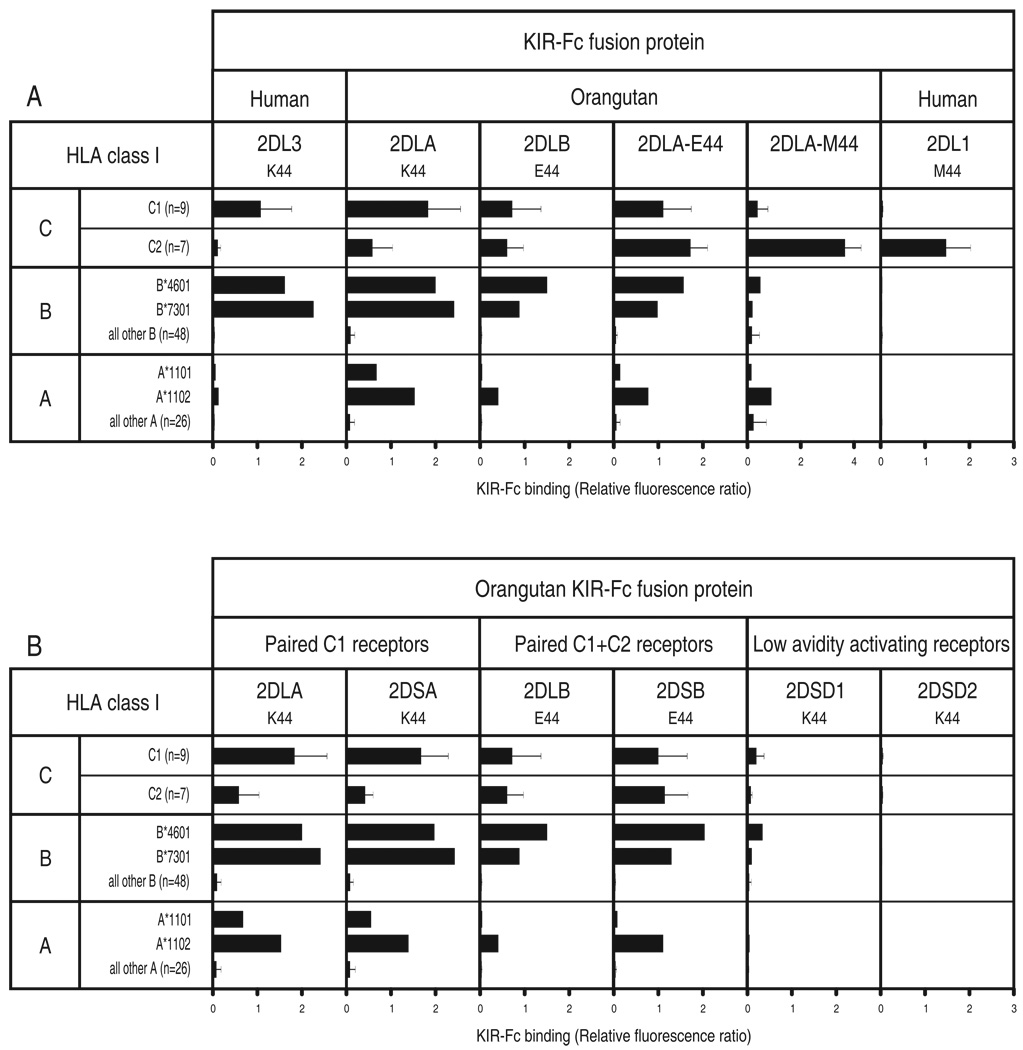
Figure 3. MHC class I binding specificities of human and orangutan lineage III KIR.
Shown are results of assays to measure the binding of KIR-Fc fusion proteins to beads each coated with one of 95 HLA class I allotypes. A) Compares human and orangutan inhibitory lineage III KIR, including mutants of Popy2DLA having either glutamate or methionine at position 44. B) Compares activating and inhibitory lineage III orangutan KIR. For all KIR the residue at position 44 is given. For each combination of bead and KIR-Fc, the binding was normalized to that obtained with the pan HLA class I mAb W6/32, thus controlling for variation in the amount of HLA class I bound to each bead. All KIR-Fc fusion proteins folded properly as assessed by the capacity to bind conformation-dependent antibodies. Mean values and standard deviation are shown for various groups of allotypes. A minimum of three independent assays was performed for each fusion protein. Shown are the averaged data for all experiments. Data for each HLA allotype:KIR-Fc combination is shown in Supplemental Figure S1.
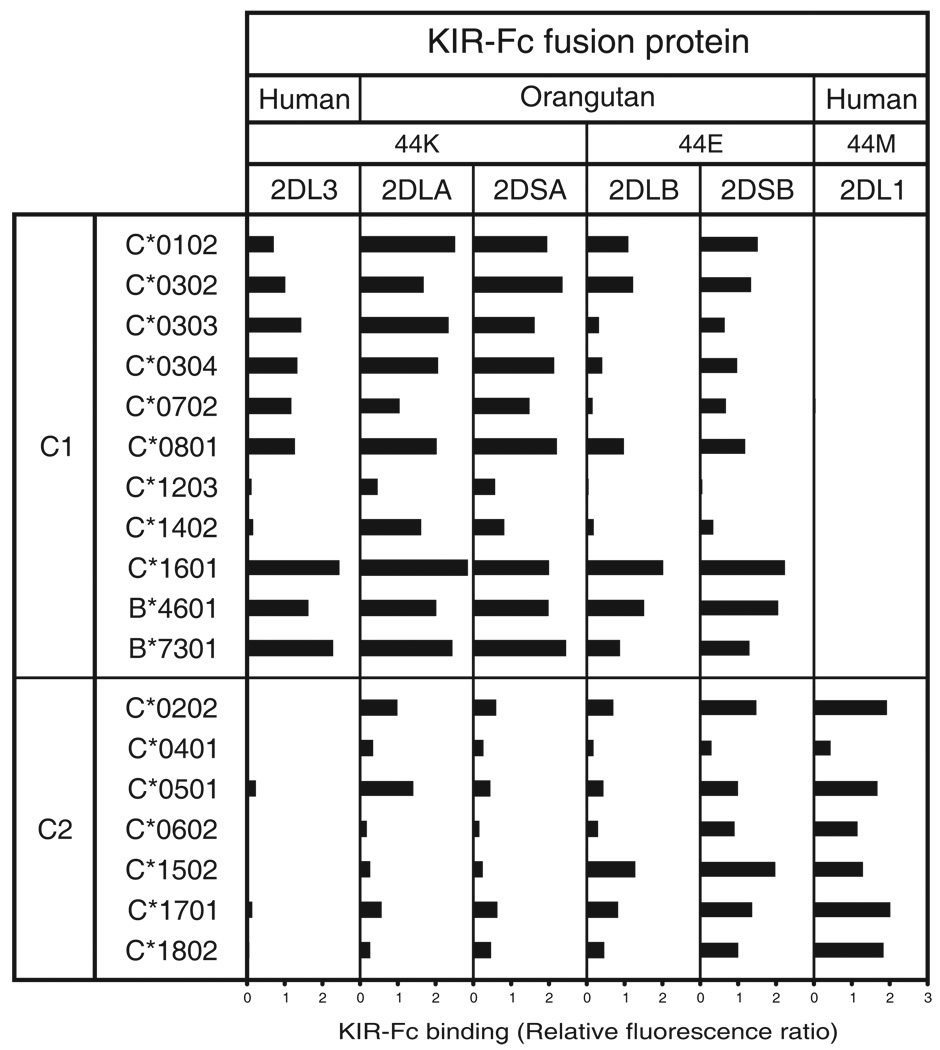
Figure 4. Popy2DLB and Popy2DSB have novel specificity for C1+C2.
Shown are the results of assays to measure the binding of human and orangutan KIR-Fc fusion proteins to nine C1+ HLA-C allotypes, two C1+ HLA-B allotypes and seven C2+ HLA-C allotypes. Assays were performed as described in the legend to Fig. 3. Shown are the averaged data for all experiments.
KIR2DL3-Fc and Popy2DLA-Fc both bound to C1+HLA-C allotypes and to the two C1+HLA-B allotypes (B*4601 and B*7301) (40, 44), with Popy2DLA being the stronger C1 receptor overall (Fig. 3A). Popy2DLA also bound to C2+HLA-C*0202, C2+HLA-C*0501 (Fig. 4) and the two HLA-A11 subtypes, with higher binding to A*1102 than to A*1101 (Fig. 3A). Previously, we showed that human KIR2DS4, a divergent, activating lineage III KIR, binds C*0202, C*0501, A*1101 and A*1102 (27), and that KIR2DL2, a divergent allotype of KIR2DL3, with higher avidity for C1 binds to C2+C*0202 and C*0501 (44).
Popy2DLB also bound to C1+HLA-B, C1+HLA-C and A*11, but with less avidity than Popy2DLA (Fig. 3A). Further distinguishing Popy2DLB from Popy2DLA was its broader, more even, reactivity with C2+HLA-C. Thus Popy2DLB binding to the C*0202 and C*0501 allotypes was lower than for Popy2DLA, but it was accompanied by increased avidity for other C2-bearing allotypes, such as C*1502, C*1701 and C*1802 (Fig. 4). As a consequence of these opposing effects, the mean avidities of Popy2DLB for the C1 and C2 epitopes are comparable, the strongest reactions being with the two C1+HLA-B allotypes (Fig. 3A). In this direct binding assay, Popy2DLB is seen to be a weaker receptor than Popy2DLA, but to have a broader specificity for both the C1 and C2 epitopes of HLA-C. These results are consistent with those obtained in the cytotoxicity assay (Fig. 2).
Mutant Popy2DLA-E44 combines the strength of Popy2DLA with the specificity of Popy2DLB, having stronger reactions with C1, C2 and A11. In contrast, the Popy2DLA-M44 mutant is specific for C2, but retains weak C1 and A11 binding (Fig. 3A). Although human KIR2DL1 is a weaker receptor than Popy2DLA-M44, it has exquisite specificity for C2, giving no detectable cross-reactivity with any C2− HLA class I. This sharp C2 specificity of KIR2DL1 cannot be completely attributed to methionine 44, but must involve contribution from one or more of the additional 30 residues that distinguish the ligand-binding D1 and D2 domains of KIR2DL1 and Popy2DLA (Fig. 1).
Variability in KIR binding to HLA-C correlates with the pattern of allotypic substitution. Within the C1 group, the strongest (C*1601) and weakest (C*1203) binding allotypes (Fig. 4) differ only by substitutions at positions 73, 152, and 156. Modeling these differences on the crystallographic structure of C*0304 bound to KIR2DL2 (61), indicated that residue 73 (threonine in C*1601/ alanine in C*1203) may contact KIR directly, while residues 152 (alanine in C*1601/ glutamate in C*1203) and 156 (glutamine in C*1601/ tryptophan in C*1203) influence the peptides bound. Moreover, the alanine 152, glutamine 156 motif is uniquely present in HLA-C*16 and HLA-A*11 and no other HLA class I (Supplemental Figure S2), which could explain why A*11 is the only HLA-A allotype bound by Popy2DLA. Of the C2+ allotypes, HLA C*0401, the prototypical C2 ligand (23), stands out for its reproducibly low binding to KIR2DL1 and Popy2DLB (Fig. 4). This difference correlates with the unique substitution of tryptophan for arginine at position 14 in a loop known to influence KIR interaction (27).
Recent observation that MHC class I bound peptides can alternatively interact with KIR to mediate strong inhibition, weak inhibition or antagonize inhibition (62), suggest an explanation for the ‘discrepancy’ between the binding and cytotoxicity assays with C*0401. If C*0401-bound peptides are not antagonistic but predominantly ones that interact weakly with KIR, then in aggregate they could develop good inhibition while individually binding weakly to KIR. Alternatively, relatively few C*0401 molecules might be sufficient to elicit a potent inhibition, or C*0401 could preferentially attach to the beads in a manner that prevents binding of KIR, but not the W6/32 antibody used to normalize for differences in abundance of HLA on the beads. The absolute level of W6/32 binding to beads varies over a range in which the weakest (B*5101) is 23% that of the strongest, and C*0401 is the second weakest (31%) (Supplemental Figure S3). No correlations could be discerned between KIR-Fc binding and amount of HLA class I protein as assessed by W6/32 binding. For example, C*1701, C*0202 and C*0501, are the strongest C2 ligands for KIR-Fc proteins and bind W6/32 (41%, 47%, and 49%, respectively) at levels that groups them with C*0401 in the bottom 20% of the class of 95 HLA class I variants.
Orangutan has paired activating and inhibitory KIR with similar specificity and high avidity for MHC class I
Four lineage III orangutan KIR were predicted to encode activating receptors (49) that engage the DAP12 signaling adaptor (63). Of these Popy2DSA, 2DSD1 and 2DSD2 have lysine 44 and the potential to be C1 receptors, whereas Popy2DSB has glutamate 44 and the potential to be a C1+C2 receptor (Fig. 1). Fc-fusion proteins were made from the extracellular domains of these PopyKIR2DS and tested for binding to the panel of HLA class I coated beads (Fig. 3B). Popy2DSA-Fc bound to C1+ HLA-B, C1+ HLA-C and A*11 like 2DLA-Fc (Fig. 3B, Fig. 4), consistent with the amino acid sequence identity of their ligand-binding domains (Fig. 1). Popy2DSA and Popy2DLA are seen to be a pair of activating and inhibitory receptors with identical specificity and avidity for C1. Popy2DSB-Fc bound to C1+ HLA-B, C1+ HLA-C, C2+ HLA-C and A*11 like Popy2DLB-Fc (Fig. 3B, Fig. 4). Thus Popy2DSB and Popy2DLB also form a pair of activating and inhibitory receptors with comparable C1+C2 specificity and avidity, consistent with the high sequence similarity in their extracellular domains (Fig. 1). Popy2DSD1 and Popy2DSD2 (which differ only at two positions and are probably allotypes of the same gene) have a D2 domain related to that of Popy2DLB, but a divergent D1 domain that is more closely related to the D1 of human KIR2D than to D1 of the other orangutan KIR2D (Fig. 1). Popy2DSD1 exhibited only very weak reactions with C1, consistent with presence of lysine 44, and Popy2DSD2 gave no detectable reactions with any HLA class I.
Humans have no paired activating and inhibitory KIR with similar avidity and specificity for HLA class I. Although three pairs of structurally similar receptors are present, in each of these the activating receptor has acquired substitutions that reduce its avidity for the ligand recognized by the inhibitory receptor. KIR2DS1 is a weaker C2 receptor than KIR2DL1, and binding of C1 by KIR2DS2 and Bw4 by KIR3DS1 is undetectable (20, 22, 23, 64, 65). In this regard, the orangutan Popy2DSD receptors are similar to their human counterparts. However, in contrast to humans, orangutans have two pairs of high avidity MHC class I-specific activating and inhibitory KIR.
The KIR binding site recognition is inherently limited to a minority of HLA class I variants
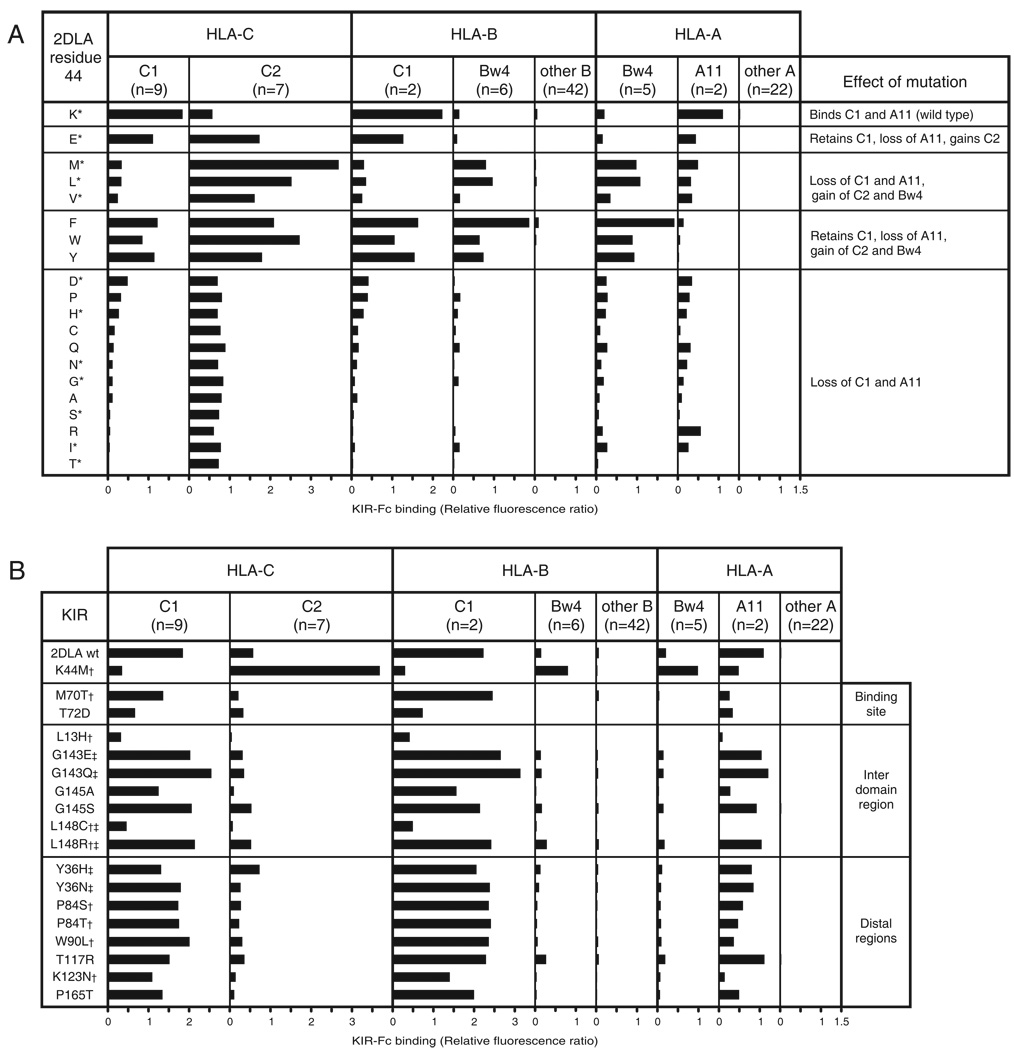
Residue 44 in the D1 domain was designated the specificity-determining residue because mutagenesis at position 44 was sufficient to swap the C1 and C2 specificities of human KIR2D (59, 60). Extending this concept, we find that glutamate 44 determines the distinctive C1+C2 specificity (Figs. 2 and 3). To explore the evolutionary potential of position 44 variation, we made a full set of 19 mutants, in which the wild-type lysine 44 of Popy2DLA was replaced by all the other natural amino acids. KIR-Fc fusion proteins made from these mutants were tested for binding to beads coated with HLA class I (Fig. 5A).
Figure 5. Mutation at position 44 in Popy2DLA produces only four different HLA class I specificities.
Binding assays were as described in the Fig. 3 legend. A) Compares the binding of wild-type Popy2DLA, having lysine 44, to 19 Popy2DLA mutant proteins, each containing a different amino acid at position 44. Mutation had one of four effects on the HLA class I specificity of Popy2DLA as is summarized on the right under ‘Effect of mutation’. Natural substitutions occurring at position 44 are shown in Supplemental Figure S4 and are indicated by asterisks (*). The binding of each position 44 mutant to individual allotypes is shown in Supplemental Figure S5. B) Natural selection produced sequence variation at many positions, additional to 44, in the D1 and D2 domains. Positions of positive selection in all primates are denoted by a dagger (†); in orangutan only by a double dagger (‡). KIR-Fc fusion proteins made from Popy2DLA mutants carrying these natural substitutions were compared to Popy2DLA for binding to HLA-A, B and C. The locations of these mutational sites within the structure of KIR2D are given in the final column. Mutations at these positions could increase or decrease receptor avidity, but did not alter receptor specificity. Shown are the averaged data for all experiments.
The position 44 mutants segregated into four groups according to their patterns of reactivity with the 95 HLA-A, B and C variants (Fig. 5A). The first group contains only glutamate, which decreased avidity for C1 and A11, while gaining avidity for C2. The second group, comprising hydrophobic methionine, leucine and valine, lost much avidity for C1 and A11, while gaining high-avidity for C2 binding and variable reactivity for HLA-A and -B allotypes carrying the Bw4 epitope, the ligand for human KIR3DL1(39). This trend continues in the third group, comprising aromatic phenylalanine, tryptophan and tyrosine. This group has C1+C2 reactivity like glutamate 44, but exhibits complete loss of A11 binding and much increased binding to Bw4. The fourth group, the majority, consists of 12 mutants causing loss or abrogation of binding to C1 and A11, with no gain in reactivity to any other HLA class I. In summary, substitution at position 44 in Popy2DLA had only four effects, all associated with reduced reactivity for C1 and A11: conversion of C1 specificity to C2; broadening of C1 specificity to C1+C2; broadening of the C1 specificity to C1+C2+Bw4; and loss, or abrogation, of C1 and A11 reactivity with no other accompanying effect.
It is striking that the only HLA class I variants recognized by the panel of mutant KIR are those known to be ligands for human KIR, and that all four epitopes recognized by human KIR -- C1, C2, Bw4 and A3/11 -- are represented in the HLA class I variants recognized by Popy2DLA and its position 44 mutants. Lysine is the only residue that confers strong avidity and specificity for C1, and of three residues that convert C1 specificity to C2, the strongest receptor is formed by methionine 44, the residue present in the human C2-specific receptor, KIR2DL1. Thus the position 44 residues in the human receptors appear optimized for maximal avidity for C1 and C2. Furthermore, glutamate 44, the natural residue in Popy2DLB is the only residue at position 44 that gives specificity for C1+C2 without additional strong reactions with the Bw4 epitope of MHC-A and –B.
As controls for the position 44 mutants, we studied a set of seventeen Popy2DLA mutants incorporating natural substitutions at twelve other positions where there is good evidence for positive diversifying natural selection (data not shown). Whereas the position 44 mutations altered both the specificity and avidity of the interaction with HLA class I (Fig. 5A), mutations at these other positions only affected the avidity for C1 and A11 (Fig. 5B), none of them exhibiting altered specificity for C2, Bw4 or other HLA class I. These mutations localize to three regions: the ligand-binding site, the hinge region between D1 and D2, and distal regions outside these two areas (61, 66). Whereas mutations in the distal regions had relatively minor effects, mutation at position 72 in the binding site significantly reduced avidity for C1 and A11 and seven mutations in the inter-domain regions caused a range of effects embracing both increased and decreased avidity. Notable was leucine 148, where arginine increased avidity, while cysteine greatly reduced avidity. That substitution at 11 of the 12 positions (the exception being position 84) either increased or decreased the avidity for C1, shows how natural variation at these positions modulates receptor function, consistent with them having been sites of natural selection.
In conclusion, the results from these two sets of mutants demonstrate that KIR recognition is restricted to binding the C1, C2, A3/11 and Bw4 epitopes of MHC class I (Fig. 5). No mutation in either the specificity-determining residue or other positions of natural selection produced reactivity with any HLA class I variant that is not recognized by the naturally occurring human KIR and carries one of these epitopes.
Elements of the four epitopes recognized by lineage II and III human KIR are represented in the specificity of Popy2DLA
Direct binding of KIR2D-Fc to HLA class I coated beads is a robust, highly reproducible assay that combines low background with sensitivity to a wide range of signal. These qualities allowed us to distinguish strong, weak, and negative reactions of human and orangutan KIR, which are jointly displayed in Figure 6 using a logarithmic scale for the binding of KIR-Fc. Evident from this analysis are the more specific reactions of human KIR2DL1 and KIR2DL3 compared to orangutan Popy2DLA and Popy2DLB. KIR2DL1 has the highest discrimination, reacting almost exclusively with C2+HLA-C. Next is KIR2DL3, which reacts principally with C1+HLA-B and HLA–C, but also with some C2+HLA-C, and weakly with HLA-A11 and Bw4. Popy2DLB has strong reactions with C1 and C2, associated with weaker binding to A*11 and Bw4. Popy2DLA has the broadest reaction pattern, which includes C1 and C2, A*03 and A*11, Bw4+HLA-A and –B, and several Bw6+HLA-B allotypes. Although Figure 6 contains data obtained with a single concentration of KIR-Fc, titrations showed that the cross-reactions were observed over a range of concentration, while no reactions with any of the ‘negative’ HLA class I were observed, even at high concentration (data not shown).
Figure 6. Orangutan KIR exhibit specificity and cross-reactivity with all four HLA-A, B, and C epitopes that are ligands for human KIR.
Binding assays were as described in the Fig. 3 legend, with the difference that the data is here displayed on a logarithmic scale to visualize both strong and weak reactions. Shown are the individual values for selected allotypes and means and standard deviations for several groups of allotypes. The averaged data for all experiments is presented.
From this comparison of orangutan and human KIR we see the evolutionary progression, by which an ancestral and flexible KIR that was broadly reactive with epitopes of MHC-A,-B and C evolved to produce modern human lineage II and III KIR with more specialized specificities for A3/A11, Bw4, C1+C2, C1 and C2. Orangutan Popy2DLA retains the broader reactions predicted to be the hallmark of such an ancestor.
Bw4 binding to KIR is influenced by polymorphism at positions 80 and 152 in MHC class I
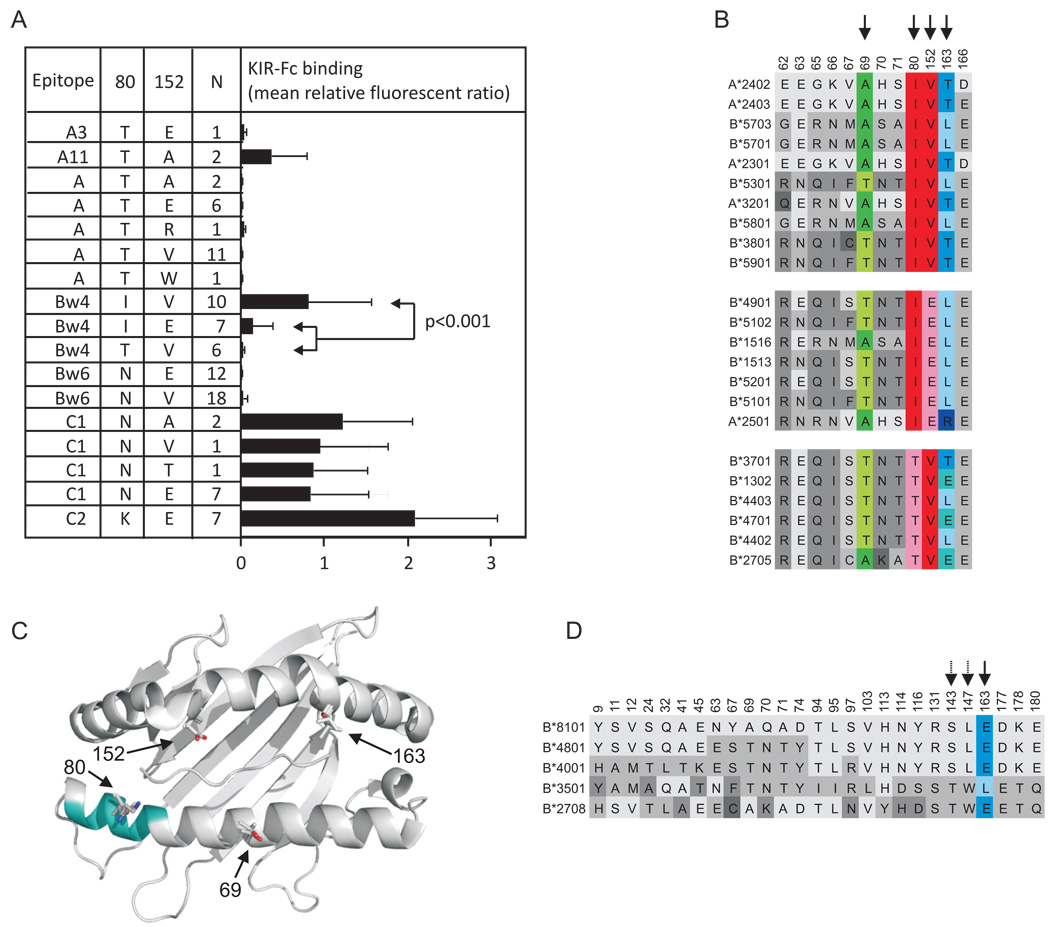
The set of 23 Bw4+ HLA class I allotypes divides into two groups based upon the isoleucine/threonine dimorphism at position 80 within the Bw4 sequence motif (residues 77–83) (67). The four Bw4+HLA-A and six Bw4+HLA-B allotypes that bind Popy2DLA, and some of its position 44 mutants, all have isoleucine 80, a residue reported to influence Bw4-recognition by human KIR3DL1 (38) and progression to AIDS of HIV infections (68, 69). The binding allotypes do not, however, constitute all the Bw4+ allotypes having isoleucine 80, showing that this residue is not sufficient to confer KIR reactivity. We therefore sought other polymorphisms in Bw4+HLA-A and -B that correlate with KIR binding.
The combination of residues at positions 80 and 152 correlates more strongly with KIR binding than either position alone (Fig. 7A). Isoleucine 80 and valine 152 gave strong binding, isoleucine 80 and glutamate 152 weak binding, and threonine 80 and valine 152 no binding. In addition, the more strongly binding Bw4+ variants tended to have alanine at position 69 and threonine at position 163 (Fig. 7B). Polymorphism at position 69 may affect KIR binding directly while polymorphisms at positions 152 and 163 are more likely to exert their effect on peptide binding (Fig. 7C). In contrast to the effects on Bw4 binding to KIR, polymorphism at position 152 had little effect on C1 binding (Fig. 7A). Five Bw6+ HLA-B allotypes bound weakly but reproducibly to Popy2DLA and mutants (Figure 6). The residues enriched in this subset of HLA-B allotypes are glutamate 163, serine 143 and leucine 147 (Fig. 7D, Supplemental Figure S2), again indicating that influences on peptide binding are responsible for these weak but informative cross-reactions.
Figure 7. Polymorphism at positions 80 and 152 affect the binding of Popy2DLA and Popy2DLA mutants to Bw4+ HLA-A and –B.
A) Binding assays were as described in the Fig. 3 legend. Shown here are mean values for eight KIR-Fc: Popy2DLA and its mutants having glutamate, methionine, leucine, valine, tryptophan, phenylalanine and tyrosine at position 44. The target HLA class I molecules were grouped according to the presence or absence of epitopes recognized by human KIR and the sequence motif at positions 80 and 152. Statistical significance of binding differences among the Bw4 allotypes was assessed by two-tailed t-test. B) Shown are amino-acid substitutions among the Bw4+ HLA-A and –B allotypes tested for binding to KIR-Fc. They are listed in order of decreasing binding strength, and also separated into blocks according to the motif at positions 80 and 152. Positions 69, 80, 152 and 163, which influence binding strength, are indicated by arrows and color. C) Shows a cartoon representation of the HLA-A*2402 structure (2BCK). Positions 69, 152, and 163 are indicated by arrows and the side chains present in A*2402, B*5301 (1A1O), and B*5101 (1E27) are shown; the latter structures were overlaid onto A*2402, but only these side chains are shown. The four observed residues at position 80 are shown; the position of the Bw4 epitope being indicated by cyan shading. D) Shows the amino-acid substitutions within the group of five Bw6+ HLA allotypes that exhibited weak but significant binding to KIR-Fc. They are listed in order of decreased binding. Comparison with the non-binding Bw6+ allotypes showed that glutamate 163 preferentially occurred in those that bound. Weaker association was seen with serine 143 and leucine 147 (see Supplemental Figure S2). Shown are the averaged data for all experiments.
Discussion
Inhibitory receptors for HLA class I play a major role in the education, repertoire development and response of human NK cells (16–19). These functions are performed by two types of receptor that are structurally disparate and of independent origin, but have co-evolved to complement each other. Interaction between CD94:NKG2A and its HLA-E ligand (70) is highly conserved in the human population (71) and gives NK cells a remarkably similar education in different individuals (19). In contrast, the interactions between KIR and their HLA-A, B and C ligands are extraordinarily variable, serving to diversify NK cell repertoire and response within the human population. Correlating with this difference, HLA-E is a much older gene than HLA-A, B or C (72).
Two of the four epitopes recognized by inhibitory KIR, A3/11 and Bw4, are carried by minority subsets of HLA-A and –B allotypes. The other two epitopes, C1 and C2, are principally carried by HLA-C allotypes, all of which have either C1 or C2. As a consequence of this distribution, only around half of the population has A3/11 and/or Bw4, whereas all members of the population have C1 and/or C2. By this criterion HLA-C dominates HLA-A and -B as a source of KIR ligands. MHC-C is also the most recently evolved MHC class I gene, its presence and divergence correlating with expansion of the lineage III KIR that comprises the MHC-C receptors (34, 49). These observations are consistent with MHC-C having evolved under natural selection to become a specialized source of ligands for NK cell receptors.
In phylogeny, MHC-C is specific to humans and great apes (hominids). Whereas natural selection drove MHC-C to fixation in a common ancestor of humans and chimpanzees, it is only present on ~50% of orangutan haplotypes (46), suggesting that MHC-C is less important for orangutans than for humans, and that there is balancing selection between the presence and absence of Popy-C. From these considerations and sequence comparisons of MHC-C and KIR in orangutan and human, we proposed that the orangutan system of KIR recognition of MHC-C resembles a half-way intermediate in the evolution of the human system (49). The results obtained from the investigation described here make a strong case for this hypothesis being correct.
Whereas different subsets of human and chimpanzee MHC-C allotypes carry C1 and C2, orangutan Popy-C allotypes carry only C1. Correspondingly, orangutans have an inhibitory C1 receptor (Popy2DLA) like human KIR2DL3, but lack an inhibitory C2 receptor like human KIR2DL1. Thus C1 and C1-specific KIR existed in the common ancestor of human and orangutan (>13 mya) (73), whereas C2 and C2-specific KIR evolved after separation of human and orangutan ancestors, but before the separation of human and chimpanzee ancestors (6.5–10 mya) (74). Consequently, C1-mediated regulation of NK cells was functioning in hominoids for up to 6 million years before the emergence of C2-mediated regulation. Observation that KIR2DL2/3-expressing NK cells selectively emerged in a culture system of NK cell differentiation (75) and in patients recovering from bone marrow transplant (76), raises the possibility that NK cell receptor ontogeny reflects this phylogeny. Consistent with the hypothesis, developing NK cells express conserved CD94:NKG2A (71) before they express variable KIR (77–79).
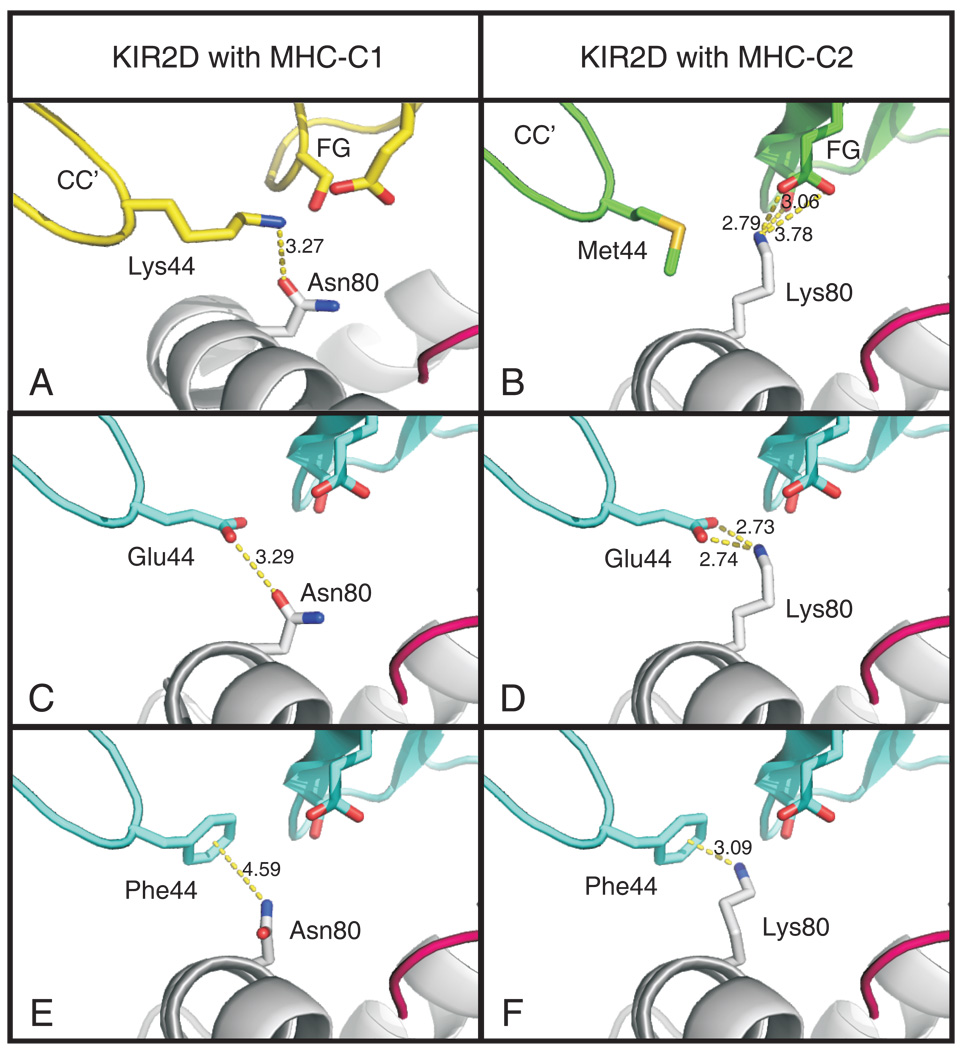
Experimental mutagenesis, in either the human (60) or orangutan system has shown that only two point mutations are needed to convert the C1-KIR interaction to the C2-KIR interaction: an asparagine to lysine substitution at position 80 in MHC-C, and a lysine to methionine substitution at position 44 in KIR. That natural evolution followed this route is highly improbable, because whichever mutation occurred first would have produced a non-functional intermediate -- either an orphan C2 receptor without a ligand, or an orphan C2 ligand without a receptor -- that could not be subject to positive natural selection. Suggesting a solution to this dilemma is the novel C1+C2 specificity of orangutan Popy2DLB, which binds to both C1 and C2 and has glutamate 44, a specificity-determining residue absent from human KIR. Modeling from the crystallographic structures of C1 bound to KIR2DL2 (61) (Fig. 8A) and C2 bound to KIR2DL1 (66) (Fig. 8B), suggests how glutamate 44 could alternatively hydrogen bond with either asparagine 80 of C1 (Fig. 8C) or lysine 80 of C2 (Fig 8D).
Figure 8. Modeling the interactions of C1+C2 specific KIR with the C1 and C2 epitopes.
The parts played by lysine 44 in binding C1 and methionine 44 in binding C2 were defined by crystal structures(61, 66), and are shown in panels A and B, respectively. The structure of HLA-C*0304 bound to KIR2DL2 (1EFX) was used to model the interaction of glutamate 44, which occurs naturally in Popy2DLB and 2DSB, with asparagine 80 of the C1 epitope, and is shown in panel C. Analogously, the structure of HLA-C*0401 bound to KIR2DL1 (1MI9) was used to model the interaction of glutamate 44 with lysine 80 of the C2 epitope (panel D). Because aromatic residues at position 44 conferred strong C1+C2 reactivity (see Fig 5), we modeled the interaction of phenylalanine 44 with C1 (panel E) and C2 (panel F), respectively. In each panel the distances between residues participating in binding are indicated.
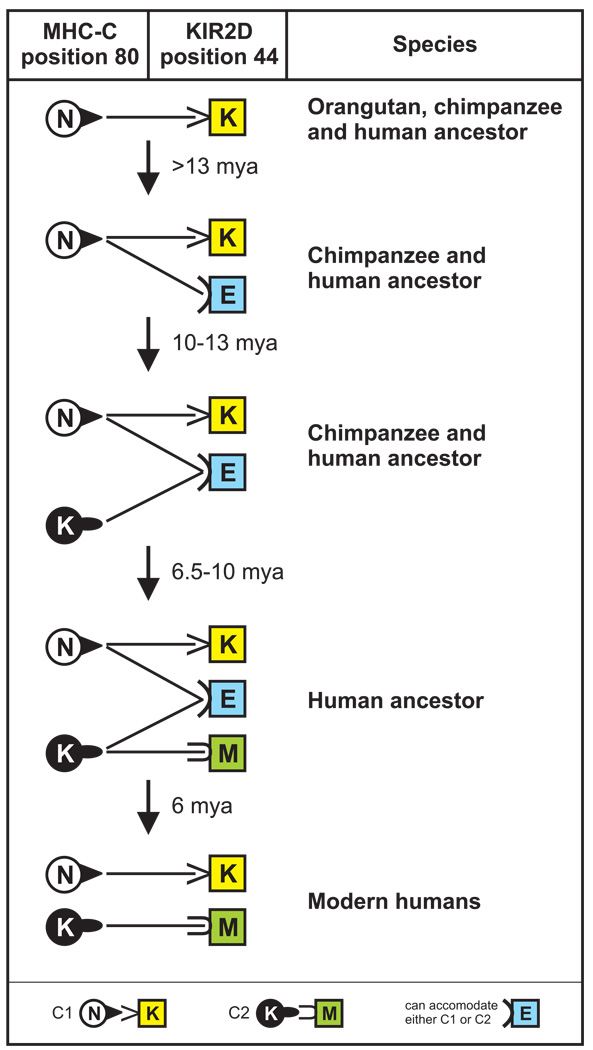
For the orangutan, which lacks C2, Popy2DLB can function only as a C1 receptor, but it has the potential to engage C2 should it arise. Consequently, Popy2DLB has the requisite properties of an evolutionary intermediate that would allow C2-mediated regulation to evolve from C1-mediated regulation without loss of function. This pathway, involving three mutations and four steps, is outlined in Figure 9. The first mutation replaces lysine 44 of the ancestral C1-specific KIR with glutamate to give a C1+C2 receptor like Popy2DLB. Initially this receptor will only function with C1 ligands, but it sets the stage for C2 evolution by the second mutation. Here, asparagine 80 of MHC-C is replaced by lysine, converting C1 to C2. The emergent C2 can immediately engage the C1+C2 receptor and be subject to positive selection that increases its frequency. Establishment of C2 in the population sets the stage for the third mutation, which replaces lysine 44 of a C1 receptor with methionine, thereby producing a C2-specific receptor like human KIR2DL1. (Mutation from glutamate [GAG] to methionine [ATG] to evolve the C2-specific receptor is an unlikely possibility, because two nucleotide substitutions are required.) Finally, in the presence of highly specific C1 and C2 receptors, there is selection against the broadly reactive C1+C2 receptor and its loss from the population, as apparently occurred in humans. In contrast to humans, the chimpanzee retains a lineage III KIR, Pt-KIR2DL9, with glutamate 44 (43). However this receptor has specificity for C2, not C1+C2, that is due to the unique substitution of cysteine for phenylalanine at position 45 (data not shown). In the chimpanzee it appears that selection against the C1+C2 receptor has not led to elimination of glutamate 44-containing receptors of but to a narrowing of their specificity to C2.
Figure 9. Predicted pathway for the evolution of the C2-KIR interaction from the C1-KIR interaction.
In principal this evolution could have been achieved with just two substitutions: lysine for asparagine at MHC-C position 80, and methionine for lysine at KIR position. This pathway is, however, improbable because it necessitates a non-functional intermediate: either a C2-receptor without a ligand, or a C2 ligand without a receptor. More likely is the four-step process shown in this schematic, which involves three mutations and maintains function throughout the process. Circles depicting MHC-C contain N for C1, or K for C2. Squares depicting KIR2DL contain K for lysine, E for glutamate, or M for methionine at position 44. A line connecting a circle to square denotes a functional ligand-receptor interaction. The hominoid ancestor had both C1 and the C1 receptor with lysine 44. Mutating lysine 44 to glutamate in step 1 produced the C1+C2 receptor, present in the common ancestor of orangutan, chimpanzee and human, and retained in modern orangutan. The C1+C2 receptor first functioned with the C1 ligand, while having the potential to engage C2. This potential was realized in step 2, when C2 evolved from C1 by mutation of asparagine 80 to lysine. This set the stage, in a common ancestor of chimpanzee and human, for the evolution of a C2-specific receptor in step 3, with mutation of lysine to methionine at KIR position 44. With specific C1 and C2 receptors in place there was selection in a human ancestor to eliminate the broadly reactive C1+C2 receptor in step 4, leaving modern humans with the just C1-specific and C2-specific receptors.
Like the human species, orangutan has two inhibitory and several activating lineage III KIR. Unlike the human situation, where all the activating KIR have reduced avidities for HLA class I ligands, both the inhibitory receptors, Popy2DLA and Popy2DLB, are paired with activating receptors (Popy2DSA and Popy2DSB, respectively) that have the identical, strong avidity and specificity for MHC class I. In addition, the orangutan has two activating receptors (Popy2DSD1 and Popy2DSD2) that have little avidity for MHC class I and lack equivalent inhibitory receptors. This comparison shows that the universal attenuation of the human activating KIR does not have general applicability to other species, but neither is the attenuation of activating KIR a human-specific phenomenon. That orangutan has an even balance between activating and inhibitory MHC-C reactive KIR, as does the chimpanzee (Moesta et al, manuscript submitted), shows that as humans evolved away from the ancestral state the balance became skewed towards favoring high avidity inhibitory receptors and low avidity activating receptors.
The benefit of paired activating and inhibitory receptors to the orangutan and chimpanzee could be the education of NK cell populations that are balanced in a manner that reduces autoimmunity while giving effective response to infection, malignancy and pregnancy (80). Characterizing human evolution has been selection for a large brain (twice the size of a chimpanzee brain), which necessitated an increased blood supply to the placenta during pregnancy (81). As outlined previously (82, 83), this selective pressure on the role of NK cells in reproduction could have led to attenuation of the activating receptors and formation of the uniquely human KIR A and B haplotypes, the latter favoring reproductive success, the former, termination of infection. A possible consequence of the skewed balance between activating and inhibitory KIR is that humans have higher propensity for autoimmunity than other hominid species. Indeed, susceptibility to several autoimmune diseases is associated with the attenuated human activating KIR (13).
Analysis of the fine-specificity of KIR binding to MHC class I shows that Popy2DLA binds strongly to C1, but also cross-reacts with C2, Bw4 and A3/11. Thus elements of all four human specificities are seen in this one orangutan KIR, showing how the four human epitopes and their cognate KIR could have evolved from an ancestral interaction between one KIR and one epitope. That human and orangutan KIR recognize the same subset of HLA-A, B and C allotypes, prompted us to explore the potential repertoire of KIR specificities for MHC class I. Saturation mutagenesis at position 44 of Popy2DLA showed that seven residues changed MHC class I specificity, while twelve reduced or eliminated C1 binding with no compensatory binding to other epitopes. Mutation at other sites of natural selection had no impact on receptor specificity, but acted to increase or decrease avidity for C1. Variation at the specificity-determining residue was thus seen to achieve four different specificities: C1+A11 (lysine), C1+C2 (glutamate), C2+Bw4 (methionine, leucine, valine), C1+C2+Bw4 (phenylalanine, tryptophan, tyrosine). Only the aromatic residues have not been observed naturally at position 44 in KIR (Supplemental figure S4).
Structural modeling indicates how the benzene ring in these aromatic residues can form hydrogen bonds with asparagine 80 of C1 (Fig. 8E) and lysine 80 of C2 (Fig. 8F) (84). Again it is striking that the same subset of HLA class I variants are recognized by the natural and mutant KIR, with 21 of 24 HLA-A, and 37 of 48 HLA-B allotypes tested being strictly non-permissive for KIR interaction. The restricted ability of KIR to bind different MHC class I isoforms fits with crystallographic observations that KIR conformation is little affected by binding to ligands indicating “that KIR and HLA associate essentially as rigid bodies”, a situation contrasting with the conformational adaptations that the αβ TCR make in binding to MHC class I (59).
HLA-A, B and C provide ligands for both KIR expressed by NK cells and the αβ TCR expressed by CD8 T cells. The sites on HLA class I to which these two receptors bind are overlapping and both include elements of the bound peptide (59). Because NK cells and T cells have different functions in immunity and reproduction they inevitably impose different and competing selection pressures on MHC class I variants. Thus variants that make the best NK cell response are unlikely to make the best CD8 T cell responses, and vice versa. Although MHC class I have interacted with αβTCR for > 400 million years (85), their interactions with variable KIR3DL emerged much more recently, during the 45–82 million years since separation of simian and prosimian ancestors (32, 86, 87). Thus KIR first interacted with forms of MHC class I that had been selected for their capacity to present antigens to T cells. That only a minority of modern HLA-A and –B allotypes are ligands for KIR may reflect balancing selection between allotypes selected on the basis of NK cell or T cell advantage. Essential to such balance is the inflexibility of KIR recognition of MHC class I, which complements the adaptability of TCR recognition (59). The modern HLA-B and –C genes originated by duplication of an ancestral gene resembling MHC-B, with selective divergence of the MHC-C daughter gene to focus on KIR binding and regulation of the NK cell response. This specialization and dominance of HLA-C in NK cell regulation has allowed HLA-A and –B to evolve more freely under pressure from T-cell immunity.
Supplementary Material
Acknowledgements
We thank Yerkes Regional Primate Center at Emory University for the orangutan blood samples.
Footnotes
This work was supported by National Institutes of Health Grant AI031168 (to P.P.) and a Ruth L. Kirschstein National Research Service Award from National Institutes of Health (to A.M.O.A.). We also acknowledge the support of the Yerkes Center Base Grant RR000165.
References
- 1.Ciccone E, Pende D, Viale O, Di Donato C, Tripodi G, Orengo AM, Guardiola J, Moretta A, Moretta L. Evidence of a natural killer (NK) cell repertoire for (allo) antigen recognition: definition of five distinct NK-determined allospecificities in humans. J. Exp. Med. 1992;175:709–718. doi: 10.1084/jem.175.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colonna M, Spies T, Strominger JL, Ciccone E, Moretta A, Moretta L, Pende D, Viale O. Alloantigen recognition by two human natural killer cell clones is associated with HLA-C or a closely linked gene. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7983–7985. doi: 10.1073/pnas.89.17.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Botet M, Moretta L, Strominger J. NK-cell receptors and recognition of MHC class I molecules. Immunol. Today. 1996;17:212–214. doi: 10.1016/0167-5699(96)30009-1. [DOI] [PubMed] [Google Scholar]
- 4.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 5.D'Andrea A, Chang C, Franz-Bacon K, McClanahan T, Phillips JH, Lanier LL. Molecular cloning of NKB1. A natural killer cell receptor for HLA-B allotypes. J. Immunol. 1995;155:2306–2310. [PubMed] [Google Scholar]
- 6.Lanier L. NK cell receptors. Annu. Rev. Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 7.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati MS, Vitale M, Bottino C, Moretta L, Moretta A, Long EO. Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra-and intracellular domains. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 8.Trowsdale J, Moffett A. NK receptor interactions with MHC class I molecules in pregnancy. Semin. Immunol. 2008;20:317–320. doi: 10.1016/j.smim.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu. Rev. Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 10.Purdy AK, Campbell KS. Natural killer cells and cancer: regulation by the killer cell Ig-like receptors (KIR) Cancer Biol. Ther. 2009;8:2211–2220. doi: 10.4161/cbt.8.23.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uhrberg M. Shaping the human NK cell repertoire: an epigenetic glance at KIR gene regulation. Mol. Immunol. 2005;42:471–475. doi: 10.1016/j.molimm.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 12.Boyton RJ, Altmann DM. Natural killer cells, killer immunoglobulin-like receptors and human leucocyte antigen class I in disease. Clin. Exp. Immunol. 2007;149:1–8. doi: 10.1111/j.1365-2249.2007.03424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin. Immunol. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 15.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari MC, Moretta L. Receptors for HLA class-I molecules in human natural killer cells. Annu. Rev. Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 16.Andersson S, Fauriat C, Malmberg JA, Ljunggren HG, Malmberg KJ. KIR acquisition probabilities are independent of self-HLA class I ligands and increase with cellular KIR expression. Blood. 2009;114:95–104. doi: 10.1182/blood-2008-10-184549. [DOI] [PubMed] [Google Scholar]
- 17.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagne F, Ugolini S, Vivier E. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, Moretta L, Moretta A. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur. J. Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 21.Fauriat C, Ivarsson MA, Ljunggren HG, Malmberg KJ, Michaelsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood. 2010;115:1166–1174. doi: 10.1182/blood-2009-09-245746. [DOI] [PubMed] [Google Scholar]
- 22.Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, Gauthier L, Romagne F, Ferracci G, Arosa FA, Moretta A, Sun PD, Ugolini S, Vivier E. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J. Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 24.Dohring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A. J. Immunol. 1996;156:3098–3101. [PubMed] [Google Scholar]
- 25.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, Rowland-Jones S, Braud VM. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur. J. Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 26.Pende D, Biassoni R, Cantoni C, Verdiani S, Falco M, di Donato C, Accame L, Bottino C, Moretta A, Moretta L. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J. Exp. Med. 1996;184:505–518. doi: 10.1084/jem.184.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, Gleimer M, Hammond JA, Guethlein LA, Bushnell DA, Robinson PJ, Parham P. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J. Exp. Med. 2009;206:2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parham P, Abi-Rached L, Matevosyan L, Moesta AK, Norman PJ, Older Aguilar AM, Guethlein LA. Primate-Specific Regulation of Natural Killer Cells. J. Med. Primatol. 2010 doi: 10.1111/j.1600-0684.2010.00432.x. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond JA, Guethlein LA, Abi-Rached L, Moesta AK, Parham P. Evolution and survival of marine carnivores did not require a diversity of killer cell Ig-like receptors or Ly49 NK cell receptors. J. Immunol. 2009;182:3618–3627. doi: 10.4049/jimmunol.0803026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guethlein LA, Abi-Rached L, Hammond JA, Parham P. The expanded cattle KIR genes are orthologous to the conserved single-copy KIR3DX1 gene of primates. Immunogenetics. 2007;59:517–522. doi: 10.1007/s00251-007-0214-x. [DOI] [PubMed] [Google Scholar]
- 31.Barrow AD, Trowsdale J. The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunol. Rev. 2008;224:98–123. doi: 10.1111/j.1600-065X.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- 32.Averdam A, Petersen B, Rosner C, Neff J, Roos C, Eberle M, Aujard F, Munch C, Schempp W, Carrington M, Shiina T, Inoko H, Knaust F, Coggill P, Sehra H, Beck S, Abi-Rached L, Reinhardt R, Walter L. A novel system of polymorphic and diverse NK cell receptors in primates. PLoS Genet. 2009;5:e1000688. doi: 10.1371/journal.pgen.1000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cadavid LF, Lun CM. Lineage-specific diversification of killer cell Ig-like receptors in the owl monkey, a New World primate. Immunogenetics. 2009;61:27–41. doi: 10.1007/s00251-008-0342-y. [DOI] [PubMed] [Google Scholar]
- 34.Guethlein LA, Older Aguilar AM, Abi-Rached L, Parham P. Evolution of Killer Cell Ig-Like Receptor (KIR) Genes: Definition of an Orangutan KIR Haplotype Reveals Expansion of Lineage III KIR Associated with the Emergence of MHC-C. J. Immunol. 2007;179:491–504. doi: 10.4049/jimmunol.179.1.491. [DOI] [PubMed] [Google Scholar]
- 35.Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. 2004;172:356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- 36.Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J. Exp. Med. 1999;189:1093–1100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu YR, Tian XH, Wang Y, Feng MF. Rapid production of human KIR2DL4 extracellular domain and verification of its interaction with HLA-G. Biochemistry (Mosc.) 2006;71 Suppl 1:S60–S64. 64–65. doi: 10.1134/s0006297906130104. [DOI] [PubMed] [Google Scholar]
- 38.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J. Exp. Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J. Exp. Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barber LD, Percival L, Valiante NM, Chen L, Lee C, Gumperz JE, Phillips JH, Lanier LL, Bigge JC, Parekh RB, Parham P. The inter-locus recombinant HLA-B*4601 has high selectivity in peptide binding and functions characteristic of HLA-C. J. Exp. Med. 1996;184:735–740. doi: 10.1084/jem.184.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biassoni R, Falco M, Cambiaggi A, Costa P, Verdiani S, Pende D, Conte R, Di Donato C, Parham P, Moretta L. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by "group 2" or "group 1" NK clones. J Exp Med. 1995;182:605–609. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG, Canavez F, Cooper SL, Valiante NM, Lanier LL, Parham P. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12:687–698. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- 43.Moesta AK, Abi-Rached L, Norman PJ, Parham P. Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig-like receptor recognition of the MHC-C1 and MHC-C2 epitopes. J. Immunol. 2009;182:3628–3637. doi: 10.4049/jimmunol.0803401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J. Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 45.Trundley AE, Hiby SE, Chang C, Sharkey AM, Santourlidis S, Uhrberg M, Trowsdale J, Moffett A. Molecular characterization of KIR3DL3. Immunogenetics. 2006;57:904–916. doi: 10.1007/s00251-005-0060-7. [DOI] [PubMed] [Google Scholar]
- 46.Adams EJ, Thomson G, Parham P. Evidence for an HLA-C-like locus in the orangutan Pongo pygmaeus. Immunogenetics. 1999;49:865–871. doi: 10.1007/s002510050566. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook JG, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, Parham P, Carrington M, Beck S. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guethlein LA, Flodin LR, Adams EJ, Parham P. NK Cell Receptors of the Orangutan (Pongo pygmaeus): A Pivotal Species for Tracking the Coevolution of Killer Cell Ig-Like Receptors with MHC-C. J. Immunol. 2002;169:220–229. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- 50.Winter CC, Long EO. Binding of Soluble KIR-Fc Fusion Proteins to HLA Class I. Natural Killer Cell Protocols: Cellular and Molecular Methods. 2000:239–250. doi: 10.1385/1-59259-044-6:239. [DOI] [PubMed] [Google Scholar]
- 51.El-Awar N, Lee J, Terasaki PI. HLA antibody identification with single antigen beads compared to conventional methods. Hum. Immunol. 2005;66:989–997. doi: 10.1016/j.humimm.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 52.Pei R, Lee JH, Shih NJ, Chen M, Terasaki PI. Single human leukocyte antigen flow cytometry beads for accurate identification of human leukocyte antigen antibody specificities. Transplantation. 2003;75:43–49. doi: 10.1097/00007890-200301150-00008. [DOI] [PubMed] [Google Scholar]
- 53.Barnstable CJ, Bodmer WF, Brown G, Galfre G, Milstein C, Williams AF, Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 54.Brodsky FM, Parham P. Evolution of HLA antigenic determinants: species cross-reactions of monoclonal antibodies. Immunogenetics. 1982;15:151–166. doi: 10.1007/BF00621948. [DOI] [PubMed] [Google Scholar]
- 55.Brodsky FM, Parham P. Monomorphic anti-HLA-A,B,C monoclonal antibodies detecting molecular subunits and combinatorial determinants. J. Immunol. 1982;128:129–135. [PubMed] [Google Scholar]
- 56.Robertson MJ, Cochran KJ, Cameron C, Le JM, Tantravahi R, Ritz J. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp. Hematol. 1996;24:406–415. [PubMed] [Google Scholar]
- 57.Carr WH, Pando MJ, Parham P. KIR3DL1 Polymorphisms That Affect NK Cell Inhibition by HLA-Bw4 Ligand. J Immunol. 2005;175:5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 58.Shimizu Y, DeMars R. Production of human cells expressing individual transferred HLA-A,-B,-C genes using an HLA-A,-B,-C null human cell line. J. Immunol. 1989;142:3320–3328. [PubMed] [Google Scholar]
- 59.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol. Immunol. 2002;38:1007–1021. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]
- 60.Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J. Immunol. 1997;158:4026–4028. [PubMed] [Google Scholar]
- 61.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 62.Fadda L, Borhis G, Ahmed P, Cheent K, Pageon SV, Cazaly A, Stathopoulos S, Middleton D, Mulder A, Claas FH, Elliott T, Davis DM, Purbhoo MA, Khakoo SI. Peptide antagonism as a mechanism for NK cell activation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10160–10165. doi: 10.1073/pnas.0913745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lanier LL, Bakker AB. The ITAM-bearing transmembrane adaptor DAP12 in lymphoid and myeloid cell function. Immunol. Today. 2000;21:611–614. doi: 10.1016/s0167-5699(00)01745-x. [DOI] [PubMed] [Google Scholar]
- 64.Carr WH, Rosen DB, Arase H, Nixon DF, Michaelsson J, Lanier LL. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J. Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vales-Gomez M, Reyburn HT, Erskine RA, Strominger J. Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14326–14331. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat. Immunol. 2001;2:452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- 67.Wan AM, Ennis P, Parham P, Holmes N. The primary structure of HLA-A32 suggests a region involved in formation of the Bw4/Bw6 epitopes. J. Immunol. 1986;137:3671–3674. [PubMed] [Google Scholar]
- 68.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O'Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 69.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O'Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borrego F, Masilamani M, Marusina AI, Tang X, Coligan JE. The CD94/NKG2 family of receptors: from molecules and cells to clinical relevance. Immunol. Res. 2006;35:263–278. doi: 10.1385/IR:35:3:263. [DOI] [PubMed] [Google Scholar]
- 71.Shum BP, Flodin LR, Muir DG, Rajalingam R, Khakoo SI, Cleland S, Guethlein LA, Uhrberg M, Parham P. Conservation and variation in human and common chimpanzee CD94 and NKG2 genes. J. Immunol. 2002;168:240–252. doi: 10.4049/jimmunol.168.1.240. [DOI] [PubMed] [Google Scholar]
- 72.Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunol. Rev. 2001;183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x. [DOI] [PubMed] [Google Scholar]
- 73.Stewart CB, Disotell TR. Primate evolution - in and out of Africa. Curr. Biol. 1998;8:R582–R588. doi: 10.1016/s0960-9822(07)00367-3. [DOI] [PubMed] [Google Scholar]
- 74.Benton MJ, Donoghue PC. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- 75.Miller JS, McCullar V. Human natural killer cells with polyclonal lectin and immunoglobulinlike receptors develop from single hematopoietic stem cells with preferential expression of NKG2A and KIR2DL2/L3/S2. Blood. 2001;98:705–713. doi: 10.1182/blood.v98.3.705. [DOI] [PubMed] [Google Scholar]
- 76.Shilling HG, McQueen KL, Cheng NW, Shizuru JA, Negrin RS, Parham P. Reconstitution of NK cell receptor repertoire following HLA-matched hematopoietic cell transplantation. Blood. 2003;101:3730–3740. doi: 10.1182/blood-2002-08-2568. [DOI] [PubMed] [Google Scholar]
- 77.Cooley S, Xiao F, Pitt M, Gleason M, McCullar V, Bergemann TL, McQueen KL, Guethlein LA, Parham P, Miller JS. A subpopulation of human peripheral blood NK cells that lacks inhibitory receptors for self-MHC is developmentally immature. Blood. 2007;110:578–586. doi: 10.1182/blood-2006-07-036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Freud AG, Becknell B, Roychowdhury S, Mao HC, Ferketich AK, Nuovo GJ, Hughes TL, Marburger TB, Sung J, Baiocchi RA, Guimond M, Caligiuri MA. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22:295–304. doi: 10.1016/j.immuni.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 79.Freud AG, Caligiuri MA. Human natural killer cell development. Immunol. Rev. 2006;214:56–72. doi: 10.1111/j.1600-065X.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- 80.Barclay AN, Hatherley D. The counterbalance theory for evolution and function of paired receptors. Immunity. 2008;29:675–678. doi: 10.1016/j.immuni.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat. Rev. Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 82.Gendzekhadze K, Norman PJ, Abi-Rached L, Graef T, Moesta AK, Layrisse Z, Parham P. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A. 2009;106:18692–18697. doi: 10.1073/pnas.0906051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parham P. The genetic and evolutionary balances in human NK cell receptor diversity. Semin. Immunol. 2008;20:311–316. doi: 10.1016/j.smim.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Levitt M, Perutz MF. Aromatic rings act as hydrogen bond acceptors. J. Mol. Biol. 1988;201:751–754. doi: 10.1016/0022-2836(88)90471-8. [DOI] [PubMed] [Google Scholar]
- 85.Collins EJ, Riddle DS. TCR-MHC docking orientation: natural selection, or thymic selection? Immunol. Res. 2008;41:267–294. doi: 10.1007/s12026-008-8040-2. [DOI] [PubMed] [Google Scholar]
- 86.Seiffert ER, Simons EL, Clyde WC, Rossie JB, Attia Y, Bown TM, Chatrath P, Mathison ME. Basal anthropoids from Egypt and the antiquity of Africa's higher primate radiation. Science. 2005;310:300–304. doi: 10.1126/science.1116569. [DOI] [PubMed] [Google Scholar]
- 87.Tavare S, Marshall CR, Will O, Soligo C, Martin RD. Using the fossil record to estimate the age of the last common ancestor of extant primates. Nature. 2002;416:726–729. doi: 10.1038/416726a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.