Abstract
Background. Uraemic toxins in the 8 to 60 kDa molecular weight range have been attracting increasing attention in dialysis therapy. However, there are no available standardized methods to evaluate their removal. Using new filtering membranes, we evaluated SDS–PAGE of spent dialysate to assess cut-off ranges and removal capacities into dialysate, while also measuring classical markers of dialyser function.
Methods. Eighteen dialysis patients were washed out for 2 weeks with FX 100 (Helixone®), followed by randomization to Xevonta Hi 23 (Amembris®) or FX dialysers for 2 weeks, then crossed over for an additional 2 weeks, and finally placed on Xenium 210 (Purema®) for 2 weeks. SDS–PAGE scanning of the removed proteins contained in the spent dialysate was performed during all dialysis sessions. Total mass of urea, creatinine, total proteins, beta 2 microglobulin (β2m), retinol-binding protein (RBP) and albumin were measured. The reduction rates of serum urea, creatinine, β2m, leptin, RBP, alpha 1-antitrypsin, albumin and total proteins were also determined.
Results. SDS–PAGE scanning identified four major protein peaks (10–18, 20–22.5, 23–30 and 60–80 kDa molecular weight) and showed clear differences in the amounts of removed proteins between the dialysers, particularly in the 20–22.5, 23–30 and 60–80 kDa ranges. Total mass of removed β2m, RBP and albumin were in agreement with SDS–PAGE, while serum assays showed differing results.
Conclusions. SDS–PAGE scanning provided a good characterization of protein patterns in the spent dialysate; it extended and agreed with protein determinations and allowed a better assessment of dialyser performance in removing 10 to 80 kDa molecular weight substances. It also identified differences between the three mainly filtrating polysulfone dialysers that were not detected with blood measurements.
Keywords: haemodialysis, middle molecules, protein removal into dialysate, SDS–PAGE, spent dialysate
Introduction
Urea levels have classically been used as a marker of dialysis adequacy [1]. However, there is an impressive list of solutes that potentially contribute to uraemic toxicity [2], and this list is still growing [3]. Evidence for participation of these newly identified culprits in the development of uraemic syndrome and their association with morbidity and mortality has directed attention to other markers in the 8 to 60 kDa molecular weight range that may help to assess dialysis adequacy [4].
Molecules that are difficult to remove by dialysis, such as these larger middle molecular weight compounds and protein-bound molecules, play a significant role in uraemic toxicity [5]. Dialysis manufacturers have developed new dialysers having improved geometry and new membranes that enhance removal of middle molecular weight molecules with the aim of improving clinical outcomes [6,7]. However, there are presently no accepted methods that allow easy and precise measurement of removal capacities of uraemic retention solutes in the 8 to 60 kDa molecular weight range by these new generation dialysers. To evaluate dialyser performance, most authors determined variations in blood levels of particular proteins [8]; however, the majority of manufacturers limit information about dialyser performance to variations in blood levels of beta 2 microglobulin (β2m) which represents quite a restraint amount of information [4].
SDS–PAGE, which was first proposed by Laemmli in 1970 [9], is a reliable method for protein characterization that is easy to use and has been rendered widely available in the clinical setting. The aim of the present study was to assess the usefulness of scanning SDS–PAGE profiles of proteins contained in spent dialysate to evaluate the removal capacities of the new generation high-flux polysulfone dialysers in vivo during routine clinical haemodialysis.
Materials and methods
Patients
Eighteen stable dialysis patients treated in the dialysis centre at Néphrologie Dialyse St Guilhem in Sète were included in the study. They were dialysed three times a week with fully equipped AK200S machines (Gambro, Lund, Sweden) using ultrapure double reverse osmosis water and a measured dialysate flow of 500 ± 10 mL/min. They had been on dialysis for more than 3 months and had no active disease at the time of testing. The study was explained to the patients and they gave informed consent to participate in the protocol. This study was approved by the Comité de Protection des Personnes of Nîmes (2009.01.07 bis) and was given a registration number at the French Agency AFSSAPS 2008-A01612-53.
Study design
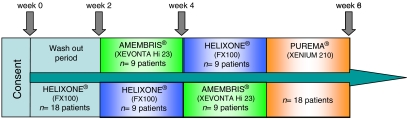
A schematic of the study design is given in Figure 1. Three new generation polysulfone dialysers were assessed. The complete in vitro characteristics of these dialysers are given in Table 1. After 2 weeks of wash out with FX 100 (Helixone® membrane, Fresenius Medical Care AG, Bad Homburg, Germany), the patients were randomized to Xevonta (Xevonta Hi 23, Amembris® membrane, B. Braun Avitum, Melsungen, Germany) (n = 9) or FX dialysers (n = 9) for 2 weeks and then crossed over for an additional 2 weeks. The eighteen patients were then switched to Xenium dialysers (Xenium 210, Purema® membrane, Baxter Healthcare Corporation, McGaw Park, IL, USA) for a final 2 weeks. The patients received routine dialysis prescription criteria that remained constant throughout the study period.
Fig. 1.
Schematic of the study protocol.
Table 1.
Dialyser characteristics provided by the manufacturers from in vitro data
| Dialyser | |||
|---|---|---|---|
| Type | FX class 100 | Xenium 210 | Xevonta Hi 23 |
| Membrane (synthetic) | Helixone® | Purema® | Amembris® |
| Wall thickness/inner diameter (μm) | 35/185 | 30/200 | 35/195 |
| Surface (m2) | 2.2 | 2.1 | 2.3 |
| In vitro KUF | 73 | 80 | 124 |
| In vitro clearances | |||
| (Qb/Qd: 300/500 mL/min) | |||
| Urea | 278 | 285 | 279 |
| Creatinine | 261 | 272 | 276 |
| Phosphate | 248 | 253 | 277 |
| Vitamin B12 (1.4 kDa) | 192 | 200 | 204 |
| Inulin (5 kDa) | 142 | NA | 144 |
| Myoglobin (16.7 kDa) | NA | 55 | NA |
| Sieving coefficient | |||
| Vitamin B12 (1.4 kDa) | NA | 0.99 | NA |
| Inulin (5 kDa) | 1 | 0.89 | 1.0 |
| β2 microglobulin (11.8 kDa) | 0.8 | NA | > 0.8 |
| Myoglobin (16.7 kDa) | NA | 0.24 | NA |
| Albumin (65 kDa) | 0.001 | < 0.01 | < 0.001 |
NA, not available.
Adsorption studies
Polysulfone membranes are classically considered as filtrating membranes. In addition to filtering, we also examined the adsorptive capacities of the new generation polysulfone membranes used in this study.
Helixone®, Purema® and Amembris® dialysers were assessed for adsorption using the technique by Mares et al. [10]. At the end of dialysis procedures, after returning the blood to the patients, the dialysers were further rinsed with 2 L of saline. After draining, the dialysers were refilled with 3 mM EDTA (EDTA/PBS, pH 7.4), and the solution (volume 144 ± 26 mL) was recirculated at 80 mL/min for 30 min at room temperature to detach and remove adhering leukocytes. The dialysers were then drained and refilled with 40% acetic acid (volume 195 ± 15 mL), which was recirculated at 80 mL/min for 30 min at room temperature. The eluate was centrifuged to remove cellular detritus and albumin. Then, β2m and total protein concentration were determined in the supernatant as described below.
Results of the protein analysis from the adsorption experiments are shown in Table 2. These data indicate that the amount of total proteins adsorbed was always < 5% of the total amount of proteins that were removed into the dialysate, confirming the filtrating abilities of the new polysulfone dialysers.
Table 2.
Adsorptive capacities of the three dialysers used in the study
| Dialyser | Adsorbed proteins (% of dialysate protein mass) | Adsorbed albumin (% of dialysate albumin mass) | Adsorbed β2m (% of dialysate β2m mass) |
|---|---|---|---|
| FX 100 (Helixone) | 1.9% | 0.5% | 1.0% |
| FX 100 (Helixone) | 2.5% | 0.3% | 0.1% |
| Xenium 210 (Purema) | 1.2% | 0.1% | 0.2% |
| Xenium 210 (Purema) | 3.5% | 2.2% | 0.8% |
| Xevonta Hi 23 (Amembris) | 0.8% | 0.2% | 1.0% |
| Xevonta Hi 23 (Amembris) | 1.4% | 0.4% | 1.4% |
| Mean | 1.9% | 0.6% | 0.7% |
| SE | 0.4% | 0.3% | 0.2% |
| Max | 3.5% | 2.2% | 1.4% |
| Min | 0.8% | 0.1% | 0.1% |
Dialysate studies
Dialysate samples and solute mass removal assessment. In order to assess total balance of the different substances from serum, continuous sampling of spent dialysate (CSSD) was performed during each dialysis treatment as previously reported by our group [11]. Samples were stored at − 80°C until analysis. Urea, creatinine, total protein, albumin (67 kDa), retinol-binding protein (RBP) (21.2 kDa) and β2m (11.8 kDa) were determined from the collected spent dialysate, and the total mass removed of a given substance was calculated by multiplying the measured concentration by the total volume of dialysate that passed through the dialyser.
The three proteins selected are representative of three of the four molecular weight ranges of interest (see the SDS–PAGE analyses of the Results section) and provided an internal control for quantification of protein content assessed by SDS–PAGE. Since some of the proteins had a concentration under the sensitivity threshold for their respective assays, the spent dialysate was precipitated with 10% trichloroacetic acid.
RBP and leptin were determined by ELISA using specific antibodies (RD Systems, Lille, France). Albumin was determined by immunoturbidimetry. The sensitivity threshold and the linearity range of the measurements of the different substances are given in Table 3.
Table 3.
Laboratory analysis methods and their sensitivity and linearity
| Substance | Measurement method | Linearity | Sensitivity | Dialysate (mean ± SD) [min–max] | Blood (mean ± SD) [min–max] |
|---|---|---|---|---|---|
| Urea (mmol/L) | UV cinetic | 0.8–50 | 0.38 | 4.6 ± 1.4 [1.3–9.3] | 12 ± 8 [1.6–31] |
| Creatinine (μmol/L) | Compensated Jaffe | 5–2000 | 2.4 | 85 ± 2 [23–173] | 406 ± 230 [73–939] |
| β2 microglobulin (mg/L) | Immunoturbidimetry | 0.5–16 | 0.06 | 6.5 ± 7.1 [0.06–37] | 13.5 ± 5.6 [3.2–23] |
| Total proteins (dialysate) (g/L) | Photometric colour | 0.01–2.00 | 0.007 | 0.10 ± 0.06 [0.01–0.44] | |
| Total proteins (dialysate) (mg/L) | Bradford | 0–500 | 0.002 | 98 ± 60 [0.002–272] | |
| Total proteins (serum) (g/L) | Photometric colour | 30–120 | 0.77 | 67 ± 7 [52–84] | |
| Albumin ‘micro’ assays (mg/L) | Immunoturbidimetry | 5–300 | 0.46 | 12 ± 14 [0.1–113] | |
| α1-Antitrypsin (g/L) | Immunoturbidimetry | 0.3–5.0 | 0.01 | 1.5 ± 0.3 [0.84–2.44] | |
| Leptin (μg/L)a | ELISA | 0.0156–1 | 0.0078 | 25 ± 47 [0.6–386] | |
| RBP (mg/L)a | ELISA | 1.56–100 | 0.224 | 13 ± 7 [1.4–44] | 178 ± 96 [55–667] |
Leptin and RBP were diluted before measurement in order to be in the linearity range.
SDS–PAGE and scanning of protein profiles
SDS–PAGE of desalted dialysate was performed according to the method described by Laemmli [9] using a Bio-Rad system (Bio-Rad laboratories, CA, USA).
Total protein concentrations were assessed from all dialysis sessions using the Bradford method adapted for the low concentration range as previously described [12]. For each patient, the dialysate obtained from six different dialyses with the same dialyser was pooled and submitted to SDS–PAGE. This decreased the variability and the sensitivity of the method, but increased the specificity of the analyses such that observed differences were more likely to be real. Approximately 10 μg of proteins in 2% SDS buffer solution were run in a 12.5% SDS–PAGE and then stained with Coomassie blue. SDS–PAGE gels were scanned with an Epson Perfection 4990 PHOTO (Epson, CA, USA), and the surfaces under the curve of optical density of the electrophoretic bands for the specified molecular weight ranges were calculated using WCIF Image J, 1.37 software (WCIF, ON, Canada). The molecular weight range of the scanning procedure was normalized by following the migration of the molecular weight markers and was divided in 500 readings. The percentage of density for each reading over the entire density value was calculated. The amount of protein contained in each molecular weight range was obtained by multiplying the percentage reading by the total protein level of the dialysate obtained with the Bradford assay and is expressed as the percentage of Coomassie blue-stained proteins.
Blood studies
Blood was sampled before and after mid-week dialysis sessions. In addition to the compounds measured in the dialysate studies, alpha 1-antitrypsin (α1AT) (55 kDa) (immunoturbidimetry) and leptin (16 kDa) (ELISA) were determined. Post-dialysis values were corrected using the formula of Bergstrom and Wehle [13].
Clearances (K) were obtained from dialysate mass (M), mean serum concentration (Cm) and time (T).
Kt/V was calculated from serum urea level as follows:
[14].
Dialysance was obtained from DiascanTM and was iteratively measured during dialysis.
Statistics
Statistical analyses were performed using SAS V9.01 package (SAS Corporation, N Cary, USA). Differences in the continuous variables among the three different dialysers were assessed using a variance analysis that takes into account the repetitive nature of the data. We used a Fisher’s test with a covariance matrix model and compound symmetry that uses patients as a random effect and the dialysers as a fixed effect. Bonferroni’s test was used to assess differences between two of the three groups.
P-values < 0.05 were considered significant. Values are expressed as means ± standard error of the mean.
Results
The dialysis characteristics and the ability of the three dialysers to remove small molecular weight substances are presented in Table 4. They were globally equivalent in their removal capacities of small molecular weight substances.
Table 4.
Dialysis characteristics and removal of small molecular weight solutes
| Amembris® | Helixone® | Purema® | |
|---|---|---|---|
| Patient characteristics | |||
| Age (years) | 79 ± 1.7 | ||
| BW (kg) | |||
| Before | 68.8 ± 2.5 | 68.8 ± 2.5 | 68.4 ± 2.4 |
| After | 66.3 ± 2.3 | 66.2 ± 2.3 | 65.8 ± 2.3 |
| WL | 2.5 ± 0.3 | 2.6 ± 0.3 | 2.6 ± 0.2 |
| Dialysis characteristics | |||
| t (min) | 222 ± 3 | 222 ± 3 | 223 ± 3 |
| Qb (mL/min) | 317 ± 2 | 318 ± 2 | 319 ± 2 |
| Qd (mL/min) | 500 ± 10 | 500 ± 10 | 500 ± 10 |
| Urea | |||
| Blood before (mmol/L) | 19.6 ± 0.9 | 18.4 ± 0.8 | 18.0 ± 0.9 |
| Blood after (mmol/L) | 4.9 ± 0.3 | 4.5 ± 0.2 | 4.7 ± 0.3 |
| Blood RR (%) | 75.1 ± 0.8 | 75.2 ± 0.8 | 74.2 ± 0.6 |
| Kt/V | 1.41 ± 0.03 | 1.41 ± 0.03 | 1.37 ± 0.03 |
| Total mass removed (mmol/session) | 528 ± 20 | 533 ± 18 | 480 ± 20 |
| K (mL/min) | 217 ± 5 | 217 ± 4 | 207 ± 2a |
| D (mL/min) | 219 ± 2 | 218 ± 2 | 213 ± 1a |
| Creatinine | |||
| Blood before (μmol/L) | 603 ± 25 | 620 ± 24 | 600 ± 25 |
| Blood after (μmol/L) | 196 ± 10 | 199 ± 9 | 197 ± 9 |
| Blood RR (%) | 67.6 ± 0.8 | 68.3 ± 0.8 | 67.1 ± 0.9 |
| Total mass removed (μmol/session) | 9645 ± 319 | 9862 ± 326 | 9292 ± 346 |
| K (mL/min) | 119 ± 3 | 115 ± 3 | 113 ± 2 |
P < 0.05.
BW, body weight; WL, weight loss during dialysis; Qb, blood flow; Qd, dialysate flow; RR, reduction ratio; K, clearance; D, ionic dialysance; t, time; V, urea distribution volume.
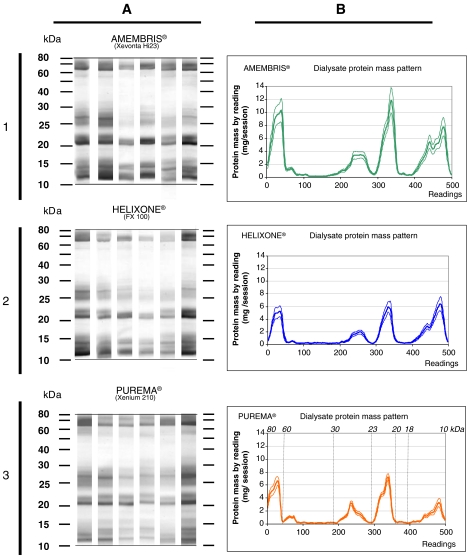
SDS–PAGE of spent dialysate
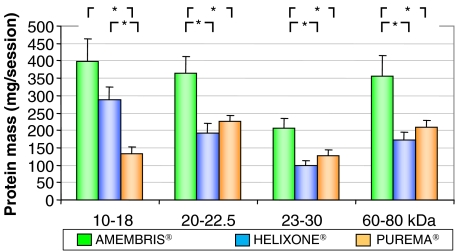
Examples of SDS–PAGE of proteins obtained from the spent dialysate from the different dialysers are presented in Figure 2A. A plotting of the density readings, presented as means ± SEM from the 80 to 10 kDa molecular weight range, is shown in Figure 2B. There were four major peaks corresponding to the 10 to 18, 20 to 22.5, 23 to 30 and 60 to 80 kDa molecular weight ranges. The variability on the density readings from patient to patient was quite low as indicated by the narrow SEM range. The amount of Coomassie blue-stained protein contained in each of these peaks, determined by scanning, is represented in Figure 3. These data indicate that Amembris® removed significantly more protein and particularly those contained in the 20 to 22.5, 23 to 30 and 60 to 80 molecular weight ranges.
Fig. 2.
(A) Examples of 12.5% SDS–PAGE of the proteins in the spent dialysate from the three dialysers tested in the study [1—Xevonta Hi 23 (Amembris®); 2—FX 100 (Helixone®); 3—Xenium 210 (Purema®)]. (B) Scanning profiles of the SDS–PAGE gels of the different dialysers. The thick line represents the mean value from 18 patients and the thin line is the ± SEM. Protein amount in milligram is represented vertically and molecular weight is represented horizontally. The units of the readings have been normalized and arbitrarily fixed from 0 to 500. The correspondence from the reading to molecular weight was not linear and is presented on top of the graph. Four different peaks were observed that were at the same molecular weight ranges (10–18, 20–22.5, 23–30 and 60–80 kDa) which had different heights for the different dialysers. Numbering of the panels corresponds as follows: 1—Xevonta Hi 23 (Amembris®); 2—FX 100 (Helixone®); 3—Xenium 210 (Purema®).
Fig. 3.
Amount of proteins removed in the four SDS–PAGE peaks. Bar chart of the amount of proteins contained in each of the four peaks observed with SDS–PAGE scanning. Note that the total amount of proteins removed with molecular weight < 30 kDa was higher than the amount contained in the 60–80 kDa molecular weight peak for the three dialysers, indicating that all the dialysers had a sharp cut-off, which prevented massive albumin loss. [Only 10% of the total proteins was albumin (see Figure 4)].
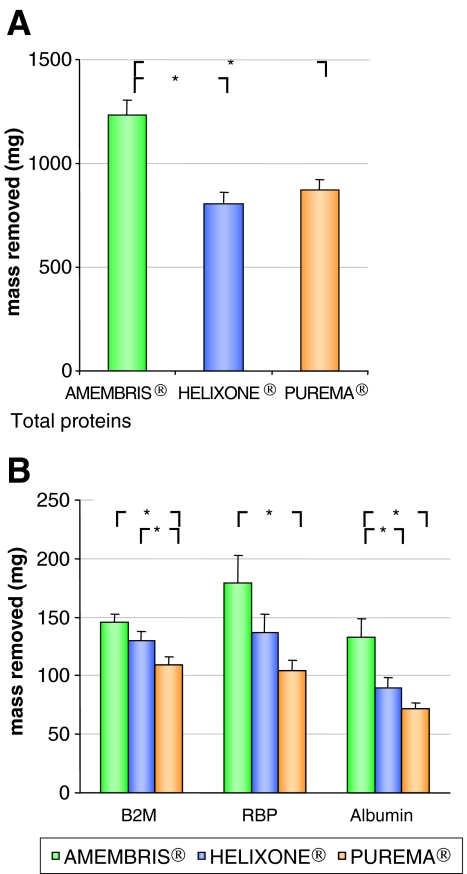
Single protein determinations and blood studies
Single protein determination in the spent dialysate confirmed the SDS–PAGE findings for the different molecular weight ranges (Figure 4). The actual numbers and comparisons are given in Table 5. Amembris® and Helixone® removed significantly more β2m than the Purema® dialyser, and Amembris® removed significantly more albumin and total proteins than the other two dialysers. Indeed, when analysing the 20–22.5 kDa molecular weight range, Amembris® removed significantly more proteins than the other two dialysers. However, when RBP was included as a serum protein having a molecular weight in this molecular weight range, there was no significant difference in removal into dialysate between the Amembris® and Helixone® dialysers. Thus, there may be differences between SDS–PAGE results and results from specific determinations of proteins having a weight in a selected molecular weight range. This difference is due, at least in part, to the fact that SDS–PAGE scanning assesses all Coomassie blue-stained proteins in that molecular weight range and not just one protein. Thus, it may be that RBP is not representative of the total amount of proteins in the molecular weight range found in the spent dialysate.
Fig. 4.
Amount of specific proteins in the spent dialysate. Bar charts of the amount of total proteins (A) and amount of β2m, retinol-binding protein (RBP) and albumin removed in the spent dialysate obtained by CSSD (B).
Table 5.
Middle molecular weight solute removal
| Dialyser |
|||
|---|---|---|---|
| Xevonta Hi 23 | FX 100 | Xenium 210 | |
| Amembris® | Helixone® | Purema® | |
| β2m | |||
| Blood before (mg/L) | 18.4 ± 0.4 | 19.3 ± 0.4 | 18.6 ± 0.4 |
| Corrected blood after (mg/L) | 6.6 ± 0.3 | 8.0 ± 0.3 | 7.5 ± 0.3a,b |
| Blood RR (%) | 64.3 ± 0.9 | 58.9 ± 1.0 | 59.9 ± 1.1a,b |
| Dialysate concentration (mg/L) | 1.29 ± 0.05 | 1.14 ± 0.07 | 0.99 ± 0.05b |
| Total dialysate mass (mg) | 146 ± 6 | 130 ± 8 | 110 ± 7b,c |
| K (mL/min) | 51 ± 3 | 54 ± 3 | 30 ± 2b,c |
| Leptin | |||
| Blood before (μg/L) | 34.7 ± 12.3 | 43.0 ± 17.5 | 26.0 ± 6.5 |
| Corrected blood after (mg/L) | 15.4 ± 5.1 | 18.2 ± 7.0 | 13.4 ± 2.9 |
| Blood RR (%) | 31.8 ± 4.7 | 27.7 ± 5.2 | 28.7 ± 4.4 |
| RBP | |||
| Blood before (mg/L) | 195.5 ± 26.6 | 160.4 ± 15.0 | 206.9 ± 20.5 |
| Corrected blood after (mg/L) | 142.2 ± 18.2 | 126.0 ± 13.6 | 163.9 ± 14.7 |
| Blood RR (%) | 22.4 ± 4.0 | 20.6 ± 4.1 | 18.2 ± 2.5 |
| Dialysate concentration (mg/L) | 1.58 ± 0.20 | 1.19 ± 0.14 | 0.90 ± 0.07b |
| Total dialysate mass (mg) | 179.4 ± 23.4 | 136.5 ± 16.3 | 104.4 ± 8.7b |
| K (mL/min) | 7.0 ± 1.2 | 4.8 ± 0.6 | 3.2 ± 0.4b |
| α1-Antitrypsin | |||
| Blood before (g/L) | 1.53 ± 0.05 | 1.50 ± 0.05 | 1.42 ± 0.04 |
| Corrected blood after (mg/L) | 1.38 ± 0.04 | 1.36 ± 0.04 | 1.29 ± 0.04 |
| Blood RR (%) | 9.4 ± 0.9 | 8.9 ± 1.0 | 8.7 ± 0.9 |
| Albumin | |||
| Blood before (g/L) | 36.9 ± 0.5 | 36.5 ± 0.5 | 36.7 ± 0.4 |
| Corrected blood after (g/L) | 33.6 ± 0.6 | 33.0 ± 0.5 | 33.5 ± 0.5 |
| Blood RR (%) | 9.0 ± 0.9 | 9.2 ± 0.9 | 8.7 ± 0.8 |
| Dialysate concentration (mg/L) | 1.19 ± 0.14 | 0.80 ± 0.09 | 0.66 ± 0.04a,b |
| Total dialysate mass (mg) | 133.1 ± 15.7 | 89.4 ± 9.3 | 71.4 ± 5.4a,b |
| K (mL/min) | 0.017 ± 0.003 | 0.017 ± 0.003 | 0.008 ± 0.001b,c |
| Total proteins | |||
| Blood before (g/L) | 64.6 ± 0.8 | 61.4 ± 0.8 | 64.1 ± 0.7a,c |
| Corrected blood after (g/L) | 58.7 ± 0.9 | 55.8 ± 0.7 | 58.5 ± 0.7a,c |
| Blood RR (%) | 9.0 ± 0.8 | 8.8 ± 1.0 | 8.6 ± 0.9 |
| Dialysate concentration (mg/L) | 10.7 ± 0.7 | 7.1 ± 0.5 | 7.6 ± 0.4a,b |
| Total dialysate mass (mg) | 1209 ± 74 | 810 ± 55 | 809 ± 52a,b |
| K (mL/min) | 0.09 ± 0.01 | 0.08 ± 0.01 | 0.05 ± 0.01b,c |
P < 0.05 Xevonta vs FX.
P < 0.05 Xevonta vs Xenium.
P < 0.05 FX vs Xenium.
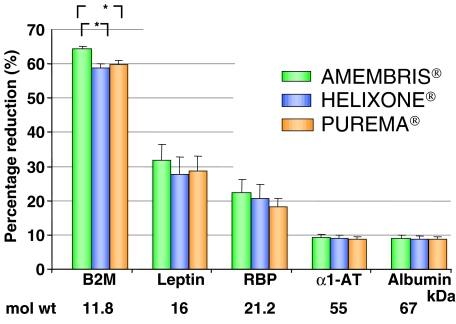
Blood levels of the selected proteins are represented in Figure 5. The three dialysers were able to decrease serum β2m levels by about 60% [Amembris® 64.3 ± 1, Helixone® 58.9 ± 1 (P < 0.01 vs Amembris®) and Purema® 59.9 ± 1 (P < 0.01 vs Amembris®)]. The per cent reduction of serum levels of compounds having higher molecular weights decreased to < 10% for α1AT and albumin, and there were no significant differences between the dialysers.
Fig. 5.
Blood measurements: reduction rate of the different serum proteins. The reduction rate for the measured proteins decreased from about 60% to < 10% as molecular weight increased from 12 to 67 kDa. β2m reduction was significantly higher with Amembris® than with the other dialysers; serum levels of the other proteins showed a similar decrease in the three dialysers.
Discussion
In the present study, we assessed the performances of three new generation, mainly filtrating, high-flux dialysers, having large surface areas, high water permeability and equivalent removal characteristics for small molecular weight compounds. The dialysers were tested in 18 patients given routine haemodialysis. We assessed both low and high molecular weight solutes (middle molecules). The protein profiles of proteins removed into the dialysate were precisely determined by gel electrophoresis techniques, and the four main peaks of proteins were quantified.
Our results show that these three dialysers had similar capacities for removal of small molecular weight solutes but differed in their abilities to remove middle molecules (molecular weight range 8 to 60 kDa) into the dialysate. SDS–PAGE scanning of the spent dialysate showed that Amembris® removed significantly more middle weight proteins during routine dialysis.
Although the results of single proteins found in spent dialysate generally agreed with those from SDS–PAGE scanning, there were differences between the two tests. For example, although Amembris® and Helixone® removed similar amounts of RBP (21 kDa molecular weight), area under the curve measurements from SDS–PAGE scanning in the 20 to 22.5 kDa range showed clear differences between the two dialysers. Since SDS–PAGE includes analysis of all the Coomassie blue-stained proteins contained in the sample, it is thought to provide a more complete assessment of protein content in the dialysate than single protein measurements.
For removal of proteins into the dialysate in the 20 to 80 kDa range, SDS–PAGE scanning again revealed differences between the three dialysers that the assessment of blood levels of single proteins did not. For instance, there were no differences in serum levels of albumin (67 kDa) between the dialysers, whereas significant differences were observed in albumin removal into the dialysate. These differences in sensitivity in measuring dialyser performance may be due to at least two factors: the different sample types (blood and spent dialysate) and the different measurements (single protein determination versus total amount of proteins included in a range of molecular weight). The analysis of the spent dialysate allowed mass estimation of removal into the dialysate, while blood analysis of selected proteins provided a concentration measurement which is affected by multiple factors (compartmental distribution of the given protein, shifts in volume distribution during dialysis and other factors). Furthermore, for some particular proteins, such as albumin, the amount removed by dialysis is so minor compared to the total pool that no visible variations in serum levels would be expected, even when clear differences in removal into the dialysate exist. It may be that the non-circulating pool and/or generation rate is such that removed protein is quickly replaced. Thus, assessment of protein removal, using blood measurements, is difficult to interpret because of the multiple factors that affect this measurement.
Alternatively, measuring the removal by determining what has left the plasma requires analyzing what is adsorbed in the dialysis membrane as well as what passes across it and is recovered in the spent dialysate. The relative participation of these two mechanisms in determining total removal may vary markedly according to membrane characteristics. PMMA is the classical example of an adsorptive membrane [15], while polysulfone is the classical example of a filtrating membrane [16–18]. Chanard et al. [16] assessed decreases in plasma radioactivity after injection of radiolabelled 131I-β2m. They showed that polysulfone was more efficient than PMMA in removing radiolabelled β2m and demonstrated clear differences in adsorption between the dialysers. The radioactivity recovered from the PMMA membrane was about 90% of that removed from the plasma, while the radioactivity recovered from the polysulfone membrane was only about 10% of that removed from the plasma, and ~ 90% had accumulated in the ultrafiltrate. These studies clearly confirm the filtrating ability of the polysulfone dialysers. Because we used three polysulfone membranes with known filtration characteristics, analysis of the spent dialysate was the best method to evaluate removal. The error induced by adsorption onto the membrane over the total mass of the proteins removed of a defined molecular weight range is minimized when using polysulfone [17,18]. In our hands, the amount of protein recovered from the dialyser membrane while assessing adsorption (see Materials and methods section and Table 2) was < 5% of the total mass of proteins that crossed the membrane and that were obtained in the spent dialysate (1.9 ± 0.4%, 0.6 ± 0.3% and 0.7 ± 0.2% for total proteins, albumin and β2m, respectively).
Proteomics have previously been proposed as a promising analytical tool for recognizing specific peptide profiles in different pathological situations [19,20], to assess uraemic toxin removal and to identify new molecules with putative influences on uraemic toxicity [21]. Weissinger et al. [21] identified 1046 and 1394 polypeptides using low- and high-flux membranes, respectively, in the ultrafiltrate of one dialysed patient. However, this technique, perfectly adapted to identifying new markers of disease, is still reserved to a few specialized laboratories and it is not, at the present time, envisaged as a toll of wide use to quantify dialysis in a repetitive way in routine clinical practice.
Although SDS–PAGE technique provides semiquantitative data, it has previously been used to test protein permeability of different dialysers at various times during dialysis sessions using silver staining and laser densitometry [22]. For example, Mann and colleagues [22] used it to identify significant differences in permeability during individual dialysis sessions. In our study, we attempted to adapt SDS–PAGE into a reliable and easy-to-use method that can be performed in any dialysis unit to assess total amounts of middle weight molecules removed during a complete dialysis session.
To make this method easier, we adopted Coomassie blue staining of the gels. Although this technique is less sensitive than silver staining, it is not influenced by as many factors, it is technically simpler, and is not as expensive. To render it more reliable, we assessed the spent dialysate obtained by the CSSD, which we have previously shown to perfectly reflect the total spent dialysate [11]. Therefore, our current results did not show the intra-dialysis variations that had previously been observed [22], allowing us to accurately evaluate dialysis performance during a complete dialysis session, regardless of intra-session variability. The sensitivity provided by our method, using a standardized optical analysis over a molecular weight scale, proved to be satisfactory since it allowed identification of clear differences between the dialysers that could not be observed using other approaches (single protein determinations and blood measurements). Finally, the reliability of the method was supported by the small range of SEM observed in our analyses.
Conclusion
In summary, the present study showed that scanning the SDS–PAGE profile of middle weight molecules contained in the spent dialysate obtained by CSSD is a reliable and easy-to-use method that allowed identification of differences in behaviour of three new generation high permeability polysulfone dialysers. Using this method, we observed (i) very high efficacy and no difference in ability to remove small molecular weight compounds; (ii) that Amembris® removed significantly more proteins at all weight ranges than Purema®, but only at higher molecular weights than Helixone®; (iii) that Amembris® and Helixone® removed significantly more β2m than Purema®, which agrees with the SDS–PAGE scanning results, and that Amembris® removed more total proteins than the other two dialysers; (iv) that β2m reduction differed among the three dialysers only when measuring serum levels of the representative proteins, indicating that serum levels may be influenced by many factors other than removal into dialysate; therefore, (v) measurement of serum levels of single proteins is not the best approach to assess uraemic toxin removal capacities by new dialysers, particularly for the middle weight molecules. Since adsorption contributes to < 5% of total protein removal and was equivalent in the three dialysers, the differences in dialyser performance were due to differences in removal into the dialysate.
Many different substances are removed from the plasma; some of these are uraemic retention solutes, some may be considered toxins [2] and some may even have beneficial properties. Therefore, to establish the benefits or drawbacks of substance removal, it will be necessary to better identify, probably using MS-based techniques, the complete array of substances removed. However, it may be easier to install and perform SDS–PAGE in a dialysis unit than other techniques that require more complex technologies. The use of SDS–PAGE scanning of spent dialysate in many dialysis units would enlarge our knowledge about how the different dialysis settings are able to remove uraemic toxins of greater importance (8–60 kDa molecular weight range).
Acknowledgments
The authors thank Priscilla Martinez and the staff of the NDSG dialysis centre for their participation and assistance during the trial. Dr Peter G Kerr’s help in reviewing the manuscript is deeply appreciated.
Conflict of interest statement. B. Braun Avitum participated in the fees generated to perform the study. The Centre de Néphrologie et de Transplantation of the Hospital ‘La Conception’ receives funding from Baxter, Fresenius, Méditor, Gambro and Hemotech for various clinical projects.
References
- 1.Lowrie EG, Laird NM, Parker TF, et al. Effect of the hemodialysis prescription of patient morbidity: report from the National Cooperative Dialysis Study. N Engl J Med. 1981;305:1176–1181. doi: 10.1056/NEJM198111123052003. [DOI] [PubMed] [Google Scholar]
- 2.Vanholder R, De Smet R, Glorieux G, et al. for the European Uremic Toxin Work Group (EUTox) Review on uremic toxins: classification, concentration, and interindividual variability. Kidney Int. 2003;63:1934–1943. doi: 10.1046/j.1523-1755.2003.00924.x. [DOI] [PubMed] [Google Scholar]
- 3. European Uremic Toxins Work Group (EUTox). Uremic Solutes Database. http://www.nephro-leipzig.de/eutoxdb/index.php (17 November 2010, last accessed date)
- 4.Vanholder R, Eloot S, Van Biesen W. Do we need new indicators of dialysis adequacy based on middle-molecule removal? Nat Clin Pract Nephrol. 2008;4:174–175. doi: 10.1038/ncpneph0750. [DOI] [PubMed] [Google Scholar]
- 5.Vanholder R, Baurmeister U, Brunet P, et al. for the European Uremic Toxin Work Group A bench to bedside view of uremic toxins. J Am Soc Nephrol. 2008;19:863–870. doi: 10.1681/ASN.2007121377. [DOI] [PubMed] [Google Scholar]
- 6.Clark WR, Ronco C. Determinants of haemodialyser performance and the potential effect on clinical outcome. Nephrol Dial Transplant. 2001;16:56–60. doi: 10.1093/ndt/16.suppl_5.56. [DOI] [PubMed] [Google Scholar]
- 7.Locatelli F, Martin-Malo A, Hannedouche T, et al. Effect of membrane permeability on survival of hemodialysis patients. J Am Soc Nephrol. 2009;20:645–654. doi: 10.1681/ASN.2008060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krieter DH, Lemke HD, Wanner C. A new synthetic dialyzer with advanced permselectivity for enhanced low-molecular weight protein removal. Artif Organs. 2008;32:547–554. doi: 10.1111/j.1525-1594.2008.00583.x. [DOI] [PubMed] [Google Scholar]
- 9.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 10.Mares J, Thongboonkerd V, Tuma Z, et al. Specific adsorption of some complement activation proteins to polysulfone dialysis membranes during hemodialysis. Kidney Int. 2009;76:404–413. doi: 10.1038/ki.2009.138. [DOI] [PubMed] [Google Scholar]
- 11.Argilés A, Ficheux A, Thomas M, et al. Precise quantification of dialysis using continuous sampling of spent dialysate and total dialysate volume measurement. Kidney Int. 1997;52:530–537. doi: 10.1038/ki.1997.364. [DOI] [PubMed] [Google Scholar]
- 12.Argilés A, Mourad G, Basset N, et al. Acute adaptative changes to unilateral nephrectomy in humans. Kidney Int. 1987;32:714–720. doi: 10.1038/ki.1987.265. [DOI] [PubMed] [Google Scholar]
- 13.Bergstrom J, Wehle B. No change in corrected beta 2-microglobulin concentration after cuprophane haemodialysis. Lancet. 1987;329:628–629. doi: 10.1016/s0140-6736(87)90266-2. [DOI] [PubMed] [Google Scholar]
- 14.Gotch FA, Sargent JA. A mechanistic analysis of the National Cooperative Dialysis Study (NCDS) Kidney Int. 1985;28:526–534. doi: 10.1038/ki.1985.160. [DOI] [PubMed] [Google Scholar]
- 15.Aoike I. Clinical Significance of protein adsorbable membranes - long-term clinical effects and analysis using a proteomic technique. Nephrol Dial Transplant. 2007;22:S13–S19. doi: 10.1093/ndt/gfm295. [DOI] [PubMed] [Google Scholar]
- 16.Chanard J, Caudwell V, Valeire J, et al. Kinetics of 131I-β2 microglobulin in hemodialysis patients: assessment using total body counting. Artif Organs. 1998;22:574–580. doi: 10.1046/j.1525-1594.1998.06197.x. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa I, Chikazawa Y, Sato K, et al. Proteomic analysis of serum, outflow dialysate and adsorbed protein onto dialysis membranes (polysulfone and PMMA) during hemodialysis treatment using SELDI-TOF-MS. Am J Nephrol. 2006;26:372–380. doi: 10.1159/000094779. [DOI] [PubMed] [Google Scholar]
- 18.Tian Q, Gomersall CD, Leung PPN, et al. The adsorption of vancomycin by polyacrylonitrile, polyamide, and polysulfone hemofilters. Artif Organs. 2008;32:81–84. doi: 10.1111/j.1525-1594.2007.00460.x. [DOI] [PubMed] [Google Scholar]
- 19.Fliser D, Novak J, Thongboonkerd V, et al. Advances in urinary proteome analysis and biomarker discovery. J Am Soc Nephrol. 2007;18:1057–1071. doi: 10.1681/ASN.2006090956. [DOI] [PubMed] [Google Scholar]
- 20.Weissinger EM, Nguyen-Khoa T, Fumeron C, et al. Effects of oral vitamin C supplementation in hemodialysis patients: a proteomic assessment. Proteomics. 2006;6:993–1000. doi: 10.1002/pmic.200500210. [DOI] [PubMed] [Google Scholar]
- 21.Weissinger EM, Kaiser T, Meert N, et al. Proteomics: a novel tool to unravel the patho-physiology of uraemia. Nephrol Dial Transplant. 2004;19:3068–3077. doi: 10.1093/ndt/gfh509. [DOI] [PubMed] [Google Scholar]
- 22.Mann H, Melzer H, Al-Bashir A, et al. Testing protein permeability of dialysis membranes using SDS-PAGE. Int J Artif Organs. 2002;25:441–446. doi: 10.1177/039139880202500515. [DOI] [PubMed] [Google Scholar]