Abstract
Organic cation transporters (OCT1-3 and OCTN1/2) facilitate cardiac uptake of endogenous compounds and numerous drugs. Genetic variants of OCTN2, for example, reduce uptake of carnitine, leading to heart failure. Whether expression and function of OCTs and OCTNs are altered by disease has not been explored in detail. We therefore studied cardiac expression, heart failure–dependent regulation, and affinity to cardiovascular drugs of these transporters. Cardiac transporter mRNA levels were OCTN2>OCT3>OCTN1>OCT1 (OCT2 was not detected). Proteins were localized in vascular structures (OCT3/OCTN2/OCTN1) and cardiomyocytes (OCT1/OCTN1). Functional studies revealed a specific drug-interaction profile with pronounced inhibition of OCT1 function, for example, carvedilol [half maximal inhibitory concentration (IC50), 1.4 μmol/L], diltiazem (IC50, 1.7 μmol/L), or propafenone (IC50, 1.0 μmol/L). With use of the cardiomyopathy model of coxsackievirus-infected mice, Octn2mRNA expression was significantly reduced (56% of controls, 8 days after infection). Accordingly, in endomyocardial biopsy specimens OCTN2 expression was significantly reduced in patients with dilated cardiomyopathy, whereas the expression of OCT1-3 and OCTN1 was not affected. For OCTN2 we observed a significant correlation between expression and left ventricular ejection fraction (r = 0.53, P < 0.0001) and the presence of cardiac CD3+ T cells (r = −0.45, P < 0.05), respectively. OCT1, OCT3, OCTN1, and OCTN2 are expressed in the human heart and interact with cardiovascular drugs. OCTN2 expression is selectively reduced in dilated cardiomyopathy patients and predicts the impairment of cardiac function.
Cardiovascular diseases are the leading cause of death. Consequently, numerous drugs targeting the cardiovascular system are given to a large number of patients. Little is known about drug concentrations at their target sites, which can be modulated by local factors, among them drug efflux and uptake transporters.1 In particular, with regard to tissues such as skeletal or cardiac muscle, knowledge about transporter expression as a prerequisite for intracellular drug concentrations is still limited. Yet, several transport proteins, among them members of the ATP-binding cassette proteins (ABC transporter), have been identified within the cardiovascular system.2 The potential relevance of transport proteins in these structures is highlighted by a recent study on transporter expression in skeletal muscle cells, indicating an impact of uptake and efflux transporters on local statin concentrations and hence on statin-mediated myotoxicity.3 Although several publications address expression of efflux-mediating ABC transport proteins in the human heart, the mode of cardiac drug uptake is unclear. In this context, we attempted to demonstrate that the cardiac expression of two uptake transporters in humans and the expression of one of those, the carnitine transporter OCTN2, were affected by cardiac disease.4,5
In the present study, we investigated the cardiac expression of uptake transporters for organic cation transporters [OCT(N)s], namely, the OCT1-3 (SLC22A1-3) and OCTN1 and OCTN2 (SLC22A4 and 5). Recent studies indicate an important pharmacologic role of these transporters.6 For example, in a functional study of the ubiquitously expressed OCT3 in knockout mice, the distribution of the OCT standard substrate 1-methyl-4-phenylpyridinium (MPP+) was selectively altered in cardiac tissue.7 Furthermore, there is evidence of an interaction of the physiologically important carnitine transporters OCTN1 and OCTN28,9 with certain drugs, thereby acting as an uptake transporter or being inhibited in their physiologic function by these compounds.6 Such interactions may have consequences for systemic and cardiac carnitine homeostasis as already discussed for the valproic acid–induced carnitine depletion.10 On the other hand, these transporters may modify drug action itself by controlling local concentrations, which has recently been shown for OCTN1 and the hERG (KCNH2) channel blocker quinidine.11
Although the tissue-specific mRNA expression of these transporters has been investigated in general studies,12 knowledge about their cardiac localization and disease-dependent regulation is still limited. In addition to drugs modifying these proteins on a functional level (eg, inhibiting l-carnitine uptake), changes in cardiac expression will result in an altered uptake of both endogenous substrates and drugs. Therefore, this study focused on disease-dependent expression of cardiac OCT(N)s and possible functional interactions with cardiovascular drugs. Taken together, we demonstrate selective regulation of OCTN2 by impaired cardiac function and studied the interaction profile of OCT(N)s and cardiovascular drugs.
Materials and Methods
Patients
In the present study, we examined the expression of OCT(N)s in the human heart. The general expression and localization were studied in explanted hearts of potential donors with no indications for cardiac dysfunction, which have been described before.13,14
We studied the cardiac expression of OCT(N)s in endomyocardial biopsy (EMB) specimens in a cohort of 83 patients. We included patients with a broad range of left ventricular ejection fraction (LVEF) as a measure of left ventricular systolic function and left ventricular end diastolic diameter (LVEDD) as a key measure of left ventricular dilatation. We aimed to encompass patients presenting with acute myocarditis (AMC) and dilated cardiomyopathy (DCM) versus noninflammatory pathogenesis. The latter were composed of non-DCMs (n = 8) and patients with unspecified symptoms [dyspnea on exertion, chest discomfort, palpitations (n = 9)] and normal or mildly reduced LVEF (>45%). Patients' EMB specimens were characterized by immunohistologic proof of inflammatory DCM and PCR proof of cardiac viral infections. For the selection of AMC and DCM patients, we focused on patients without proof of cardiac infections, patients with the most frequently detected parvovirus B19 (PVB19), and patients with enteroviral infection, which have been well characterized in animal models of coxsackievirus-induced myocarditis15 and have been associated with adverse prognosis in human disease.16,17 This selection criterion was met to ascertain possible virus-specific effects on OCTN regulation in virus-associated cardiomyopathy. The patients were randomly selected according to these entry criteria from the biomaterial database of the Sonderforschungsbereich Transregio 19 (SFB TR19). Written consent was obtained from each patient, and the protocol was approved by the Ethics Committee of the Charité – Universitätsmedizin, Berlin, Germany, within the framework of the SFB TR19. Patients' EMB specimens were obtained from the right ventricular septum and were characterized immunohistochemically for the presence of a cardiac inflammation and by PCR techniques to detect cardiotropic viruses as described elsewhere.18–21 Significant coronary disease was excluded by coronary angiography in all patients with nonischemic cardiomyopathy. The clinically suspected cardiomyopathy entities of AMC, DCM, and non-DCMs (n = 8 controls with LVEF ≤45%; ischemic cardiomyopathy: n = 5, hypertrophic cardiomyopathy: n = 1, toxic cardiomyopathy: n = 1, and tachycardiomyopathy: n = 1) were determined in accordance with widely accepted classifications,22,23 considering the chief clinical presentation, laboratory, echocardiographic, and cardiac catheterization data. AMC was suspected in cases with antecedent viral illness and duration of history up to 4 weeks before first clinical assessment. In AMC cases with preserved or mildly impaired LVEF, a clinical presentation mimicking acute coronary syndrome and arrhythmias was frequent.24,25 In AMC cases with impaired LVEF (≤45%), cardiac decompensation and progressive heart failure prevailed. Three AMC patients were admitted after cardiopulmonary resuscitation.26,27 The patients' clinical and EMB specimen characteristics are summarized in Table 1.
Table 1.
Patient Characteristics From the First Study
| Characteristics | Controls with LVEF ≤45% (n = 8) | Controls with LVEF >45% (n = 9) | AMC patients with LVEF ≤45% (n = 7) | AMC patients with LVEF >45% (n = 32) | DCM patients (n = 27) |
|---|---|---|---|---|---|
| Age, mean ± SD (range), years | 56.8 ± 15.4 (27–71) | 43.1 ± 20.3 (16–66) | 52.0 ± 10.3 (39–63) | 37.0 ± 12.8 (18–65) | 51.1 ± 13.7 (30–78) |
| Sex, No. M/F | 7/1 | 4/5 | 6/1 | 26/6 | 23/4 |
| LVEF, mean ± SD (range), % | 30.3 ± 9.5 (17–44) | 77.1 ± 10.9 (50–84)⁎ | 36.6 ± 8.1 (25–45) | 66.6 ± 9.4 (47–83)⁎ | 27.9 ± 11.4 (7–45) |
| LVEDD, mean ± SD (range), mm | 64.3 ± 8.4 (56–81) | 49.3 ± 5.2 (39–54) ⁎ | 63.3 ± 8.7 (54–78) | 52.8 ± 5.5 (40–62)⁎ | 65.4 ± 9.2 (45–82) |
| LVESD, mean ± SD (range), mm | 47.4 ± 11.9 (24–62) | 29.8 ± 6.2 (20–38) | 51.4 ± 13 (36–72) | 33.6 ± 6.8 (12–44) | 51.9 ± 11.1 (26–71) |
| Virus (B19V or EV), No. | 1⁎ | 1⁎ | 4 | 21 | 15 |
| B19V | 1 | 0 | 2 | 13 | 4 |
| EV | 0 | 0 | 1 | 3 | 10 |
| B19V and EV | 0 | 0 | 1 | 5 | 1 |
| Inflammatory DCM (immunohistology) | 1⁎ | 0⁎ | 4 | 14 | 14 |
M, male; F, female. LVEF, left ventricular ejection fraction; AMC, acute myocarditis; DCM, dilated cardiomyopathy; LVEDD, left ventricular end diastolic diameter; LVESD, left ventricular end systolic diameter.
Significantly (P < 0.05) different from the remaining patient groups (Tukey-Kramer post hoc analysis in comparisons with continuous data).
A second, independent patient group consisting of 15 DCM and six control patients was used to confirm these findings. Among these patients, coronary heart disease was excluded by angiography and ACM by myocardial biopsy. Patients were treated with angiotensin-converting enzyme inhibitors, angiotensin receptor antagonists, β-blockers, digitalis, and diuretics and received stable oral medication for 3 months before the study. From all patients we obtained six to 10 EMB specimens from the interventricular septum of the right ventricle for clinical reasons. Written consent was obtained from each patient, and the protocol was approved by the Ethics Committee of the University Hospital in Greifswald, Germany (Table 2).
Table 2.
Patient Characteristics From the Second Study
| Characteristics | Control patients | DCM patients |
|---|---|---|
| No. | 6 | 15 |
| Age, mean ± SD, years | 45.7 ± 9.1 | |
| LVEF, mean ± SD, % | 58.7 ± 9.2 | 35.9 ± 5.0 |
| LVEDD, mean ± SD, mm | 50.5 ± 2.9 | 68.6 ± 7.3 |
| Sex, No. M/F | 4/2 | 9/6 |
| Virus diagnostic | Negative | 8 Negative/7 positive |
M, male; F, female; LVEF, left ventricular ejection fraction; LVEDD, left ventricular end diastolic diameter.
Animal Studies
Viral infections are one etiologic factor for the development of DCM. Again, the consequences for expression and function of cardiac uptake transport proteins are unknown. We therefore studied murine Oct(n)transporter expression in a viral myocarditis model. Inbred mice of strains C57BL/6 (H-2b) and ABY/SnJ (H-2b) were infected with coxsackievirus B3 (CVB3) in the animal facilities of the Department of Molecular Pathology, University Hospital Tübingen, as described elsewhere.15 Four- to 5-week-old mice were infected i.p. with 5 × 104 plaque-forming units of purified CVB3. Animals were sacrificed at different time points (4, 8, 12, and 28 days) after infection. Noninfected animals of both strains were used as controls. Samples of aseptically removed hearts were snap frozen in liquid nitrogen for real-time RT-PCR analysis.
RNA Isolation and cDNA Amplification
RNA isolation from explanted hearts of patients with no indication for cardiac dysfunction and cDNA synthesis were performed as described before.5
Three to four EMB specimens from each patient of the second collective were stored in liquid nitrogen, pooled, and homogenized. An aliquot of the cell pellet was transferred to RLT-lysis buffer (RNeasy Micro Kit; Qiagen Inc., Valencia, California). Subsequently, RNA was isolated following the manufacturer's instructions for total RNA isolation from fibrous tissues (Qiagen Inc.). RNA was purified using a Charge Switch Total RNA Cell Kit (Invitrogen, Carlsbad, California), and concentration and quality were assessed using a Nanodrop ND-1000 (NanoDrop Technologies Inc., Wilmington, Delaware) and an RNA 6000 Pico LabChip on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, California). Before real-time PCR one-round T7-RNA polymerase-mediated linear amplification of 30 ng of total RNA was performed using the first part of the Two Cycle Target labeling protocol (Affymetrix, Santa Clara, California). The amplified RNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, California).
For RNA preparation from murine hearts, snap frozen hearts were homogenized in the presence of liquid nitrogen by a mortar. Subsequently, the tissue powder was lysed using 350 μL of RLT-lysis buffer following a proteinase K digestion. RNA isolation was performed according to the manufacturer's protocol (Nucleo Spin Extract II Kit; Macherey-Nagel, Düren, Germany). Finally, the RNA concentration was determined using a Nanodrop ND-1000 system.
Real-Time PCR
In explanted hearts, expression of transporter transcripts and the reference gene for 18S rRNA was measured by conventional TaqMan real-time PCR using the following assays on demand (Applied Biosystems): Hs00901881_m1 (OCT1), Hs00161893_m1 (OCT2), Hs00222691_m1 (OCT3), and Hs00268200_m1 (OCTN1). The expression of OCTN2 and 18S rRNA was measured as described before.5
The transporter expression measurement in the first patient collective was performed by a recently developed preamplification/real-time PCR technique28 using the mRNA specific assays on demand as already mentioned.
Real-time PCR analysis of OCT1, OCTN1, and OCTN2 within the EMB specimens of the second patient collective was performed using a custom-manufactured TaqMan low-density array (TLDA; Applied Biosystems) that, in addition to the target genes, contained the common housekeeping genes B2M, GUSB, TBP, 18S rRNA, HMBS, and GAPDH (due to technical problems OCT3 could not be included in this analysis). The TLDA cards were loaded with reaction mixes from control and patient samples, and PCR amplification was performed using a 7900 HT Sequence Detection System (Applied Biosystems). Thermal cycling was as follows: 2 minutes at 50°C; 10 minutes at 94.5°C, followed by 30 seconds at 97°C; and 1 minute at 59.7°C for 40 cycles.
In murine probes, transporter expression and the reference gene for 18S rRNA were measured by conventional TaqMan real-time PCR system (7900HT) using the following assays on demand: Mm00456303_m1 (Oct1), Mm00457295_m1 (Oct2), Mm00488294_m1 (Oct3), Mm00457739_m1 (Octn1), and Mm00441468_m1 (Octn2) (all Applied Biosystems). Real-time PCR data were quantified using the SDS 2.3 software package (Applied Biosystems).
Immunofluorescence Staining
Protein localization was investigated by immunofluorescence microscopy. For OCT1, a polyclonal antibody against the human transporter was used (rabbit, dilution 1:50; GenWay Biotech Inc., San Diego, California). OCT3 and OCTN1 were detected using affinity-purified polyclonal rabbit antibodies generated against the rat (OCT3) and murine (OCTN1) transporter (Alpha Diagnostics, San Antonio, Texas). OCTN2 was detected as described before (rabbit, dilution 1:150).5 Paraffin sections of 2 μm were used (prepared by standard methods). Incubation with primary antibodies was performed at 4°C overnight. After being washed with Tris-buffered saline, the sections were incubated for 2 hours with secondary antibody Alexa Fluor 488–labeled IgG (anti-rabbit IgG, Invitrogen). Nuclei were stained using a 1:1000 dilution of DAPI (Sigma Aldrich, Munich, Germany).
Cloning and Overexpression of Transporter Constructs
Functional studies for OCT1, OCT3, OCTN1, and OCTN2 were performed using transporter overexpressing MDCKII cells. Although the OCTN2 overexpressing cell line has already been described,5 OCT1, OCT3, and OCTN1 overexpressing cell lines were generated. Therefore, the coding sequence of these transporters was amplified from cDNA preparations (derived from placenta for OCT3 and OCTN1 and from liver for OCT1). Subsequently, transporter fragments were cloned into the mammalian expression vector pcDNA3.1/hygro(-) (Invitrogen) and checked for sequence variations against the reference sequences (OCT1, NM_003057; OCT3, NM_021977; OCTN1, NM_003059). Overexpression of all constructs was performed as described before for OCTN2.5 Stable overexpressing cell lines were characterized on protein and functional levels.
Functional Studies
For transport experiments, cells were cultured to confluence in 24-well plates and washed with prewarmed (37°C) PBS before uptake assays were performed. Transport studies were performed by incubating the cells with a transport buffer (140 mmol/L NaCl2, 5 mmol/L KCl, 1 mmol/L KH2PO4, 1.5 mmol/L CaCl2, 5 mmol/L glucose, and 12.5 mmol/L HEPES, pH 7.4). For studies with MDCKII-OCT1 and MDCKII-OCT3, [3H]MPP+ and unlabeled MPP+ were added to the incubation buffer at a final concentration of 500 nmol/L. [3H]-carnitine and [14C]-tetraethylammonium (all radiochemicals were from Hartmann Analytics, Braunschweig, Germany) were used for uptake assays using MDCKII-OCTN2 and MDCKII-OCTN1, respectively. The cells were incubated at 37°C for 5 minutes (OCT1, OCTN1, and OCTN2) or 90 seconds (OCT3). Afterward, the cells were washed three times with ice-cold PBS before lysing cells with 0.2% SDS containing 0.5% EDTA. An aliquot of the lysate was dissolved in 2 mL of scintillation cocktail (Rotiszint, Roth, Karlsruhe, Germany) and measured in a scintillation β-counter (type 1409, LKB-Wallac, Turku, Finland).
Statistical Analyses
For graphs and statistical analyses, GraphPad Prism software 5.02 (GraphPad, San Diego, California) was used. Gene expression data were analyzed using the relative expression values (ΔΔCT-method) and statistical tests as indicated in the respective section and figure legend. When using box plots, whiskers represent the 10th to 90th percentiles.
For functional studies, uptake inhibition was calculated as a percentage of control in the absence of drugs. Here, statistical analysis was performed by Student's t-test, and data were depicted as mean ± SD values. The half maximal inhibitory concentrations (IC50) were calculated by fitting the values to a sigmoidal dose-response curve (GraphPad Prism software 5.02). In all cases, P < 0.05 was considered significant.
Results
Cardiac Expression of OCT(N) Transporters
To characterize cardiac expression of OCT1, OCT2, OCT3, OCTN1, and OCTN2, real-time PCR was performed on RNA samples from eight nonfailing hearts. RNA expression was normalized to 18S rRNA expression and depicted in relation to the mean expression of all measured transporters (Figure 1A). Although OCT2 was not expressed in the human heart (CT values >40 for 10 ng of reverse transcribed RNA) and OCT1 was only detected in low abundance (mean expression, 0.25 ± 0.18 in relation to all transporters; mean CT value, 33.9), OCTN1 and OCT3 exhibit medium expression levels (0.59 ± 0.49 for OCTN1, mean CT value, 32.6; and 1.30 ± 0.51 for OCT3, mean CT value, 31.3). The most abundant cardiac OCT(N) was OCTN2 (10.8 ± 7.3; mean CT value, 28.4).
Figure 1.
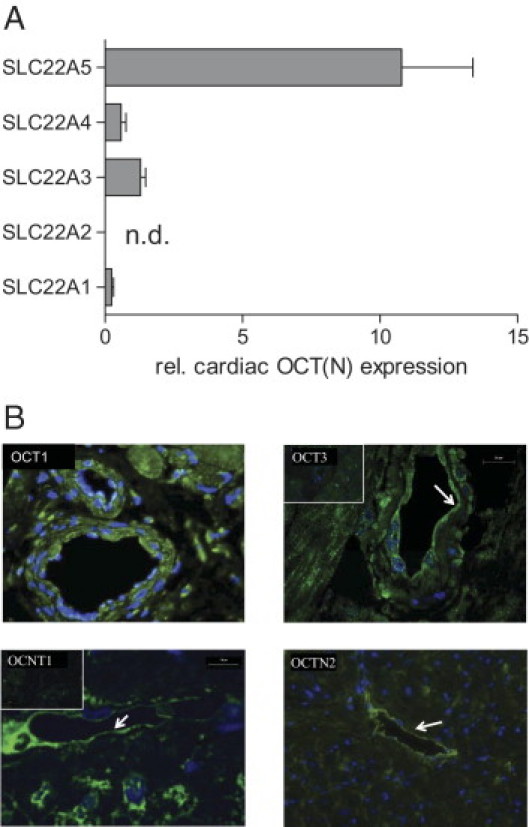
Expression of OCT(N)s in the human heart. A: mRNA expression was examined by real-time PCR and normalized to 18S rRNA expression. Values are depicted in relation (rel.) to the mean expression of all measured OCT(N)s (n.d., not detected). B: Cardiac localization of OCT1, OCT3, OCTN1, and OCTN2 (green fluorescence) as assessed by immunofluorescence staining (blue fluorescence: nuclei staining, insets: control staining, arrows: endothelial cells).
Furthermore, cardiac protein expression of OCT(N)s was examined by immunofluorescence staining, thereby detecting OCT3 and OCTN2 predominately in vascular structures (vessel wall and endothelium, respectively), whereas OCTN1 and OCT1 were also present in cardiomyocytes (Figure 1B).
Interaction with Cardiovascular Drugs
To study the interaction of cardiovascular drugs with cardiac expressed OCT(N)s, we used OCT1-, OCT3-, OCTN1-, and OCTN2-overexpressing MDCKII cell lines. Therefore, the cDNA of the respective transporter was cloned into a mammalian expression vector and transfected into MDCKII cells. Stable transfected cell clones were selected and tested for transporter overexpression by immunofluorescence staining, demonstrating the transporters to be expressed in the plasma membrane. Functional characterization using standard substrates verified the transporter overexpression (data not shown). Inhibition studies with cardiovascular drugs were performed for all transporters. OCT(N)-mediated uptake of the radiolabeled standard substrates MPP+ (OCT1 and OCT3), tetraethylammonium (TEA) (OCTN1), and l-carnitine (OCTN2) was measured in the absence and presence of a 100 μmol/L concentration of the drug. The results depicted in Table 3 demonstrate different interactions of the respective transporter. Although OCTN2-mediated carnitine uptake was only a little affected by drugs such as verapamil or amiodarone (approximately 30% inhibition), the transport activity of OCT3 and OCTN1 was modified by various drugs, and the most comprehensive interaction was observed for OCT1, which was inhibited by almost all tested drugs.
Table 3.
Inhibition of OCT(N) Function
| Agent | OCT1 | OCT3 | OCTN1 | OCTN2 |
|---|---|---|---|---|
| Amiodarone | 72.5 ± 10.5⁎ | 87.5 ± 20.5 | 68 ± 10 | 66.3 ± 16.2⁎ |
| Atenolol | 81.8 ± 10.8⁎ | 99.1 ± 27.2 | 80.1 ± 10.9 | 99.5 ± 3.3 |
| Atorvastatin | 69.5 ± 16.3⁎ | 72.9 ± 11.6⁎ | 98.4 ± 28.4 | 88.0 ± 5.4⁎ |
| Atropine | 16.9 ± 3.0⁎ | 106.5 ± 8.6 | 18.8 ± 21.3⁎ | 86.8 ± 22.4 |
| Bisoprolol | 62.8 ± 9.3⁎ | 94.0 ± 13.3 | 91.1 ± 16.6 | 93.4 ± 6.6 |
| Carvedilol | 6.7 ± 3.0⁎ | 75.3 ± 13.6⁎ | 29.8 ± 16.3⁎ | 91.8 ± 12.2 |
| Digoxin | 53.8 ± 8.4⁎ | 87.2 ± 4.6⁎ | 91.5 ± 29.7 | 92.3 ± 8.3 |
| Diltiazem | 7.2 ± 2.9⁎ | ND | 30.7 ± 18.0⁎ | ND |
| Flecainide | 18.5 ± 5.3⁎ | 83.8 ± 7.4⁎ | 32.4 ± 16.8⁎ | 98.1 ± 68.5 |
| Ipratropium bromide | 11.5 ± 0.7⁎ | 76.9 ± 1.7⁎ | 32.3 ± 8.8⁎ | 71.3 ± 2.3⁎ |
| Lidocaine | 52.2 ± 4.2⁎ | 85.2 ± 10.8⁎ | 37.5 ± 5.3⁎ | 77.6 ± 7.8⁎ |
| Metoprolol | 32.7 ± 5.7⁎ | 87.2 ± 24.6 | 86.9 ± 10.6 | 95.8 ± 15.0 |
| Molsidomine | 87.1 ± 5.3⁎ | 84.6 ± 12.2⁎ | 96.8 ± 19.1 | 92.7 ± 24.3 |
| Nadolol | 58.2 ± 16.3⁎ | 112.1 ± 19.3 | 101.1 ± 12.8 | 93.3 ± 9.0 |
| Nifedipine | 15.7 ± 8.1⁎ | 35.4 ± 17.5⁎ | 6.3 ± 5.7⁎ | 69.6 ± 16.4⁎ |
| Propafenone | 14.1 ± 5.6⁎ | 56.2 ± 4.3⁎ | 32.8 ± 17.9⁎ | 86.7 ± 15.4 |
| Propranolol | 12.8 ± 6.7⁎ | 12.6 ± 4.1⁎ | 41.1 ± 9.2⁎ | 78.9 ± 5.5⁎ |
| Sotalol | 86.2 ± 10.1 | 91.8 ± 22.8 | 91.9 ± 22.0 | 90.2 ± 17.3 |
| Spironolactone | 9.8 ± 4.6⁎ | 72.9 ± 6.5⁎ | 32.6 ± 11.2⁎ | 72.2 ± 12.0⁎ |
| Talinolol | 29.2 ± 4.5⁎ | 79.9 ± 21.1 | 89.3 ± 13.8 | 90.8 ± 5.7⁎ |
| Verapamil | 7.0 ± 3.5⁎ | 22.9 ± 14.8⁎ | 8.5 ± 7.0⁎ | 66.5 ± 38.3 |
Cells were incubated with radiolabeled standard substrates in the presence of various drugs (100 μmol/L) as described in Materials and Methods. Values (mean ± SD) represent the percentage of residual transport activity compared with solvent control. ND, not determined.
P < 0.05, Student's t-test.
The calcium channel blockers verapamil, diltiazem, and nifedipine, as well as the β-blockers carvedilol and propranolol, interact with a variety of transport proteins, thereby exhibiting low IC50 in particular for OCT1 and OCTN1. To further evaluate these drug-transporter interactions, the IC50 values were determined for the most potent inhibitors and compared with plasma concentrations reached by the respective drug (Table 4). The results indicate that IC50 values of several drugs inhibiting OCT1 are in the range of concentrations reached during drug therapy as indicated by maximum concentration (Cmax)/IC50 values above 0.1. In contrast to OCT1, most IC50 values for OCT3, OCTN1, and OCTN2 are above therapeutic concentrations for the most drugs tested.
Table 4.
Potency of OCT(N) Inhibitors
| IC50 values, μmol/L | Plasma Cmax | Reference | Cmax/IC50 | |
|---|---|---|---|---|
| Carvedilol | ||||
| OCT1 | 1.4 (0.9–2.1) | 66 ng/mL (163 nmol/L) | 29 | 0.12 |
| OCT3 | 74.2 (15.3–360.5) | <0.1 | ||
| OCTN1 | 73.1 (58.6–91.1) | <0.1 | ||
| OCTN2 | ND | |||
| Diltiazem | ||||
| OCT1 | 1.7 (1.2–2.4) | 205.6 ± 91.3 ng/mL (496 ± 220 nmol/L) | 30 | 0.29 |
| OCT3 | ND | |||
| OCTN1 | 126.4 (93.8–170.2) | <0.1 | ||
| OCTN2 | ND | |||
| Flecainide | ||||
| OCT1 | 2.5 (1.5–4.3) | 371 ± 124 ng/mL (895 ± 299 nmol/L) | 31 | 0.36 |
| OCT3 | ND | |||
| OCTN1 | 176 (125–247) | <0.1 | ||
| OCTN2 | ND | |||
| Metoprolol | ||||
| OCT1 | 52.6 (11.9–232.9) | 114.4–2543 ng/mL (dependent on CYP2D6 genotype) (0.426–9.5 μmol/L) | 32 | <0.1–0.18 |
| OCT3 | ND | |||
| OCTN1 | ND | |||
| OCTN2 | ND | |||
| Nifedipine | ||||
| OCT1 | 31.1 (15.2–63.4) | |||
| OCT3 | 102.3 (27.9–69.1) | |||
| OCTN1 | 74.6 (48.0–116.0) | |||
| OCTN2 | 59.4 (2.4–1473) | |||
| Propafenone | ||||
| OCT1 | 1.0 (0.6–1.6) | 883 ± 421 ng/mL (2.6 ± 1.2 μmol/L) | 33 | 2.6 |
| OCT3 | ND | |||
| OCTN1 | 67.5 (54.8–83.2) | <0.1 | ||
| OCTN2 | ND | |||
| Propranolol | ||||
| OCT1 | 1.3 (0.8–2.2) | 42.9 ± 19.3 ng/mL (165 ± 74 nmol/L) | 34 | 0.13 |
| OCT3 | 78.1 (20.8–292.5) | <0.1 | ||
| OCTN1 | ND | |||
| OCTN2 | ND | |||
| Spironolactone | ||||
| OCT1 | 1.2 (0.9–1.6) | |||
| OCT3 | 73.4 (27.3–196.8) | |||
| OCTN1 | 125 (98–161) | |||
| OCTN2 | 36.0 (17.7–73.2) | |||
| Talinolol | ||||
| OCT1 | 23.7 (11.5–49.0) | 147.8 ± 63.8 ng/mL (400 ± 173 nmol/L) | 35 | <0.1 |
| OCT3 | ND | |||
| OCTN1 | ND | |||
| OCTN2 | ND | |||
| Verapamil | ||||
| OCT1 | 1.2 (1.0–1.6) | 74.9 ± 25.8 ng/mL (165 ± 57 nmol/L) | 36 | 0.14 |
| OCT3 | 57.4 (5.3–623) | <0.1 | ||
| OCTN1 | 11.0 (8.1–14.9) | <0.1 | ||
| OCTN2 | 50.9 (20.3–127) | <0.1 |
Transporter overexpressing cells were incubated with probe substrates as described in Materials and Methods in the presence of various drug concentrations, ranging from 0.1 to 100 μmol/L (n = 3). IC50 values were calculated from the respective transport values by nonlinear regression analysis (brackets indicate the calculated range of IC50 values). The Cmax for the respective drugs were taken from the literature. In addition, Cmax/IC50 values were calculated to compare in vitro results with in vivo concentrations. ND, not determined.
Expression of Oct(n)s in a Murine Myocarditis Model
Previous data indicate reduced expression of OCTN2 in patients with DCM.5 Because one underlying reason for this disease may be a virus-induced inflammatory heart disease, we studied the expression of Octn2 and the other Oct(n)s in a murine myocarditis model. Therefore, C57BL/6 and A.BY/SnJ mice were infected with CVB3. Although C57BL/6 mice eliminate the virus and recover from myocarditis, the permissive A.BY/SnJ animals develop a persistent cardiac infection and chronic myocarditis. Transport protein expression was measured 4, 8, 12, and 28 days after CVB3 infection and in untreated animals of both strains (day 0). The expression of Oct3 was down-regulated in both mouse strains during infection; however, this effect only reaches significance in the permissive A.BY/SnJ mice. In contrast to permissive ABY/SnJ mice, the resistant C57BL/6 animals revealed a recovering of Oct3 expression to initial values 28 days after infection. With regard to the expression of Octn1 and 2, we observed significant differences between permissive and resistant animals. Although expression levels of both transporters remained unaffected in C57BL/6 mice during disease, Octn1 was significantly increased at days 4 to 28 after infection in A.BY/SnJ mice with a maximum at 8 days after infection. For Octn2, in A.BY/SnJ mice, the mRNA levels were significantly enhanced at 4 days after infection compared with the uninfected controls but significantly reduced from day 8 to 28 after infection [44% ± 33% of control (day 8), 44% ± 20% (day 12), and 204% ± 63% (day 28)] (Figure 2).
Figure 2.
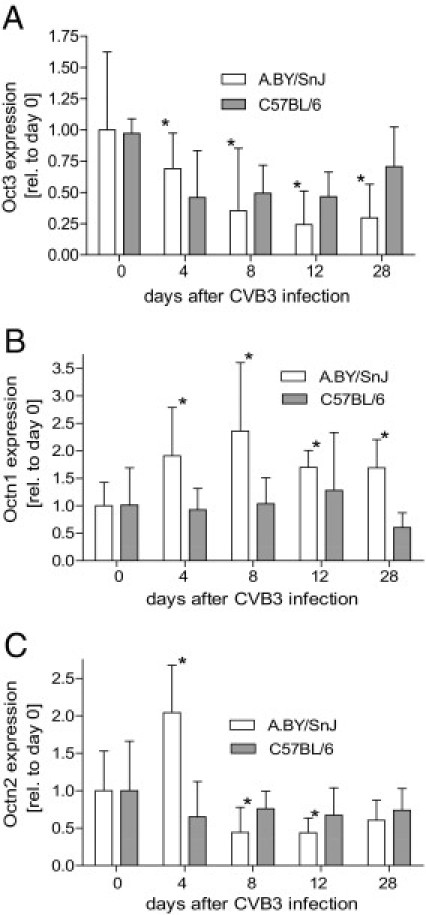
mRNA expression of Oct3 (A), Octn1 (B), and Octn2 (C) in the hearts of CVB3-infected A.BY/SnJ and C57/BL/6 mice. Oct(n) and 18S rRNA expression was determined in control animals and 4, 8, 12, and 28 days after CVB3 infection. Values are presented as mean ± SD in relation (rel.) to day 0. Statistical significance was tested versus untreated mice using the Mann-Whitney U-test (*P < 0.05).
Expression of OCT(N)s in EMB Specimens
To translate these results to humans, we investigated the OCT(N) expression in EMB specimens of two independent patient groups.
In the first group of patients, approximately 40% had inflammatory cardiac infiltrates as assessed by immunohistochemistry (IHC). Moreover, viral genomes were detected in approximately 50% of the EMB specimens (Table 1). These EMB results are consistent with published cross-sectional data.19,21,37 OCT(N) expression in these samples was quantified by preamplified real-time RT-PCR. The transporter expression was normalized to CDKN1 as both housekeeping and preamplification uniformity genes.28 All measured transporters (OCT1, OCT3, OCTN1, and OCTN2) were detectable in all samples. Concerning general parameters, such as age and sex, we observed a significantly higher expression of approximately 40% for OCT3 and for OCTN1/2 in female patients and a positive correlation between age and OCT3 (r = 0.30, P < 0.01) and OCTN1 (r = 0.28, P < 0.05) (data not shown).
With regard to cardiac disease, the expression of OCT1, OCT3, and OCTN1 was not altered, whereas the mRNA levels of OCTN2 were significantly reduced in DCM patients and in failing hearts due to nonDCMs (65% ± 20% and 61% ± 16%, respectively) compared with that of control patients with unimpaired heart function (LVEF >45). OCTN2 expression was unaffected in patients with AMC (Figure 3). In line with these findings, the OCTN2 expression was strongly correlated with functional cardiac parameters such as LVEF as measure of left ventricular systolic function (r = 0.53, P < 0.0001) and LVEDD (r = −0.45, P < 0.0001). No relationship with cardiac function parameters was observed for the other transporters (except OCT3; LVEF: r = 0.24, P < 0.05) (Figure 4). Similar findings for all transporters were found in the male subgroup, whereas no association between cardiac function and OCTN2 expression was detected in female ones (data not shown).
Figure 3.
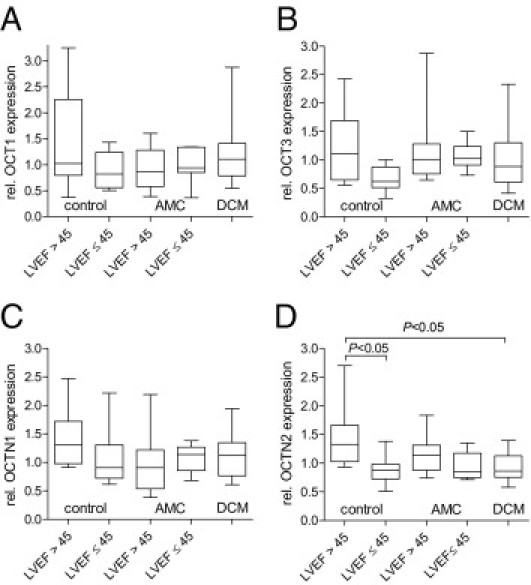
Cardiac mRNA expression of OCT1 (A), OCT3 (B), OCTN1 (C), and OCTN2 (D) in EMB specimens of 83 cardiac patients. These patients were divided into controls with unimpaired heat function (no AMC, no DCM, no virus, LVEF >45, n = 9), controls with other forms of cardiomyopathy (no AMC, no DCM, LVEF ≤45, n = 8), patients with AMC with (n = 32) and without (n = 7) LVEF >45 but no DCM, and DCM patients (n = 27). Transporter expression was examined by real-time PCR and normalized to the expression of CDKN1. Values are depicted in relation (rel.) to the median expression of all samples. Statistical testing was performed by the Kruskal-Wallis test followed by Dunn's Multiple Comparison test.
Figure 4.
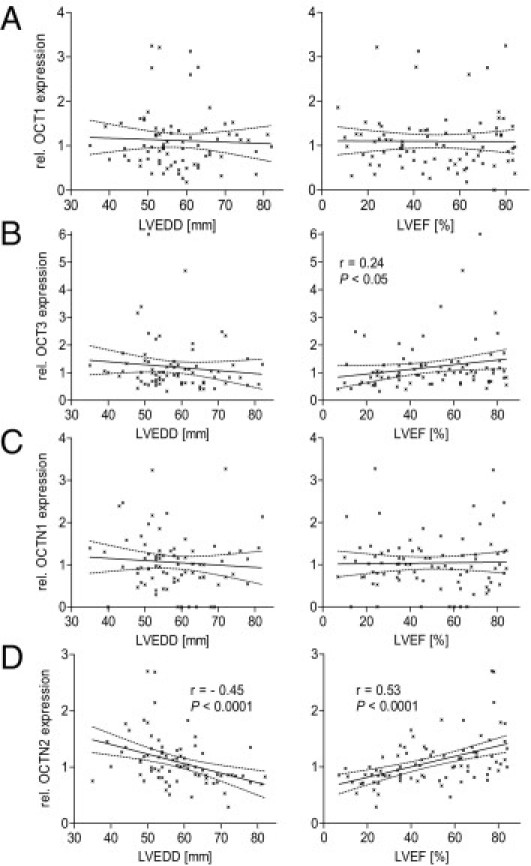
Association of cardiac mRNA expression of OCT1 (A), OCT3 (B), OCTN1 (C), and OCTN2 (D) in EMB specimens of cardiac patients and functional parameters [LVEDD (n = 78) and LVEF (n = 83)]. Transporter expression was examined by real-time PCR and normalized to the expression of CDKN1. Values are depicted in relation (rel.) to the median expression of all samples. Statistical significance was tested by the Spearman test for correlation analysis.
Besides the clinical diagnosis and the functional parameters, the EMB specimens were also characterized with regard to the presence of viral infection and myocardial inflammation. Focusing on the first parameter, no association with the cardiac OCT1, OCT3, and OCTN2 expression could be detected; however, there was a significant difference in OCTN1 expression revealing higher OCTN1 expression in enterovirus-infected hearts compared with PVB19-positive hearts (Figure 5). With regard to cardiac inflammation, OCT1, OCT3, and OCTN1 expression was unaffected (data not shown); however, we detected a trend toward a reduced OCTN2 expression in DCM patients with an immunohistologically detected inflammation compared with DCM patients without (21% ± 22% reduction in inflammatory DCM patients, P = 0.104). Although this difference was not statistically significant, a more detailed correlation between OCTN2 expression and CD3-positive T cells revealed a significant inverse relationship (r = −0.45, P < 0.05) (Figure 6B).
Figure 5.
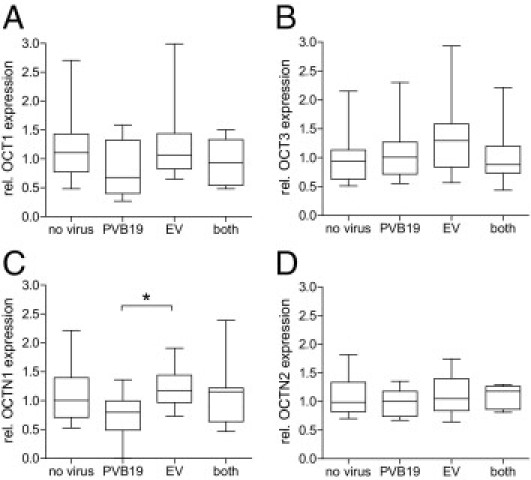
mRNA expression of OCT1 (A), OCT3 (B), OCTN1 (C), and OCTN2 (D) in EMB specimens of cardiac patients studied in dependency of the presence of a viral infection [no virus: n = 42, PVB19: n = 20, enterovirus (EV): n = 14, both virus: n = 7]. Values are depicted in relation (rel.) to the median expression of all samples. Statistical testing was performed by Kruskal-Wallis test followed by Dunn's Multiple Comparison test. *P < 0.05.
Figure 6.
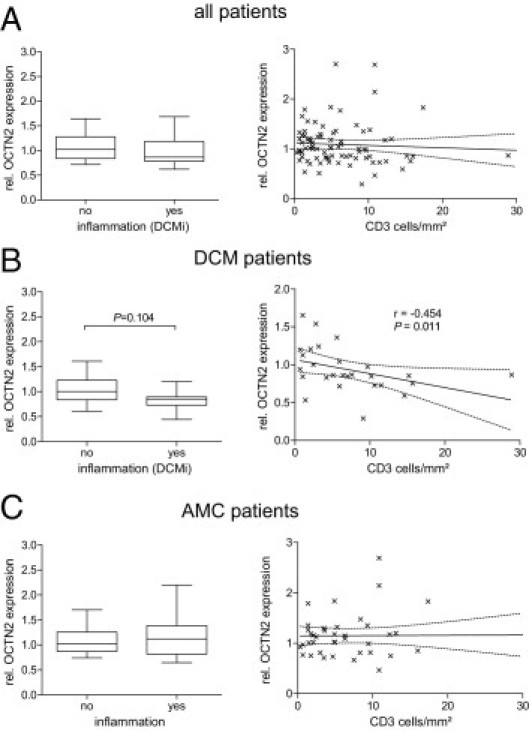
mRNA expression of OCTN2 in EMB specimens of cardiac patients with or without an immunohistologic proof of cardiac inflammation (left). Data depicted on the right show the correlation between OCTN2 expression and the cardiac CD3+ cell count (cells/mm2). Analysis was performed for all patients (n = 83, A), DCM patients (n = 27, B), and AMC patients (n = 39, C). Values are depicted in relation (rel.) to the median expression of all samples. For statistical testing, Mann-Whitney U-test and Spearman test were used.
Within the second collective, we studied the expression of OCT1, OCTN1, and OCTN2 using TLDA. OCT1 was only detectable in low abundance in five control and 12 DCM samples, with no significant difference between both groups. OCTN1 was not significantly altered by disease, whereas OCTN2 mRNA levels were significantly lowered by 47% ± 24% (P < 0.05) (Figure 7). In addition to this general observation, OCTN2 expression was associated with cardiac function as shown by its correlation with the LVEF (r = 0.44, P < 0.05) and the inverse correlation with the LVEDD (r = −0.49, P < 0.05) (Figure 7). Besides the clinical diagnosis and the functional parameters, the EMB specimens were also characterized with regard to the presence of viral infection and cardiac inflammation. Focusing on these parameters, the OCTN2 expression was strongly reduced in hearts with significant inflammation (52% compared with no inflammation), whereas the expression in the presence of a minor inflammation was unaltered compared with hearts with no inflammation (Figure 8). Concerning the presence of viral infection, the most pronounced difference in OCTN2 expression (reduction by 57%) was observed between control patients with no cardiomyopathy and virus-positive [PVB19 (n = 2), human herpesvirus 6 (n = 4), and herpes simplex virus 1 (n = 1)] DCM patients (Figure 8).
Figure 7.
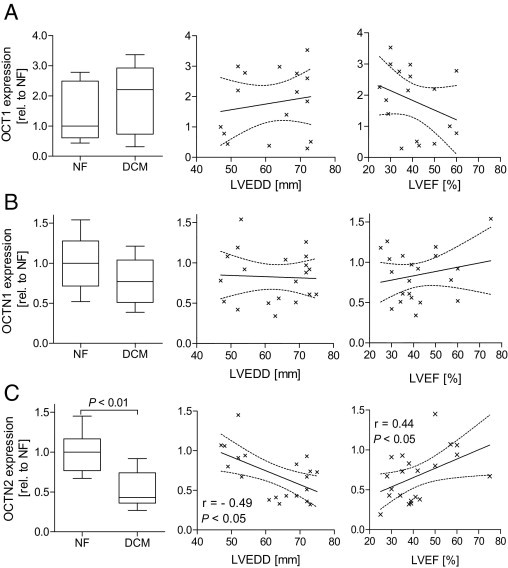
Cardiac mRNA expression of OCT1 (A), OCTN1 (B), and OCTN2 (C) in EMB specimens of patients with impaired cardiac function (DCM, n = 15) and unimpaired function (NF, n = 6). Transporter expression was examined by real-time PCR and normalized to a set of reference genes (see Materials and Methods) and depicted in relation (rel.) to the median expression of the NF probes. Left: disease-dependent expression of the transporter. Middle and right: correlation between transporter expression and cardiac function characterized by the LVEDD (middle) and LVEF (right). Statistical significance was tested by the Mann-Whitney U-test, and the Spearman test for correlation analysis.
Figure 8.
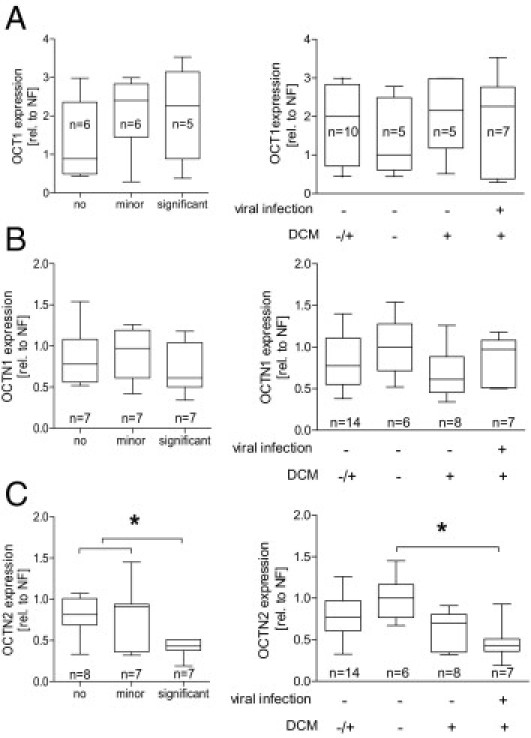
mRNA expression of OCT1 (A), OCTN1 (B), and OCTN2 (C) in EMB specimens of the second study in association with the presence of a cardiac inflammation and in association with the presence of a viral infection and DCM. Values are depicted in relation (rel.) to the median expression of the NF probes. Statistical testing was performed by Mann-Whitney U-test (left) and Kruskal-Wallis test followed by Dunn's Multiple Comparison test (right). *P < 0.05.
Discussion
In the present study, we investigated the expression and disease-dependent regulation of OCT1-3 and OCTN1 and OCTN2 in the human heart. All measured OCT(N)s except OCT2 were detectable by real-time PCR in the human heart; however, the expression levels showed a wide interindividual variation. The highest expression was detected for OCTN2, whereas OCTN1 and OCT3 exhibited moderate expression levels, and OCT1 expression was very low. These findings are in line with previous reports indicating a broad tissue distribution for OCT3, OCTN1, and OCTN2, but liver and kidney restricted expression of OCT1 and OCT2, respectively.12 With regard to its cardiac localization, immunofluorescence staining indicated a preferential localization within vascular structures for all OCT(N)s, whereas signals for OCT1 and OCTN1 were also detected in cardiomyocytes. Thereby, OCT3 was expressed in the vascular wall, whereas OCTN2 seems to be restricted to the endothelium with minor expression in cardiomyocytes. The results for OCTN2 support our previous findings5 and point to its function as an important high-affinity carnitine uptake transporter in the human heart.38 The major impact of OCTN2 for cardiac carnitine uptake is supported by the fact that intracardiac carnitine concentrations were highly elevated compared with the plasma concentrations.39 Consequently, loss of function mutations within the OCTN2 gene lead to systemic carnitine deficiency.9 In accordance with our findings, a recent study also localized OCTN1 to cardiomyocytes,11 where it is suggested to play a role for the low-affinity carnitine transport into these cells and mitochondria.40,41 However, a further study on the expression of murine OCTN1 and OCTN2 reported OCTN2 to be preferentially expressed in cardiomyocytes and OCTN1 in vascular structures.42
In addition, we developed transporter overexpressing MDCKII cell lines, a widely used model for functional studies of membrane transporters, for OCT1, OCT3, OCTN1, and OCTN2. Using these cell lines, we characterized the interaction of the individual uptake protein with various drugs as suggested by the IC50 data. Regarding OCTN2, only a limited number of compounds, such as amiodarone and verapamil, led to a moderate reduction of carnitine transport. Although these data confirm previous findings with regard to verapamil,43 an interaction with amiodarone has not been described so far. It is rather unlikely that amiodarone is a substrate for OCTN2 because of the restrictive substrate profile of this transporter44; however, it may act as an inhibitor, thereby modulating its physiologic function. A pathophysiologic role of such a drug/transporter interaction has already been discussed for valproic acid (and especially its carnitine derivates) and verapamil.10,45,46 Amiodarone has already been shown to inhibit β-oxidation as the main metabolic process dependent on carnitine by inhibition of carnitine–palmitoyltransferase I,47,48 but as described for the carnitine analog mildronate, inhibition of carnitine transport provides additional explanation for this observation.49,50 OCTN1 and OCT3 function was significantly inhibited at low concentrations by a variety of drugs. Among them, compounds such as verapamil and nifedipine, as well as propranolol and carvedilol, are the most potent inhibitors. In addition, a previous study indicated a direct OCTN1-dependent verapamil uptake, thereby modulating local drug concentrations.8 In contrast to OCTN2, the physiologic function of OCTN1 is still unclear; however, the associations between its genetic variant c.1507C>T (p.Leu503Phe; rs1050152) and the susceptibility for colitis ulcerosa or Crohn's disease point to a pathophysiologic relevance.51,52
The most pronounced drug/transporter interactions were observed for OCT1. Except sotalol, all tested compounds significantly inhibited OCT1 function. Moreover, for most compounds the inhibitory concentrations were in the range of plasma concentrations reached in pharmacotherapy as indicated by the Cmax/IC50 quotient higher than 0.1.53 As for the other OCT(N)s, not only calcium channel blockers but also drugs such as the β-blocker carvedilol and the aldosterone receptor antagonist spironolactone are potent inhibitors; however, in contrast to the transporters mentioned before, OCT1 inhibition by these compounds was much more effective as shown by very low IC50 values. This interaction profile identifies new potential inhibitors of this transporter and confirms in part previous studies, which have already demonstrated an interaction of OCT1 and drugs such as spironolactone and verapamil.54 In addition to possible effects on its physiologic function, a direct transport mediated by OCT1 may be relevant for cardiac drug uptake, especially because OCT1 could be located to cardiac blood vessels. A general impact of OCT1 on pharmacokinetic parameters has already been demonstrated for the OCT1 substrate metformin.55,56
Beside functional interactions, disease-dependent regulation of cardiac OCT(N)s can influence their physiologic and pharmacologic activity. To examine this point, we used the murine infection model of CVB3-induced myocarditis because enteroviruses are discussed as one major underlying reason for human inflammatory cardiomyopathy.57 We investigated Oct(n) expression in C57BL/6 mice, which eliminate CVB3 infection during the acute phase of myocarditis and are thus resistant to chronic myocarditis, and in A.BY/SnJ mice, which are permissive to CVB3 persistence and chronic myocarditis.15 Thereby, we found a decreased expression of Oct3 in both mouse strains with a comparably higher reduction in permissive mice probably due to the stronger immune response in ABY/SnJ mice. Interestingly, we found a differentially regulated expression of Octn1 and Octn2. Octn2 expression was significantly reduced during the acute (days 8 and 12) and the chronic (day 28) phases of myocarditis in the permissive mice, whereas the second carnitine transporter, Octn1, was up-regulated in these animals, which may represent a compensatory mechanism. In contrast, both transporters remained unchanged in resistant C57BL/6 mice. These findings indicate regulatory effects of the expression of Octn2 and Oct3 dependent on the severity of cardiac infection and inflammation.
On the basis of these observations, we studied the OCT(N) expression in EMB specimens of cardiac patients. Here, we could detect a significantly higher expression of OCT3, OCTN1, and OCTN2 in female patients and an age-associated expression for OCT1 and OCTN1.
As in the murine model, we found a reduced OCTN2 expression in patients with depressed LVEF, which is in accordance with our data from a previous study on this transporter demonstrating a reduced OCTN2 expression in DCM patients.5 In contrast, all further investigated transport proteins were not significantly regulated by cardiac disease. In addition to these findings, OCTN2 expression was significantly correlated with functional cardiac parameters, such as LVEF and the LVEDD, indicating an association among left ventricular systolic function, left ventricular dilatation, and carnitine uptake, which in turn would have consequences for cardiac energy metabolism (eg, reduced energy production by β-oxidation and enhanced glycolysis in hearts with DCM). A reduced cardiac carnitine pool may also have consequences for the cytosolic pool of free CoA, because by the transfer of acetyl and acyl groups to carnitine, this molecule is able to deliver free CoA.58 Moreover, carnitine can act as a scavenger for free acyl groups, thereby protecting the cell against radical stress. In line with these conclusions are observations in hearts with hypertrophic cardiomyopathy. Here, a reduced β-oxidation and enhanced carnitine plasma concentrations explained by reduced cardiac uptake were observed.59,60
Concerning the immunohistologic proof of cardiac inflammation and the presence of viral genomes, no significant differences could be observed for OCT1 and OCT3. However, OCTN2 expression tended to lower expression levels in DCM patients with immunohistologically detected inflammatory DCM compared with DCM patients without cardiac inflammation. This result was further underlined by the observation that the OCTN2 expression shows an inverse correlation with the cardiac CD3+ T-cell count in DCM patients and the findings of the second independent study. In addition, similar results for OCTN2 were observed in the murine myocarditis model, indicating that inflammatory mechanisms are involved in the down-regulation of OCTN2 in cardiomyopathy. With regard to the presence of the tested viral genomes, only OCTN1 exhibited significant differences between PVB19 and enterovirus-positive EMB specimens.
These findings are in accordance with previous reports on OCTN2 and inflammatory bowel disease. Here, a reduced expression of the carnitine transporter was described in intestinal probes of patients with colitis ulcerosa and Crohn's disease, whereas the expression of OCTN1 remained unchanged.61 A promoter polymorphism (-207G>C, rs2631367), affecting the promoter activity of SLC22A5 (OCTN2) by elimination of a heat shock protein binding site, has been identified as a risk factor for inflammatory bowel disease.51 The mechanism of this down-regulation is still unclear; however, as shown in patients with genetic variants, reduced OCTN2 expression is associated with a decrease in carnitine uptake. In addition, cardiac cytokine release may affect OCTN2 expression. In this context, an up-regulation of cardiac transforming growth factor-β has been described in DCM patients62 and in the animal model used in our study.63 In line with this observation, we found a down-regulation of OCTN2 expression by transforming growth factor-β (data not shown). Interestingly, recent publication indicate opposite effect for other cytokines, such as interferon-γ and tumor necrosis factor-α.62,64 With new data pointing to immunosuppressive effects of l-carnitine,65 impaired cardiac uptake as a consequence of decreased OCTN2 expression may have additional consequences for inflammatory processes.
Taken together, our data indicate a 50% reduction in cardiac OCTN2 expression in patients with DCM. This effect is comparable to patients heterozygous for OCTN2 loss of function variants. Because these variants are rare, studies on these patients are limited; however, selected groups (patients of middle age) of heterozygous carriers have been characterized by an impaired cardiac function.66 In line with these findings, data on heterozygous Octn2-deficient mice indicate an increased risk for cardiac dysfunction in the presence of additional risk factors.67
In summary, the present study demonstrates a selective disease-dependent regulation of the high-affinity carnitine transporter OCTN2 in patients with cardiomyopathy, whereas the other OCT(N)s remain unaffected. In view of the crucial role of this protein for carnitine uptake and subsequent β-oxidation, decreased OCTN2 function may evolve as one biomarker of cardiomyopathy.
Footnotes
Supported by a grant from the Deutsche Forschungsgemeinschaft (SFB TR19).
Current address of M.N., Department of Internal Medicine–Cardiology, University Hospital of Marburg and Gieβen, Philipps University of Marburg, Marburg, Germany; of K.K., University of North Carolina, Eshelman School of Pharmacy, Chapel Hill, North Carolina
References
- 1.Klaassen C.D., Aleksunes L.M. Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev. 2010;62:1–96. doi: 10.1124/pr.109.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solbach T.F., Konig J., Fromm M.F., Zolk O. ATP-binding cassette transporters in the heart. Trends Cardiovasc Med. 2006;16:7–15. doi: 10.1016/j.tcm.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Knauer M.J., Urquhart B.L., Meyer Zu Schwabedissen H.E., Schwarz U.I., Lemke C.J., Leake B.F., Kim R.B., Tirona R.G. Human skeletal muscle drug transporters determine local exposure and toxicity of statins. Circ Res. 2010;106:297–306. doi: 10.1161/CIRCRESAHA.109.203596. [DOI] [PubMed] [Google Scholar]
- 4.Grube M., Kock K., Oswald S., Draber K., Meissner K., Eckel L., Bohm M., Felix S.B., Vogelgesang S., Jedlitschky G., Siegmund W., Warzok R., Kroemer H.K. Organic anion transporting polypeptide 2B1 is a high-affinity transporter for atorvastatin and is expressed in the human heart. Clin Pharmacol Ther. 2006;80:607–620. doi: 10.1016/j.clpt.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Grube M., Meyer Zu Schwabedissen H.E., Prager D., Haney J., Moritz K.U., Meissner K., Rosskopf D., Eckel L., Bohm M., Jedlitschky G., Kroemer H.K. Uptake of cardiovascular drugs into the human heart: expression, regulation, and function of the carnitine transporter OCTN2 (SLC22A5) Circulation. 2006;113:1114–1122. doi: 10.1161/CIRCULATIONAHA.105.586107. [DOI] [PubMed] [Google Scholar]
- 6.Koepsell H., Lips K., Volk C. Polyspecific organic cation transporters: structure, function, physiological roles, and biopharmaceutical implications. Pharm Res. 2007;24:1227–1251. doi: 10.1007/s11095-007-9254-z. [DOI] [PubMed] [Google Scholar]
- 7.Zwart R., Verhaagh S., Buitelaar M., Popp-Snijders C., Barlow D.P. Impaired activity of the extraneuronal monoamine transporter system known as uptake-2 in Orct3/Slc22a3-deficient mice. Mol Cell Biol. 2001;21:4188–4196. doi: 10.1128/MCB.21.13.4188-4196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yabuuchi H., Tamai I., Nezu J., Sakamoto K., Oku A., Shimane M., Sai Y., Tsuji A. Novel membrane transporter OCTN1 mediates multispecific, bidirectional, and pH-dependent transport of organic cations. J Pharmacol Exp Ther. 1999;289:768–773. [PubMed] [Google Scholar]
- 9.Nezu J., Tamai I., Oku A., Ohashi R., Yabuuchi H., Hashimoto N., Nikaido H., Sai Y., Koizumi A., Shoji Y., Takada G., Matsuishi T., Yoshino M., Kato H., Ohura T., Tsujimoto G., Hayakawa J., Shimane M., Tsuji A. Primary systemic carnitine deficiency is caused by mutations in a gene encoding sodium ion-dependent carnitine transporter. Nat Genet. 1999;21:91–94. doi: 10.1038/5030. [DOI] [PubMed] [Google Scholar]
- 10.Tein I., DiMauro S., Xie Z.W., De Vivo D.C. Valproic acid impairs carnitine uptake in cultured human skin fibroblasts: an in vitro model for the pathogenesis of valproic acid-associated carnitine deficiency. Pediatr Res. 1993;34:281–287. doi: 10.1203/00006450-199309000-00008. [DOI] [PubMed] [Google Scholar]
- 11.McBride B.F., Yang T., Liu K., Urban T.J., Giacomini K.M., Kim R.B., Roden D.M. The organic cation transporter, OCTN1, expressed in the human heart, potentiates antagonism of the HERG potassium channel. J Cardiovasc Pharmacol. 2009;54:63–71. doi: 10.1097/FJC.0b013e3181abc288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimura M., Naito S. Tissue-specific mRNA expression profiles of human ATP-binding cassette and solute carrier transporter superfamilies. Drug Metab Pharmacokinet. 2005;20:452–477. doi: 10.2133/dmpk.20.452. [DOI] [PubMed] [Google Scholar]
- 13.Meissner K., Sperker B., Karsten C., Zu Schwabedissen H.M., Seeland U., Bohm M., Bien S., Dazert P., Kunert-Keil C., Vogelgesang S., Warzok R., Siegmund W., Cascorbi I., Wendt M., Kroemer H.K. Expression and localization of P-glycoprotein in human heart: effects of cardiomyopathy. J Histochem Cytochem. 2002;50:1351–1356. doi: 10.1177/002215540205001008. [DOI] [PubMed] [Google Scholar]
- 14.Dazert P., Meissner K., Vogelgesang S., Heydrich B., Eckel L., Bohm M., Warzok R., Kerb R., Brinkmann U., Schaeffeler E., Schwab M., Cascorbi I., Jedlitschky G., Kroemer H.K. Expression and localization of the multidrug resistance protein 5 (MRP5/ABCC5), a cellular export pump for cyclic nucleotides, in human heart. Am J Pathol. 2003;163:1567–1577. doi: 10.1016/S0002-9440(10)63513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klingel K., Hohenadl C., Canu A., Albrecht M., Seemann M., Mall G., Kandolf R. Ongoing enterovirus-induced myocarditis is associated with persistent heart muscle infection: quantitative analysis of virus replication, tissue damage, and inflammation. Proc Natl Acad Sci U S A. 1992;89:314–318. doi: 10.1073/pnas.89.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Why H.J., Meany B.T., Richardson P.J., Olsen E.G., Bowles N.E., Cunningham L., Freeke C.A., Archard L.C. Clinical and prognostic significance of detection of enteroviral RNA in the myocardium of patients with myocarditis or dilated cardiomyopathy. Circulation. 1994;89:2582–2589. doi: 10.1161/01.cir.89.6.2582. [DOI] [PubMed] [Google Scholar]
- 17.Fujioka S., Kitaura Y., Ukimura A., Deguchi H., Kawamura K., Isomura T., Suma H., Shimizu A. Evaluation of viral infection in the myocardium of patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol. 2000;36:1920–1926. doi: 10.1016/s0735-1097(00)00955-4. [DOI] [PubMed] [Google Scholar]
- 18.Holzmann M., Nicko A., Kuhl U., Noutsias M., Poller W., Hoffmann W., Morguet A., Witzenbichler B., Tschope C., Schultheiss H.P., Pauschinger M. Complication rate of right ventricular endomyocardial biopsy via the femoral approach: a retrospective and prospective study analyzing 3048 diagnostic procedures over an 11-year period. Circulation. 2008;118:1722–1728. doi: 10.1161/CIRCULATIONAHA.107.743427. [DOI] [PubMed] [Google Scholar]
- 19.Noutsias M., Seeberg B., Schultheiss H.P., Kuhl U. Expression of cell adhesion molecules in dilated cardiomyopathy: evidence for endothelial activation in inflammatory cardiomyopathy. Circulation. 1999;99:2124–2131. doi: 10.1161/01.cir.99.16.2124. [DOI] [PubMed] [Google Scholar]
- 20.Noutsias M., Pauschinger M., Ostermann K., Escher F., Blohm J.H., Schultheiss H., Kuhl U. Digital image analysis system for the quantification of infiltrates and cell adhesion molecules in inflammatory cardiomyopathy. Med Sci Monit. 2002;8:MT59–MT71. [PubMed] [Google Scholar]
- 21.Kuhl U., Pauschinger M., Noutsias M., Seeberg B., Bock T., Lassner D., Poller W., Kandolf R., Schultheiss H.P. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation. 2005;111:887–893. doi: 10.1161/01.CIR.0000155616.07901.35. [DOI] [PubMed] [Google Scholar]
- 22.Richardson P., McKenna W., Bristow M., Maisch B., Mautner B., O'Connell J., Olsen E., Thiene G., Goodwin J., Gyarfas I., Martin I., Nordet P. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of cardiomyopathies. Circulation. 1996;93:841–842. doi: 10.1161/01.cir.93.5.841. [DOI] [PubMed] [Google Scholar]
- 23.Elliott P., Andersson B., Arbustini E., Bilinska Z., Cecchi F., Charron P., Dubourg O., Kuhl U., Maisch B., McKenna W.J., Monserrat L., Pankuweit S., Rapezzi C., Seferovic P., Tavazzi L., Keren A. Classification of the cardiomyopathies: a position statement from the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2008;29:270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 24.Kuhl U., Pauschinger M., Bock T., Klingel K., Schwimmbeck C.P., Seeberg B., Krautwurm L., Poller W., Schultheiss H.P., Kandolf R. Parvovirus B19 infection mimicking acute myocardial infarction. Circulation. 2003;108:945–950. doi: 10.1161/01.CIR.0000085168.02782.2C. [DOI] [PubMed] [Google Scholar]
- 25.Angelini A., Calzolari V., Calabrese F., Boffa G.M., Maddalena F., Chioin R., Thiene G. Myocarditis mimicking acute myocardial infarction: role of endomyocardial biopsy in the differential diagnosis. Heart. 2000;84:245–250. doi: 10.1136/heart.84.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kindermann I., Kindermann M., Kandolf R., Klingel K., Bultmann B., Muller T., Lindinger A., Bohm M. Predictors of outcome in patients with suspected myocarditis. Circulation. 2008;118:639–648. doi: 10.1161/CIRCULATIONAHA.108.769489. [DOI] [PubMed] [Google Scholar]
- 27.D'Ambrosio A., Patti G., Manzoli A., Sinagra G., Di Lenarda A., Silvestri F., Di Sciascio G. The fate of acute myocarditis between spontaneous improvement and evolution to dilated cardiomyopathy: a review. Heart. 2001;85:499–504. doi: 10.1136/heart.85.5.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noutsias M., Rohde M., Block A., Klippert K., Lettau O., Blunert K., Hummel M., Kuhl U., Lehmkuhl H., Hetzer R., Rauch U., Poller W., Pauschinger M., Schultheiss H.P., Volk H.D., Kotsch K. Preamplification techniques for real-time RT-PCR analyses of endomyocardial biopsies. BMC Mol Biol. 2008;9:3. doi: 10.1186/1471-2199-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Mollendorff E., Neugebauer K., Fornasini G. Pharmacokinetics and bioavailability of carvedilol, a vasodilating beta-blocker. Eur J Clin Pharmacol. 1987;33:511–513. doi: 10.1007/BF00544245. [DOI] [PubMed] [Google Scholar]
- 30.Shum L., Pieniaszek H.J., Jr., Robinson C.A., Davidson A.F., Widner P.J., Benedek I.H., Flamenbaum W. Pharmacokinetic interactions of moricizine and diltiazem in healthy volunteers. J Clin Pharmacol. 1996;36:1161–1168. doi: 10.1002/j.1552-4604.1996.tb04171.x. [DOI] [PubMed] [Google Scholar]
- 31.Tjandra-Maga T.B., van Hecken A., van Melle P., Verbesselt R., de Schepper P.J. Altered pharmacokinetics of oral flecainide by cimetidine. Br J Clin Pharmacol. 1986;22:108–110. [PMC free article] [PubMed] [Google Scholar]
- 32.Jin S.K., Chung H.J., Chung M.W., Kim J.I., Kang J.H., Woo S.W., Bang S., Lee S.H., Lee H.J., Lee J. Influence of CYP2D6*10 on the pharmacokinetics of metoprolol in healthy Korean volunteers. J Clin Pharm Ther. 2008;33:567–573. doi: 10.1111/j.1365-2710.2008.00945.x. [DOI] [PubMed] [Google Scholar]
- 33.Kowey P.R., Kirsten E.B., Fu C.H., Mason W.D. Interaction between propranolol and propafenone in healthy volunteers. J Clin Pharmacol. 1989;29:512–517. doi: 10.1002/j.1552-4604.1989.tb03373.x. [DOI] [PubMed] [Google Scholar]
- 34.Ohashi K., Ebihara A., Kondo K., Usami M. Clinical pharmacokinetics and pharmacological actions of a long-acting formulation of propranolol. Arzneimittelforschung. 1984;34:507–512. [PubMed] [Google Scholar]
- 35.He J., Terhaag B., Yang L.Y., Zhang B.K., Su F.L., Zhu Y.G., Song J., Tang J., Liu X.L., Peng W.X. Determination of talinolol in human plasma by high performance liquid chromatography-electrospray ionization mass spectrometry: application to pharmacokinetic study. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;853:275–280. doi: 10.1016/j.jchromb.2007.03.035. [DOI] [PubMed] [Google Scholar]
- 36.Bauer L.A., Horn J.R., Maxon M.S., Easterling T.R., Shen D.D., Strandness D.E., Jr. Effect of metoprolol and verapamil administered separately and concurrently after single doses on liver blood flow and drug disposition. J Clin Pharmacol. 2000;40:533–543. doi: 10.1177/00912700022009152. [DOI] [PubMed] [Google Scholar]
- 37.Baccouche H., Mahrholdt H., Meinhardt G., Merher R., Voehringer M., Hill S., Klingel K., Kandolf R., Sechtem U., Yilmaz A. Diagnostic synergy of non-invasive cardiovascular magnetic resonance and invasive endomyocardial biopsy in troponin-positive patients without coronary artery disease. Eur Heart J. 2009;30:2869–2879. doi: 10.1093/eurheartj/ehp328. [DOI] [PubMed] [Google Scholar]
- 38.Tamai I., Ohashi R., Nezu J., Yabuuchi H., Oku A., Shimane M., Sai Y., Tsuji A. Molecular and functional identification of sodium ion-dependent, high affinity human carnitine transporter OCTN2. J Biol Chem. 1998;273:20378–20382. doi: 10.1074/jbc.273.32.20378. [DOI] [PubMed] [Google Scholar]
- 39.Evans A.M., Fornasini G. Pharmacokinetics of L-carnitine. Clin Pharmacokinet. 2003;42:941–967. doi: 10.2165/00003088-200342110-00002. [DOI] [PubMed] [Google Scholar]
- 40.Tamai I., Ohashi R., Nezu J.I., Sai Y., Kobayashi D., Oku A., Shimane M., Tsuji A. Molecular and functional characterization of organic cation/carnitine transporter family in mice. J Biol Chem. 2000;275:40064–40072. doi: 10.1074/jbc.M005340200. [DOI] [PubMed] [Google Scholar]
- 41.Lamhonwah A.M., Tein I. Novel localization of OCTN1, an organic cation/carnitine transporter, to mammalian mitochondria. Biochem Biophys Res Commun. 2006;345:1315–1325. doi: 10.1016/j.bbrc.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 42.Iwata D., Kato Y., Wakayama T., Sai Y., Kubo Y., Iseki S., Tsuji A. Involvement of carnitine/organic cation transporter OCTN2 (SLC22A5) in distribution of its substrate carnitine to the heart. Drug Metab Pharmacokinet. 2008;23:207–215. doi: 10.2133/dmpk.23.207. [DOI] [PubMed] [Google Scholar]
- 43.Ohashi R., Tamai I., Yabuuchi H., Nezu J.I., Oku A., Sai Y., Shimane M., Tsuji A. Na(+)-dependent carnitine transport by organic cation transporter (OCTN2): its pharmacological and toxicological relevance. J Pharmacol Exp Ther. 1999;291:778–784. [PubMed] [Google Scholar]
- 44.Grigat S., Fork C., Bach M., Golz S., Geerts A., Schomig E., Grundemann D. The carnitine transporter SLC22A5 is not a general drug transporter, but it efficiently translocates mildronate. Drug Metab Dispos. 2009;37:330–337. doi: 10.1124/dmd.108.023929. [DOI] [PubMed] [Google Scholar]
- 45.Ohnishi S., Okamura N., Sakamoto S., Hasegawa H., Norikura R., Kanaoka E., Takahashi K., Horie K., Sakamoto K., Baba T. Role of Na+/L-carnitine transporter (OCTN2) in renal handling of pivaloylcarnitine and valproylcarnitine formed during pivalic acid-containing prodrugs and valproic acid treatment. Drug Metab Pharmacokinet. 2008;23:293–303. doi: 10.2133/dmpk.23.293. [DOI] [PubMed] [Google Scholar]
- 46.Rigault C., Dias J.V., Demarquoy J., Le Borgne F. Characteristics of L-carnitine import into heart cells. Biochimie. 2008;90:542–546. doi: 10.1016/j.biochi.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Fromenty B., Fisch C., Labbe G., Degott C., Deschamps D., Berson A., Letteron P., Pessayre D. Amiodarone inhibits the mitochondrial beta-oxidation of fatty acids and produces microvesicular steatosis of the liver in mice. J Pharmacol Exp Ther. 1990;255:1371–1376. [PubMed] [Google Scholar]
- 48.Kennedy J.A., Unger S.A., Horowitz J.D. Inhibition of carnitine palmitoyltransferase-1 in rat heart and liver by perhexiline and amiodarone. Biochem Pharmacol. 1996;52:273–280. doi: 10.1016/0006-2952(96)00204-3. [DOI] [PubMed] [Google Scholar]
- 49.Dambrova M., Liepinsh E., Kalvinsh I. Mildronate: cardioprotective action through carnitine-lowering effect. Trends Cardiovasc Med. 2002;12:275–279. doi: 10.1016/s1050-1738(02)00175-5. [DOI] [PubMed] [Google Scholar]
- 50.Liepinsh E., Vilskersts R., Skapare E., Svalbe B., Kuka J., Cirule H., Pugovics O., Kalvinsh I., Dambrova M. Mildronate decreases carnitine availability and up-regulates glucose uptake and related gene expression in the mouse heart. Life Sci. 2008;83:613–619. doi: 10.1016/j.lfs.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 51.Peltekova V.D., Wintle R.F., Rubin L.A., Amos C.I., Huang Q., Gu X., Newman B., Van Oene M., Cescon D., Greenberg G., Griffiths A.M., George-Hyslop P.H., Siminovitch K.A. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 52.Silverberg M.S. OCTNs: will the real IBD5 gene please stand up. World J Gastroenterol. 2006;12:3678–3681. doi: 10.3748/wjg.v12.i23.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giacomini K.M., Huang S.M., Tweedie D.J., Benet L.Z., Brouwer K.L., Chu X., Dahlin A., Evers R., Fischer V., Hillgren K.M., Hoffmaster K.A., Ishikawa T., Keppler D., Kim R.B., Lee C.A., Niemi M., Polli J.W., Sugiyama Y., Swaan P.W., Ware J.A., Wright S.H., Wah Y.S., Zamek-Gliszczynski M.J., Zhang L. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahlin G., Karlsson J., Pedersen J.M., Gustavsson L., Larsson R., Matsson P., Norinder U., Bergstrom C.A., Artursson P. Structural requirements for drug inhibition of the liver specific human organic cation transport protein 1. J Med Chem. 2008;51:5932–5942. doi: 10.1021/jm8003152. [DOI] [PubMed] [Google Scholar]
- 55.Shu Y., Sheardown S.A., Brown C., Owen R.P., Zhang S., Castro R.A., Ianculescu A.G., Yue L., Lo J.C., Burchard E.G., Brett C.M., Giacomini K.M. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest. 2007;117:1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shu Y., Brown C., Castro R.A., Shi R.J., Lin E.T., Owen R.P., Sheardown S.A., Yue L., Burchard E.G., Brett C.M., Giacomini K.M. Effect of genetic variation in the organic cation transporter 1: OCT1, on metformin pharmacokinetics. Clin Pharmacol Ther. 2008;83:273–280. doi: 10.1038/sj.clpt.6100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knowlton K.U. CVB infection and mechanisms of viral cardiomyopathy. Curr Top Microbiol Immunol. 2008;323:315–335. doi: 10.1007/978-3-540-75546-3_15. [DOI] [PubMed] [Google Scholar]
- 58.Hoppel C. The role of carnitine in normal and altered fatty acid metabolism. Am J Kidney Dis. 2003;41:S4–S12. doi: 10.1016/s0272-6386(03)00112-4. [DOI] [PubMed] [Google Scholar]
- 59.Nakamura T., Sugihara H., Kinoshita N., Yoneyama S., Azuma A., Nakagawa M. Can serum carnitine levels distinguish hypertrophic cardiomyopathy from hypertensive hearts. Hypertension. 2000;36:215–219. doi: 10.1161/01.hyp.36.2.215. [DOI] [PubMed] [Google Scholar]
- 60.Nakamura T., Sugihara H., Kinoshita N., Ito K., Adachi Y., Hirasaki S., Matsuo A., Azuma A., Kodo N., Nakagawa M. Serum carnitine concentrations in patients with idiopathic hypertrophic cardiomyopathy: relationship with impaired myocardial fatty acid metabolism. Clin Sci (Lond) 1999;97:493–501. [PubMed] [Google Scholar]
- 61.Wojtal K.A., Eloranta J.J., Hruz P., Gutmann H., Drewe J., Staumann A., Beglinger C., Fried M., Kullak-Ublick G.A., Vavricka S.R. Changes in mRNA expression levels of solute carrier transporters in inflammatory bowel disease patients. Drug Metab Dispos. 2009;37:1871–1877. doi: 10.1124/dmd.109.027367. [DOI] [PubMed] [Google Scholar]
- 62.Dobaczewski M., Chen W., Frangogiannis N.G. Transforming growth factor (TGF)-beta signaling in cardiac remodeling. J Mol Cell Cardiol. 2010 doi: 10.1016/j.yjmcc.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; [Epub ahead of press]
- 63.Lang C., Sauter M., Szalay G., Racchi G., Grassi G., Rainaldi G., Mercatanti A., Lang F., Kandolf R., Klingel K. Connective tissue growth factor: a crucial cytokine-mediating cardiac fibrosis in ongoing enterovirus myocarditis. J Mol Med. 2008;86:49–60. doi: 10.1007/s00109-007-0249-3. [DOI] [PubMed] [Google Scholar]
- 64.Fujiya M., Inaba Y., Musch M.W., Hu S., Kohgo Y., Chang E.B. Cytokine regulation of OCTN2 expression and activity in small and large intestine. Inflamm Bowel Dis. 2010;17:907–916. doi: 10.1002/ibd.21444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fortin G., Yurchenko K., Collette C., Rubio M., Villani A.C., Bitton A., Sarfati M., Franchimont D. L-carnitine, a diet component and organic cation transporter OCTN ligand, displays immunosuppressive properties and abrogates intestinal inflammation. Clin Exp Immunol. 2009;156:161–171. doi: 10.1111/j.1365-2249.2009.03879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koizumi A., Nozaki J., Ohura T., Kayo T., Wada Y., Nezu J., Ohashi R., Tamai I., Shoji Y., Takada G., Kibira S., Matsuishi T., Tsuji A. Genetic epidemiology of the carnitine transporter OCTN2 gene in a Japanese population and phenotypic characterization in Japanese pedigrees with primary systemic carnitine deficiency. Hum Mol Genet. 1999;8:2247–2254. doi: 10.1093/hmg/8.12.2247. [DOI] [PubMed] [Google Scholar]
- 67.Takahashi R., Asai T., Murakami H., Murakami R., Tsuzuki M., Numaguchi Y., Matsui H., Murohara T., Okumura K. Pressure overload-induced cardiomyopathy in heterozygous carrier mice of carnitine transporter gene mutation. Hypertension. 2007;50:497–502. doi: 10.1161/HYPERTENSIONAHA.107.088609. [DOI] [PubMed] [Google Scholar]


