Abstract
5-hydroxymethylcytosine (5hmC) is a modified base present at low levels in diverse cell types in mammals1–5. 5hmC is generated by the TET family of Fe(II) and 2-oxoglutarate-dependent enzymes through oxidation of 5-methylcytosine (5mC)1,2,4–7. 5hmC and TET proteins have been implicated in stem cell biology and cancer1,4,5,8,9, but information on the genome-wide distribution of 5hmC is limited. Here we describe two novel and specific approaches to profile the genomic localization of 5hmC. The first approach, termed GLIB (glucosylation, periodate oxidation, biotinylation) uses a combination of enzymatic and chemical steps to isolate DNA fragments containing as few as a single 5hmC. The second approach involves conversion of 5hmC to cytosine 5-methylenesulphonate (CMS) by treatment of genomic DNA with sodium bisulphite, followed by immunoprecipitation of CMS-containing DNA with a specific antiserum to CMS5. High-throughput sequencing of 5hmC-containing DNA from mouse embryonic stem (ES) cells showed strong enrichment within exons and near transcriptional start sites. 5hmC was especially enriched at the start sites of genes whose promoters bear dual histone 3 lysine 27 trimethylation (H3K27me3) and histone 3 lysine 4 trimethylation (H3K4me3) marks. Our results indicate that 5hmC has a probable role in transcriptional regulation, and suggest a model in which 5hmC contributes to the ‘poised’ chromatin signature found at developmentally-regulated genes in ES cells.
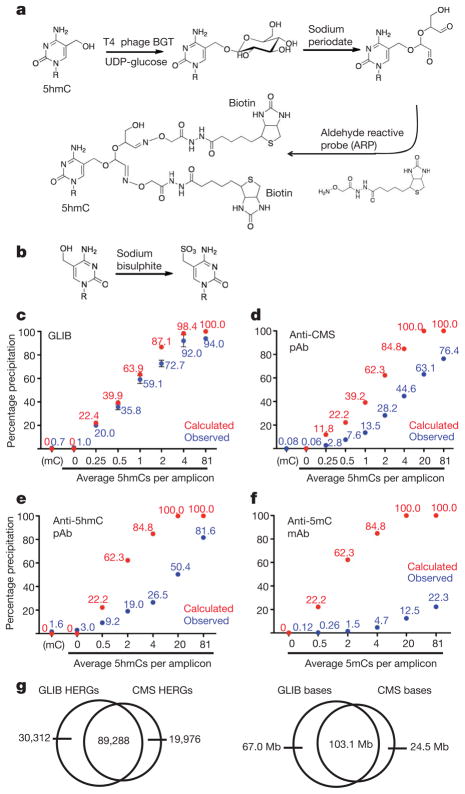
We developed two independent methods for precipitation of 5hmC in genomic DNA. The GLIB method (Fig. 1a) entails addition of a glucose molecule to each 5hmC with T4 phage β-glucosyltransferase (BGT)3 (Supplementary Fig. 1a). The glucose moiety is oxidized with sodium periodate, which converts the vicinal hydroxyl groups to aldehydes10, and further modified with aldehyde-reactive probe, which adds two biotin molecules to each 5hmC (Fig. 1a). A related strategy, which uses a custom-synthesized UDP-glucose analogue (UDP-6-N3-glucose), was recently used to profile 5hmC distribution in mouse brain11. The second method uses an antibody against cytosine 5-methylenesulphonate (CMS)5, produced by reaction of 5hmC with sodium bisulphite (Fig. 1b)12. Anti-CMS antibodies are more sensitive and less density-dependent than anti-5hmC in DNA dot blot assays5. Both methods are specific for DNA containing 5hmC (Supplementary Fig. 1b)5.
Figure 1. Comparison of 5hmC enrichment methodsa.
a, The GLIB method. Glucose is added to 5hmC by BGT, oxidized with sodium periodate to yield aldehydes, and reacted with the aldehyde reactive probe (ARP), yielding two biotins at the site of every 5hmC. b, 5hmC is converted to CMS by sodium bisulphite. c–f, Precipitation of PCR amplicons containing (1) varying amounts of 5hmC by GLIB methodology (c), anti-CMS methodology (d), or anti-5hmC antibody (e); or (2) varying amounts of 5mC by anti-5mC antibody (f). pAb, polyclonal antibody; mAb, monoclonal antibody. Between 1 and 6 independent experiments per method, mean percentage input precipitated ± s.d. is indicated. g, Overlap between HERGs identified by the GLIB and anti-CMS methodologies. Left panel, number of HERGs; right panel, number of base pairs contained within HERGs.
We examined the ability of GLIB-treated (biotinylated) and bisulphite-treated 5hmC-containing DNA to be pulled down by streptavidin and anti-CMS antisera, respectively. Using varying ratios of dCTP:dhmCTP, we generated 201 base pairs PCR amplicons with differing incorporation of cytosine and 5hmC in identical sequence contexts (Supplementary Table 1). At each dhmCTP:dCTP ratio, the fraction of amplicons that contain no 5hmC, and therefore should not be precipitated, can be calculated using the binomial equation (Supplementary Table 2). Observed and calculated pull-down efficiencies were very similar (Fig. 1c): even at low densities of 5hmC, more than 90% of DNA fragments calculated to contain a single 5hmC were precipitated after GLIB treatment. Anti-CMS pull-down showed increased density dependence compared to GLIB, but had very low background, such that there was still a strong preference for precipitation of sparsely hydroxymethylated amplicons over unmodified ones (Fig. 1d). The performance of a commercial polyclonal anti-hmC antiserum was inferior to that of anti-CMS, in terms of higher background pull-down of unmodified DNA (3.0% versus 0.06%) as well as greater density dependence (Fig. 1e). By testing PCR amplicons with varying 5mC, we confirmed that the methyl-DNA immunoprecipitation (MeDIP) technique, which uses a monoclonal antibody to 5mC, is extremely density-dependent (Fig. 1f).
We applied the GLIB and anti-CMS techniques to enrich 5hmC-containing regions in genomic DNA using genomic DNA with low, intermediate and high levels of 5hmC (Supplementary Fig. 1c, left panel). For the GLIB and anti-CMS pull-downs, the amount of specifically precipitated genomic DNA was proportional to the relative amount of 5hmC (Supplementary Fig. 1c). The GLIB technique did not produce mutations (Supplementary Table 3), the biotinylated DNA could be efficiently eluted by heating with formamide (Supplementary Fig. 1d), and the biotinylated adduct had a minimal inhibitory effect on PCR at 5–25% hmC density (delay of approximately 0.1 cycles per converted 5hmC residue; Supplementary Fig. 1f). There was no PCR delay with CMS-containing PCR amplicons except at very high CMS levels (Supplementary Fig. 1e), consistent with our previous report that CMS inhibits PCR predominantly at biologically irrelevant sequences where multiple CMS adducts occur in a row13.
We investigated the genome-wide localization of 5hmC in murine V6.5 ES cells. For GLIB-treated DNA, we chose Helicos single molecule DNA sequencing, which does not require an amplification step and thus avoids PCR bias14,15. For CMS-enriched genomic DNA, we used an Illumina instrument, as longer read lengths are needed for efficient alignment of bisulphite-treated DNA to the genome16. With the GLIB method, 119,600 regions of the genome, averaging 1,422 bp in length, showed a substantially higher density of reads in the +BGT as opposed to the −BGT sample; with the CMS method, comparison of enriched to input DNA identified 109,264 enriched regions (average length 1,168 bp). There was high overlap in the enriched regions, here designated 5hmC-enriched regions of the genome (HERGs) (Fig. 1g). Comparing the number of HERGs retrieved by using different fractions of aligned reads yielded a curve that approached an asymptote, suggesting that a majority of hydroxymethylated regions had been identified (Supplementary Fig. 2a).
To determine whether HERGs overlapped with methylated DNA regions, we identified 62,991 5mC-enriched regions of the genome (MERGs) by MeDIP. The resulting 5mC profile does not represent a complete map of 5mC in mouse ES cells, but rather is biased towards regions of dense methylation. Statistics pertaining to the GLIB, anti-CMS and MeDIP enrichments are shown in Supplementary Figs 2b–d, the corresponding annotations are provided in Supplementary Tables 4–9, and reads and enrichment for the Hoxb locus are provided in Supplementary Table 10. As expected, both HERGs and MERGs contained a high frequency of CG sequences relative to the genome at large (Supplementary Fig. 3a). Intriguingly, HERGs also contained relatively high levels of CAG sequences, the most frequent site of non-CpG methylation in human ES cells16, and we confirmed that the TET1 catalytic domain is capable of hydroxylating 5mC in CHG and CHH (H = A, T or C) contexts in vitro (Supplementary Fig. 3b).
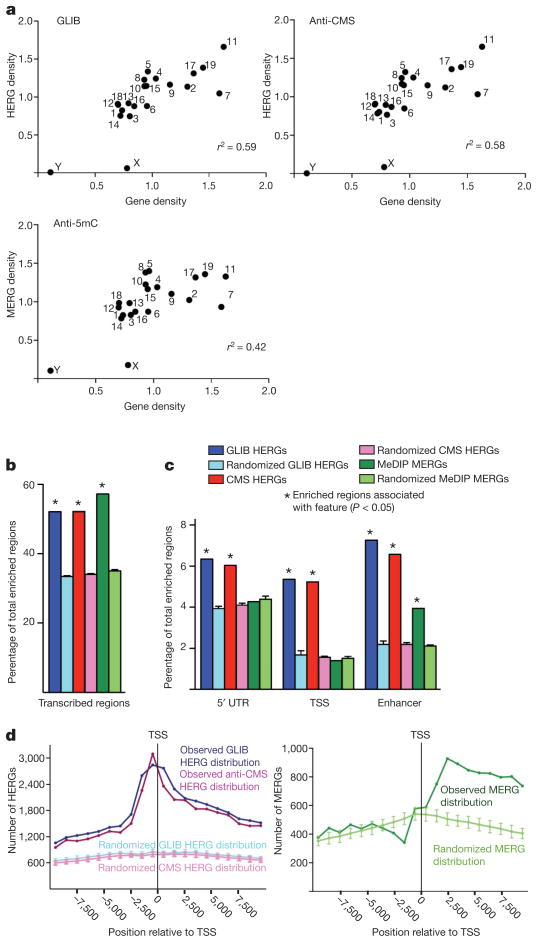
Analysis of the GLIB and anti-CMS HERG sets gave very similar results. We observed a strong correlation between the densities of HERGs and genes on a given chromosome; this trend was less pronounced for MERGs (Fig. 2a). When we compared the distribution of HERGs and MERGs to the distribution of DNA fragments of equivalent length distributed randomly across the genome, both 5hmC and 5mC were enriched within transcribed regions, particularly exons, which are known to be sites of high CpG density17 as well as high DNA methylation18 (Fig. 2b and Supplementary Fig. 3c). However, only 5hmC was enriched at transcription start sites (TSSs) and within the 5′ untranslated regions (UTRs) of genes (Fig. 2c). Moreover, 5hmC was relatively more enriched in enhancers (defined by H3K4me1 in the absence of H3K4me3)19 than 5mC, strongly indicating a connection between 5hmC and regulatory elements (Fig. 2c). Plotting each HERG as a single point relative to the nearest TSS, we found that 5hmC is heavily enriched both 5′ and 3′ of the TSS, whereas 5mC is enriched primarily 3′ of the TSS (Fig. 2d). These results show a unique distribution of 5hmC in regulatory elements of genes, one that is not explained simply by the distribution of 5mC, the substrate for TET enzymes.
Figure 2. Genomic distribution of 5hmC or 5mC enriched regions of the genome.
a, Correlation of HERG or MERG density on each chromosome (y-axis) with gene density in the same chromosome (x-axis). Density is defined as frequency divided by chromosome length. b, c, Both HERGs and MERGs are enriched in transcribed regions (b), whereas HERGs are preferentially enriched at enhancers and the start sites of genes (c). The percentage of HERGs or MERGs mapping to the indicated genomic feature (darker bar) is compared with the percentage of randomly chosen sequences mapping to that feature (lighter bar). 5′ UTR, 5′ untranslated region. TSS, transcription start site (−800 bp to +200 bp relative to start of transcription). See Supplementary Methods for detailed definition of how HERGs or MERGs were classified as mapping to genomic features. d, Distribution of HERGs and MERGs relative to the TSS. The centre of each HERG was plotted relative to the nearest TSS in 1,000 bp increments from −10 kb to +10 kb surrounding the TSS.
The enrichment of 5hmC at the TSS suggested a role for 5hmC in transcriptional regulation. To evaluate this possibility, we used published data sets on gene expression20,21 and histone modification22,23 profiles in mouse ES cells to compare the sets of genes with 5hmC or 5mC at their start sites (Supplementary Tables 11–13) to the set of all genes in the genome. 5hmC is preferentially found at promoters with high or intermediate CpG content (Supplementary Fig. 4a), even though high CpG promoters are hypomethylated in ES cells16,18,24. This distribution is consistent with the possibility that TET proteins are preferentially recruited to high CpG regions through their CpG-binding CXXC domains6,25.
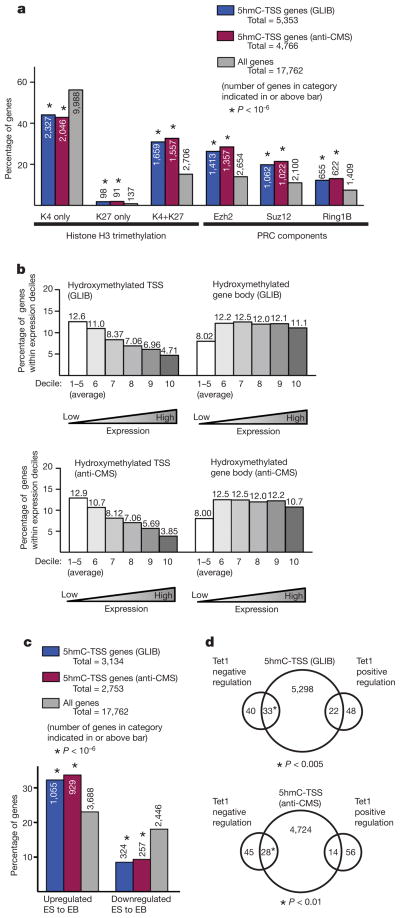
In ES cells, genes with ‘bivalent’ H3K27 and H3K4 trimethylation are transcriptionally inactive but poised for expression upon differentiation to embryoid bodies20,26,27. We found that genes with 5hmC at their start sites were disproportionately likely to contain bivalent domains at their promoters; likewise, a majority (~60%) of genes reported to contain bivalent domains have 5hmC at their start sites (Fig. 3a). 5hmC was less likely to be found at genes with the activating ‘H3K4me3 only’ mark than is predicted by chance. Moreover, genes with 5hmC at their start sites showed lower expression in murine ES cells than other genes (Fig. 3b) and were more likely to be upregulated upon embryoid body differentiation (Fig. 3c). The correlation of 5hmC with bivalent domains holds even after adjusting for the known relation between promoter CpG content and bivalency22 (Supplementary Fig. 5). Although 5mC at the TSS also correlates with lower gene expression in murine ES cells (Supplementary Fig. 4b), 5mC is not enriched at the promoters of genes with bivalent domains28 (Supplementary Fig. 4c), and genes with high levels of 5mC did not tend to be upregulated upon embryoid body differentiation (Supplementary Fig. 4d). Thus 5hmC is preferentially enriched at the promoters of genes with bivalent histone marks in ES cells, indicating that 5hmC may contribute functionally to the ‘poised’ but inactive state of these genes in ES cells.
Figure 3. Properties of HERGs at transcription start sites.
a, The percentage of genes with 5hmC at the TSS (blue and red bars) reported to contain histone H3 trimethylation (left) or PRC components (right) at their promoters is compared to the fraction of all genes (grey bars) with these promoter marks22. Number of genes in each category is indicated. b, HERGs are enriched at the TSSs of genes with low expression in ES cells. All genes were ranked by level of expression in ES cells21 and sorted into deciles from lowest to highest. The per cent of genes within the decile category with 5-hmC enriched at the TSS (left) or within gene bodies (right) are shown for each methodology. The first five deciles, which are comprised of genes lacking statistically significant expression, are pooled and averaged in this analysis. c, HERGs are enriched at the TSS of genes upregulated upon differentiation to embryoid bodies (EB)26. The percentage of genes with 5-hmC at their TSS (blue bars) that are substantially upregulated or downregulated upon differentiation to EB is compared with the percentage of total genes similarly regulated (grey bars). Number of genes in each category is indicated. d, Overlap between genes with 5hmC at the TSS and genes positively or negatively regulated by Tet1 (ref. 8).
Genes with 5hmC at their start sites are also disproportionately enriched in the set of genes whose promoters bind polycomb repressor complex (PRC) components, and in a majority of genes with the ‘H3K27me3’ only mark (Fig. 3a). There is a statistically significant correlation between genes that had 5hmC at the TSS and genes that were upregulated upon small interfering RNA-mediated Tet1 depletion8 (therefore, negatively regulated by Tet1) (Fig. 3d), indicating that 5hmC in the promoter region has a negative role in the transcription of some genes in ES cells. Unlike 5mC, however, 5hmC is not substantially enriched at sites of heterochromatic H3K9 or H4K20 trimethylation22 (data not shown).
Collectively, our results support a model in which 5hmC and 5mC have different roles in transcription. Like 5mC28, 5hmC at promoters is predictive of lower levels of gene expression. However, 5hmC is uniquely associated with a ‘poised’ chromatin configuration and with genes that are upregulated upon differentiation, and may thus be involved in priming loci for rapid activation in response to appropriate signals. Activation of lineage-specific genetic loci upon differentiation could occur via a postulated 5mC ‘demethylation’ pathway (5mC to 5hmC to cytosine)1 or through recruitment of transcriptional regulators that specifically recognize 5hmC and are activated in response to differentiation signals. The ability to profile 5hmC even at sparsely hydroxymethylated loci will allow a careful evaluation of these possibilities in differentiating cells.
METHODS SUMMARY
GLIB precipitation
V6.5 ES cells were lysed and proteins digested by treatment with Proteinase K at 55 °C. DNA was purified by phenol-chloroform extraction and then precipitated with ethanol. RNA was removed with RNase A (Qiagen). Samples were treated with 20 ng BGT per 1 μg DNA at 30 °C for 3 h (50 mM HEPES pH 8.0, 25 mM MgCl2, 50 μM UDPG for 3 h at 30 °C), then oxidized with 23 mM sodium periodate 16 h at 22 °C in 0.1 M sodium phosphate pH 7.0. Periodate was quenched by the addition of 46 mM sodium sulphite at room temperature for 10 min, then exchanged into 1×PBS and incubated with 2 mM Aldehyde Reactive Probe (Invitrogen) for 1 h at 37 °C. DNA was sequenced with a HeliScope Single Molecule Sequencer. See Supplementary Methods for detailed protocol.
CMS precipitation
The generation of the anti-CMS antibody is described elsewhere5. DNA fragments were ligated with methylated adaptors and treated with sodium bisulphite (Qiagen). The DNA was then denatured for 10 min at 95 °C (0.4 M NaOH, 10 mM EDTA), neutralized by addition of cold 2 M ammonium acetate pH 7.0, incubated with anti-CMS antiserum in 1× immunoprecipitation buffer (10 mM sodium phosphate pH 7.0, 140 mM NaCl, 0.05% Triton X-100) for 2 h at 4 °C, and then precipitated with Protein G beads. Precipitated DNA was eluted with Proteinase K, purified by phenol-chloroform extraction, and amplified by 4–6 cycles PCR using Pfu TurboCx hotstart DNA polymerase (Stratagene). DNA sequencing was carried out using Illumina/Solexa Genome Analyzer II and HiSeq sequencing systems.
Acknowledgments
We thank B. Ren for assistance in next generation sequencing using the Illumina platform. We thank M. Guttman for making his RNASeq data set available to us. W.A.P. is supported by a predoctoral graduate research fellowship from the National Science Foundation, and Y.H. by a postdoctoral fellowship from the Leukemia and Lymphoma Society. R.L. is supported by a California Institute for Regenerative Medicine Training Grant. This study was supported by the National Institute of Health grants RC1 DA028422, R01 AI44432 and 1 R01 HD065812-01A1 and a grant from the California Institute of Regenerative Medicine (to A.R.), a pilot grant from Harvard Catalyst, The Harvard Clinical and Translational Science Center (NIH Grant 1 UL1 RR 025758-02) and NIH K08 HL089150 (to S.A.), and a grant from the Mary. K. Chapman Foundation (to J.R.E.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions W.A.P., Y.B. and S.A. devised the GLIB method. W.A.P., S.A., H.R.H. and E.M.M. optimized the GLIB method. Y.H. generated the anti-CMS antiserum, and Y.H. and W.A.P. optimized the anti-CMS pull-down. W.A.P. and Y.H. grew ES cells. W.A.P. prepared GLIB samples for sequencing, Y.H. prepared CMS samples, H.R.H. performed MeDIPs. Helicos sequencing and mapping was performed by P.K. and P.M.M., Illumina sequencing and mapping was performed by R.L. and J.R.E., and U.J.P. was responsible for bioinformatic analysis. M.K. performed the anti-5hmC dot blot. W.A.P. and M.T. performed anti-5hmC pull-downs. H.R.H. and S.M. performed and optimized in vitro tests of Tet substrate specificity. W.A.P., S.A. and A.R. wrote the manuscript. S.A. and A.R. coordinated research.
Author Information Data have been deposited at GEO under accession number GSE28682. Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loenarz C, Schofield CJ. Oxygenase catalyzed 5-methylcytosine hydroxylation. Chem Biol. 2009;16:580–583. doi: 10.1016/j.chembiol.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nature Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 11.Song CX, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayatsu H, Shiragami M. Reaction of bisulfite with the 5-hydroxymethyl group in pyrimidines and in phage DNAs. Biochemistry. 1979;18:632–637. doi: 10.1021/bi00571a013. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris TD, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 15.Bowers J, et al. Virtual terminator nucleotides for next-generation DNA sequencing. Nature Methods. 2009;6:593–595. doi: 10.1038/nmeth.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010 November 24; doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 21.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, et al. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- 26.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 28.Fouse SD, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]