We used up- and down-regulation of the nuclear receptor steroidogenic factor-1 (SF-1, NR5A1, Ad4BP) to identify new components of adrenal function and steroidogenesis.
Abstract
Context:
Steroidogenic factor-1 (SF-1, NR5A1, Ad4BP) is a master regulator of adrenal development and steroidogenesis. Defects in several known targets of SF-1 can cause adrenal disorders in humans.
Objective:
We aimed to identify novel targets of SF-1 in the human adrenal. These factors could be important regulators of adrenal development and steroidogenesis and potential candidates for adrenal dysfunction.
Design:
A gene discovery strategy was developed based on bidirectional manipulation of SF-1. Overexpression or knockdown of SF-1 in NCI-H295R human adrenocortical cells was used to identify a subset of positively-regulated SF-1 targets.
Results:
This approach identified well-established SF-1 target genes (STAR, CYP11A) and several novel genes (VSNL1, ZIM2, PEG3, SOAT1, and MTSS1). Given its role in cholesterol metabolism, sterol O-acyltransferase 1 (SOAT1, previously referred to as acyl-Coenzyme A:cholesterol acyltransferase 1, ACAT) was studied further and found to be expressed in the developing human fetal adrenal cortex. We hypothesized that impaired SOAT1 activity could result in adrenal insufficiency through reduced cholesteryl ester reserves or through toxic destruction of the adrenal cells during development. Therefore, mutational analysis of SOAT1 in a cohort of 43 patients with unexplained adrenal insufficiency was performed but failed to reveal significant coding sequence changes.
Conclusions:
Our reverse discovery approach led to the identification of novel SF-1 targets and defined SOAT1 as an important factor in human adrenal steroidogenesis. SF-1–dependent up-regulation of SOAT1 may be important for maintaining readily-releasable cholesterol reserves needed for active steroidogenesis and during episodes of recurrent stress.
Steroidogenic factor-1 (SF-1, NR5A1, Ad4BP) is a key transcriptional regulator of many aspects of adrenal and reproductive development, steroidogenesis, and metabolism (1). More than 30 SF-1 responsive genes have been identified, most of which play central roles in adrenal and/or reproductive function (2). Here, we describe a reverse discovery approach in an attempt to identify novel SF-1 targets, which we hypothesize could be important regulators of endocrine development and steroidogenesis. Using an experimental strategy based on bidirectional manipulation of SF-1 through overexpression or knockdown in a human adrenal cell line we have identified a subset of positively regulated SF-1 targets and investigated the potential role of one of these genes as a cause of adrenal insufficiency in humans.
Materials and Methods
Experimental design for bidirectional manipulation of SF-1
A strategy was devised to transiently coexpress green fluorescent protein (GFP) and either SF-1 cDNA (“overexpression”) or SF-1–specific small hairpin RNA (shRNA) (“knockdown”) in NCI-H295R human adrenocortical cells to allow enrichment for successfully transfected and viable cells through fluorescence-activated cell sorting (FACS) (overview of strategy in Supplemental Methods, published on The Endocrine Society's Journals Online web site at http://jcem.endojournals.org/). SF-1 overexpression was performed using the full-length coding sequence of wild-type (WT) human SF-1 cloned into a pIRES2-AcGFP1-Nuc vector (Clontech-Takara Bio Europe, Saint-Germain-en-Laye, France). The G35E mutation that impairs SF-1 DNA-binding and function in vivo (3) and in vitro (4) was used as experimental control. SF-1 knockdown was performed using the SureSilencing shRNA Plasmid for Human NR5A1 with GFP marker kit (KH05887G, SABiosciences, Frederick, MD), which includes a mismatch control.
Transfection and FACS
Plasmids (10 μg per 5 × 106 cells) were transfected into NCI-H295R cells using Amaxa Nucleofector II (Lonza Cologne AG, Cologne, Germany), Nucleofector kit R, and program T-020. Forty-eight hours after transfection, cells were harvested, prepared, and submitted to FACS in a MoFlo XDP sorter (Beckman Coulter, High Wycombe, UK) (protocol available on request). Viable GFP-expressing cells were either frozen to −80 C for protein analysis or pooled and resuspended in TRIzol reagent (Invitrogen, Paisley, UK) for RNA extraction.
Microarray analysis
Quality control of extracted RNA was performed with the 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Samples were processed using the Affymetrix GeneChip WT Sense Target Labeling kit (Affymetrix, High Wycombe, UK) according to manufacturer's instructions, starting with 200 ng total RNA. Four independent overexpression experiments and five independent knockdown experiments were performed and samples of labeled fragmented cDNA were hybridized to GeneChip Human Gene 1.0 ST Arrays (Affymetrix). Based on quality control of array data (R/Bioconductor and Partek Genomics Suite), two overexpression arrays (paired SF-1 WT and control) and one knockdown array (mismatch control) were excluded. Differential gene expression analysis was performed using the limma package in R/Bioconductor. A Benjamini-Hochberg-corrected P value cut-off of 0.05 was used to select significant differentially expressed genes.
Validation by immunoblotting and quantitative RT-PCR (qRT-PCR)
SF-1 expression in transfected cells was assessed by immunoblot (Western) analyses with an anti-SF-1 antibody (07-618; Upstate Millipore, Watford, UK). Optical densities of blots were quantified, normalized by β-actin expression (antibody AC-15, ab6276; Abcam, Cambridge, UK), and graphically represented in relation to basal/control.
SF-1–dependent changes in transcript levels of target genes were assessed by qRT-PCR. First-strand cDNA was generated using SuperScript II reverse transcriptase (Invitrogen) and quantitative PCR performed in a DNA Engine Opticon 2 Real-Time PCR System (Bio-Rad, Hemel Hempstead, UK) using RT2 SYBR Green Master Mix and qPCR Primer Assays for steroidogenic acute regulatory protein (STAR), CYP11A1, SOAT1, and B2M (β-2 microglobulin, endogenous control) (all SABiosciences). Data were analyzed using the 2−ΔΔCT method (5).
Confirmation of SF-1 responsiveness of novel targets
Based on analysis for putative SF-1–binding sites (MatInspector, www.genomatix.de) (6), the 1.0-kb, 4.8-kb, and 1.7-kb upstream sequences of VSNL1, SOAT1, and MTSS1, respectively, were PCR amplified and individually cloned into the pGL4.10[luc2] luciferase reporter (Promega, Southampton, UK). Expression vectors (pCMX) containing SF-1 [WT and mutant G35E (Mut)] cDNAs and our protocol for transient gene expression assays in NCI-H295R cells have been described previously (7).
Detection of sterol O-acyltransferase 1 (SOAT1) in human fetal adrenal glands
Human fetal adrenal tissue from 6 to 9 weeks postconception (wpc) and control tissue (heart; 8 wpc) were provided by the Medical Research Council/Wellcome Trust–funded Human Developmental Biology Resource with Research Ethics Committee approval and informed consent. Expression of SOAT1 transcript was assessed by qRT-PCR using the StepOnePlus Real-time PCR System, TaqMan Gene Expression Assays for human SOAT1 (Hs00162077_m1), and human glyceraldehyde-3-phosphate dehydrogenase as endogenous control (4333764T; all Applied Biosystems, Warrington, UK). Data were analyzed with StepOne software v2.1.
Immunofluorescence
Four-micron sections of adrenal tissue were incubated overnight with mouse polyclonal antihuman SOAT1 antibody (H00006646-B01, Abnova, Taiwan; 1:400 dilution). Alexa Fluor 555 antimouse IgG antibody (Invitrogen) was used for detection. Images were collected on Zeiss Axiophot and 710 confocal microscopes (Carl Zeiss, Hertfordshire, UK).
Mutational analysis
After institutional board approval and with informed consent, direct sequencing of the entire coding region of SOAT1 (NM_003101.4) was undertaken in a cohort of 43 patients with adrenal insufficiency of unknown etiology, with or without associated features (patients characteristics and amplification/sequencing protocols provided in Supplemental Methods). The Human Random Control-1 British Caucasian DNA Panel (Health Protection Agency Culture Collections, UK) was used for the analysis of previously unreported nonsynonymous changes.
Results
Validation of the experimental strategy
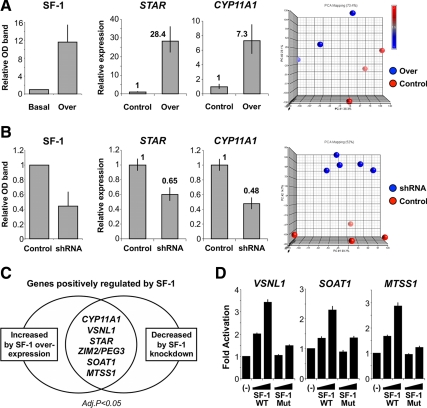
Before microarray analysis, the ability of overexpression or knockdown of SF-1 to identify known target genes was assessed using the well-established SF-1 regulated genes STAR and CYP11A1 (P450 side-chain cleavage enzyme) (2). Overexpression of SF-1 resulted in increased SF-1 protein at 48 h and increased STAR and CYP11A1 mRNA (Fig. 1A). Principal component analysis showed distinct differences in the genomic profiles resultant from SF-1 overexpression compared with control samples (Fig. 1A). In contrast, knockdown of SF-1 (NR5A1) using shRNA (Fig. 1B) resulted in decreased SF-1 protein, reduced STAR and CYP11A1 mRNA, and altered genomic profiles. Thus, this bidirectional strategy to manipulate SF-1 appeared robust.
Fig. 1.
Bidirectional manipulation of SF-1 in NCI-H295R adrenocortical cells. A, Transient overexpression of wild-type (WT) SF-1 (over) induced a 12-fold increase in SF-1 protein levels 48 h after transfection in relation to basal, as assessed by Western blotting. Transfection of a plasmid bearing the G35E mutation in SF-1 cDNA sequence that impairs SF-1 transactivation function was used as experimental control and led to overexpression of the mutated protein to similar levels (data not shown). WT SF-1 overexpression led to increased mRNA levels of well-established target genes CYP11A1 and STAR, assessed by qRT-PCR, in comparison with control. Global gene expression was analyzed using Affymetrix's Human Gene 1.0ST arrays and principal component analysis (PCA) showed distinct differences in the genomic profile resultant from WT SF-1 overexpression compared with control samples (3 arrays each, PCA performed using Partek Genomics Suite). B, Conversely, transient knockdown of SF-1 using SF-1–specific small hairpin RNA (shRNA) resulted in approximately 55% decrease in SF-1 protein levels in comparison to mismatch shRNA control at 48 h, reduced mRNA levels of STAR and CYP11A1, and distinct array genomic profiles (five SF-1–specific shRNA arrays, four mismatch controls). C, Aiming to identify novel SF-1 target genes in the adrenal, subsequent analysis focused on those genes for which significant changes in expression levels were determined by both SF-1 overexpression and knockdown (P < 0.05, Benjamini-Hochberg correction for multiple comparisons). A subset of seven genes up-regulated by SF-1 overexpression and down-regulated by SF-1 knockdown included well-established SF-1 targets STAR and CYP11A1 and five novel putative positively-regulated SF-1 target genes: visinin-like 1 (VSNL1); zinc finger, imprinted 2 (ZIM2); paternally expressed 3 (PEG3); sterol O-acyltransferase 1 (SOAT1, also referred to as acyl-Coenzyme A:cholesterol acyltransferase 1); and metastasis suppressor 1 (MTSS1). ZIM2 (Entrez Gene ID 23619) and PEG3 (Entrez Gene ID 5178) share the same locus and are represented by the same transcript cluster in Affymetrix's Human Gene 1.0ST arrays. D, SF-1 responsiveness of VSNL1, SOAT1, and MTSS1 was confirmed in luciferase assays. Promoter constructs were generated based upon in silico identification of SF-1–binding sites within the promoters of these genes [VSNL1 (1.0 kb), SOAT1 (4.8 kb), and MTSS1 (1.7 kb)]. Dose-dependent activation of all three promoters by wild-type SF-1 (WT) was seen. The amplitude of activation was similar to that of a well-established SF-1 target gene, Cyp11a1 (data not shown). This activation was diminished when the functionally-impaired G35E mutant SF-1 (Mut) was used (−, empty expression vector, followed by 50 and 100 ng per well of WT or Mut SF-1 expression vectors; data expressed as mean ± sem of at least three independent experiments, each performed in triplicate).
Identification of SF-1 targets
Overexpression of SF-1 resulted in significant up-regulation of 570 genes (P < 0.05, Benjamini-Hochberg correction for multiple comparisons) (Supplemental Table 1, A and B). Knockdown of SF-1 using shRNA resulted in significant down-regulation of 23 genes (P < 0.05, Benjamini-Hochberg correction) (Supplemental Table 2). Overlap of these datasets identified seven positively regulated genes at six loci (CYP11A1, VSNL1, STAR, ZIM2/PEG3, SOAT1, and MTSS1) (Fig. 1C) as well as fifteen negatively regulated genes at fourteen loci (Supplemental Table 3). SF-1 responsiveness of VSNL1, SOAT1, and MTSS1 was confirmed by in silico analysis of SF-1–binding sites within the promoters of these genes [VSNL1 (1.0 kb), SOAT1 (4.8 kb) and MTSS1 (1.7 kb)] as well as by showing SF-1–dependent activation of these promoters linked to luciferase (Fig. 1D).
Investigation of SOAT1 as a novel SF-1 target in steroidogenesis
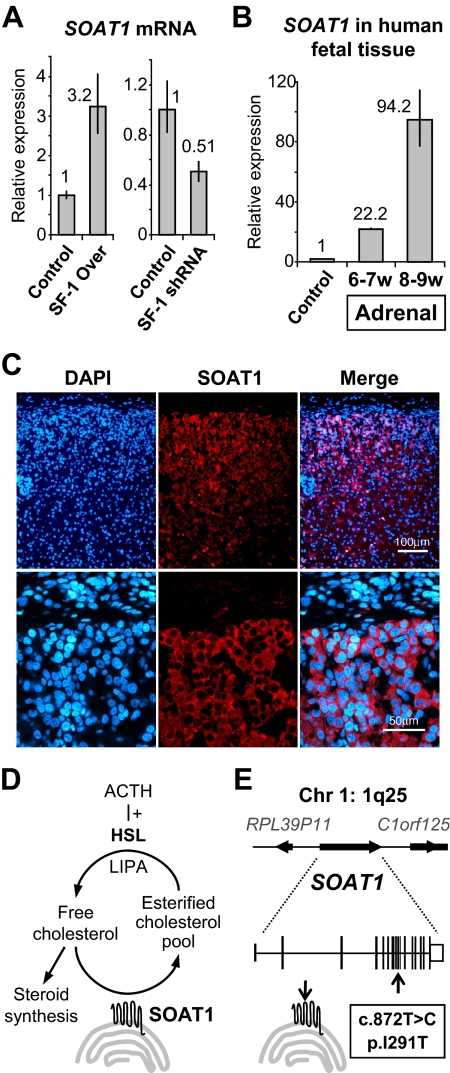
SF-1 regulation of SOAT1 was studied further, as this gene is involved in cholesterol metabolism (Supplemental Table 4). Overexpression and knockdown of SF-1 resulted in up-regulation or down-regulation of SOAT1, respectively, by qRT-PCR as well as microarray analysis (Fig. 2A). Furthermore, SOAT1 transcript levels were higher in the fetal adrenal gland at a stage of active steroidogenesis compared with control (Fig. 2B), and immunofluorescence showed strong SOAT1 signal in the developing definitive zone of the fetal adrenal cortex (Fig. 2C).
Fig. 2.
A, Increased and decreased levels of SOAT1 mRNA 48 h after SF-1 overexpression or knockdown, respectively, were confirmed by qRT-PCR [data represented according to the 2−ΔΔCT method (5)]. B, Expression of SOAT1 mRNA in human fetal adrenal tissue was assessed by qRT-PCR. Results showed a marked increase in SOAT1 expression from 6–7 to 8–9 wpc in comparison to control tissue (heart, 8 wpc), reflecting active steroidogenesis in the fetal adrenal cortex. C, Immunofluorescence confirmed strong expression of SOAT1 in the outer layers of the human fetal adrenal cortex at 8 wpc but not in overlying nonsteroidogenic capsule cells (DAPI, 4′,6-diamidino-2-phenylindol, was used to visualize nuclei). The omission of primary antibody resulted in no signal (data not shown). D, Cartoon representation of the actions of SOAT1 (HSL, hormone-sensitive lipase; LIPA, lipase A). E, Cartoon representation of the SOAT1 locus at 1q25 and of the allelic variant identified.
A role for SOAT1 in human steroidogenesis
SOAT1 (also known as acyl-Coenzyme A:cholesterol acyltransferase 1 and previously referred to as ACAT) is an important regulator of cholesteryl ester formation in the adrenal gland (8). These stored pools of esterified cholesterol can be readily liberated by the action of hormone-sensitive lipase after ACTH stimulation and protect the adrenal cells from the potentially damaging effects of free cholesterol (Fig. 2D) (9). We hypothesized that impaired SOAT1 activity could result in adrenal insufficiency in humans through reduced cholesteryl ester reserves or through toxic destruction of the adrenal cells during development. Mutational analysis of the SOAT1 gene was therefore undertaken in a cohort of 43 patients with adrenal insufficiency of unknown etiology (Supplemental Methods). A novel heterozygous allelic variant in SOAT1 was identified in 4/84 alleles (4.8%) (c.872T>C; p.I291T; Ensembl GRCh37 transcript ENST00000367619) (Fig. 2E), but a similar variant occurred in 4/186 (2.2%) control alleles (χ2 = 0.25). No potential disease-causing mutations were found in SOAT1, nor in another candidate gene (C2CD2) identified from the SF-1 overexpression dataset (Supplemental Data).
Discussion
In recent years, substantial progress has been made in our understanding of the molecular basis of human adrenal development and function. Nevertheless, a significant proportion of cases of primary adrenal failure currently remain unexplained (10). As several targets of SF-1 have been shown to cause adrenal dysfunction in humans (11, 12), we developed a gene discovery strategy based on the bidirectional manipulation of SF-1 in human adrenal cells, hypothesizing that novel SF-1 targets identified in this system could be potential candidates for adrenal disorders.
Our strategy for SF-1 overexpression or knockdown appeared robust at detecting positively-regulated SF-1 targets, as CYP11A1 and STAR were two of seven genes identified in the overlapping dataset. The five novel putative targets of SF-1 identified were as follows: visinin-like 1 (VSNL1); zinc finger, imprinted 2 (ZIM2); paternally expressed 3 (PEG3); metastasis suppressor 1 (MTSS1); and, interestingly, sterol O-acyltransferase 1 (SOAT1).
Subsequent studies focused on SOAT1 (OMIM: 102642), a regulator of cholesteryl ester formation, because ACTH-induced mobilization of stored ester pools via hormone-sensitive lipase is an important mechanism to generate free cholesterol for adrenal steroidogenesis (8, 9). Furthermore, SOAT1 is essential for intracellular cholesterol homeostasis, maintaining appropriate levels of unesterified cholesterol within cells for membrane stability (13). We show here that SOAT1 is under SF-1 regulation in adrenal cells in vitro and that SOAT1 expression in vivo coincides spatially and chronologically with active steroidogenesis in the human fetal adrenal cortex.
Naturally-occurring (ald, AKR, Soat1ald) or targeted disruption of Soat1 in mice leads to marked lipid depletion in the adrenal cortex and variable abnormalities of cholesterol esterification and corticosterone synthesis (14, 15). However, substantial interspecies differences exist in the mechanisms of cholesterol generation as well as in the expression and activity of this enzyme (16, 17), as recently demonstrated by differences in antiatherogenic effects elicited by SOAT1 inhibitors between species (18). We therefore hypothesized that impaired SOAT1 activity could result in adrenal insufficiency in humans, either through reduced cholesteryl ester reserves or through toxic destruction of the adrenal cells during development. Although SOAT1 changes are not a common cause of adrenal dysfunction in this heterogeneous cohort of patients studied, it is possible that SOAT1 allelic variants may explain less severe cases of adrenal dysfunction or have more prominent effects in systems more susceptible to subtle changes in steroidogenic output such as the developing testis.
In conclusion, our reverse discovery approach based on the bidirectional manipulation of SF-1 in adrenal cells led to the identification of novel SF-1 targets in this system and defined SOAT1 as an important factor in human adrenal steroidogenesis. We hypothesize that SF-1–dependent up-regulation of SOAT1 is important for maintaining readily-releasable pools of cholesterol esters needed at times of active steroidogenesis or during episodes of recurrent stress.
Supplementary Material
Acknowledgments
We thank Joao Metelo and Ayad Eddaoudi at the Flow Cytometry Core Facility at the University College London Institute of Child Health for supporting fluorescence-activated cell sorting; Priya Banerjee for microarray processing; Sonia Shah at the Bloomsbury Centre for Bioinformatics for microarray data analysis; Dianne Gerrelli for tissue samples; and Ronald Evans, Meera Ramayya, and J. Larry Jameson for plasmids. Human embryonic material was provided by the Medical Research Council (MRC)/Wellcome Trust-funded Human Developmental Biology Resource (www.hdbr.org) (MRC, G0700089; The Wellcome Trust, GR082557). We also thank the many pediatricians and physicians who have referred patients, especially Annie Procter at the All Wales Medical Genetics Service.
This work was supported by a Capes scholarship (4798066, Capes/Brazil) (to B.F.-d.-S.) and a Wellcome Trust Senior Research Fellowship in Clinical Science (GR079666) (to J.C.A.).
The array data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under series accession no. GSE26529.
Disclosure Summary: The authors have nothing to declare.
Footnotes
- FACS
- Fluorescence-activated cell sorting
- GFP
- green fluorescent protein
- Mut
- mutant G35E
- qRT-PCR
- quantitative RT-PCR
- SF-1
- steroidogenic factor-1
- shRNA
- small hairpin RNA
- SOAT1
- sterol O-acyltransferase 1
- STAR
- steroidogenic acute regulatory protein
- wpc
- weeks postconception
- WT
- wild-type.
References
- 1. Parker KL, Schimmer BP. 1997. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev 18:361–377 [DOI] [PubMed] [Google Scholar]
- 2. Schimmer BP, White PC. 2010. Minireview: steroidogenic factor 1: its roles in differentiation, development, and disease. Mol Endocrinol 24:1322–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Achermann JC, Ito M, Ito M, Hindmarsh PC, Jameson JL. 1999. A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat Genet 22:125–126 [DOI] [PubMed] [Google Scholar]
- 4. Ito M, Achermann JC, Jameson JL. 2000. A naturally occurring steroidogenic factor-1 mutation exhibits differential binding and activation of target genes. J Biol Chem 275:31708–31714 [DOI] [PubMed] [Google Scholar]
- 5. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 6. Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933–2942 [DOI] [PubMed] [Google Scholar]
- 7. Ferraz-de-Souza B, Martin F, Mallet D, Hudson-Davies RE, Cogram P, Lin L, Gerrelli D, Beuschlein F, Morel Y, Huebner A, Achermann JC. 2009. CBP/p300-interacting transactivator, with Glu/Asp-rich C-terminal domain, 2, and pre-B-cell leukemia transcription factor 1 in human adrenal development and disease. J Clin Endocrinol Metab 94:678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang TY, Li BL, Chang CC, Urano Y. 2009. Acyl-coenzyme A:cholesterol acyltransferases. Am J Physiol Endocrinol Metab 297:E1–E9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kraemer FB. 2007. Adrenal cholesterol utilization. Mol Cell Endocrinol 265- 266:42–45 [DOI] [PubMed] [Google Scholar]
- 10. Lin L, Gu WX, Ozisik G, To WS, Owen CJ, Jameson JL, Achermann JC. 2006. Analysis of DAX1 (NR0B1) and steroidogenic factor-1 (NR5A1) in children and adults with primary adrenal failure: ten years' experience. J Clin Endocrinol Metab 91:3048–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bose HS, Sugawara T, Strauss JF, 3rd, Miller WL. 1996. The pathophysiology and genetics of congenital lipoid adrenal hyperplasia. N Engl J Med 335:1870–1878 [DOI] [PubMed] [Google Scholar]
- 12. Kim CJ, Lin L, Huang N, Quigley CA, AvRuskin TW, Achermann JC, Miller WL. 2008. Severe combined adrenal and gonadal deficiency caused by novel mutations in the cholesterol side chain cleavage enzyme, P450scc. J Clin Endocrinol Metab 93:696–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Warner GJ, Stoudt G, Bamberger M, Johnson WJ, Rothblat GH. 1995. Cell toxicity induced by inhibition of acyl coenzyme A:cholesterol acyltransferase and accumulation of unesterified cholesterol. J Biol Chem 270:5772–5778 [DOI] [PubMed] [Google Scholar]
- 14. Meiner VL, Cases S, Myers HM, Sande ER, Bellosta S, Schambelan M, Pitas RE, McGuire J, Herz J, Farese RV., Jr 1996. Disruption of the acyl-CoA:cholesterol acyltransferase gene in mice: evidence suggesting multiple cholesterol esterification enzymes in mammals. Proc Natl Acad Sci USA 93:14041–14046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meiner VL, Welch CL, Cases S, Myers HM, Sande E, Lusis AJ, Farese RV., Jr 1998. Adrenocortical lipid depletion gene (ald) in AKR mice is associated with an acyl-CoA:cholesterol acyltransferase (ACAT) mutation. J Biol Chem 273:1064–1069 [DOI] [PubMed] [Google Scholar]
- 16. Gwynne JT, Strauss JF., 3rd 1982. The role of lipoproteins in steroidogenesis and cholesterol metabolism in steroidogenic glands. Endocr Rev 3:299–329 [DOI] [PubMed] [Google Scholar]
- 17. Lee RG, Willingham MC, Davis MA, Skinner KA, Rudel LL. 2000. Differential expression of ACAT1 and ACAT2 among cells within liver, intestine, kidney, and adrenal of nonhuman primates. J Lipid Res 41:1991–2001 [PubMed] [Google Scholar]
- 18. Nissen SE, Tuzcu EM, Brewer HB, Sipahi I, Nicholls SJ, Ganz P, Schoenhagen P, Waters DD, Pepine CJ, Crowe TD, Davidson MH, Deanfield JE, Wisniewski LM, Hanyok JJ, Kassalow LM. 2006. Effect of ACAT inhibition on the progression of coronary atherosclerosis. N Engl J Med 354:1253–1263 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.