Abstract
Traditionally, well-defined three-dimensional structure was thought to be essential for protein function. However, myriad biological functions are performed by highly dynamic, intrinsically disordered proteins (IDPs). IDPs often fold upon binding their biological targets and frequently exhibit “binding diversity” by targeting multiple ligands. We sought to understand the physical basis of IDP binding diversity and herein report that the cyclin-dependent kinase (Cdk) inhibitor, p21Cip1, adaptively binds to and inhibits the various Cdk/cyclin complexes that regulate eukaryotic cell division. Based on results from NMR spectroscopy, and biochemical and cellular assays, we show that structural adaptability of a helical sub-domain within p21 termed LH enables two other sub-domains termed D1 and D2 to specifically bind conserved surface features of the cyclin and Cdk subunits, respectively, within otherwise structurally distinct Cdk/cyclin complexes. Adaptive folding upon binding is likely to mediate the diverse biological functions of the thousands of IDPs present in eukaryotes.
Introduction
The traditional view of protein structure-function relationships posits that a well-defined three-dimensional (3D) structure is required for function1,2. However, it is now well appreciated that many biological functions are performed by highly dynamic proteins or protein domains that, in isolation, lack secondary and/or tertiary structure under physiological conditions3. Such proteins are termed intrinsically disordered (or unstructured) proteins (hereafter referred to as IDPs). IDPs exist in organisms from all kingdoms of life4 and are most prevalent in eukaryotes4. IDPs exhibit distinct, functionally relevant features compared to globular proteins. First, IDPs frequently fold upon binding to their biological targets5–7. Second, IDPs often interact with numerous biological targets, a phenomenon termed “binding diversity”7. The concept that the intrinsic flexibility affords functional advantages to IDPs by enabling binding diversity has been widely discussed3,7,8; however, the physical basis for this phenomenon is poorly understood. To understand the mechanism(s) underlying IDP binding diversity, we investigated the structural and dynamic features of the cell cycle inhibitor, p21Cip1 (p21)9, which interacts with and inhibits multiple cyclin-dependent kinase (Cdk)/cyclin complexes.
Progression of the mammalian cell cycle is regulated by numerous Cdks and their associated regulatory subunits termed cyclins10, hereafter referred to as the Cdk/cyclin repertoire. Cell cycle initiation via progression from G1 to S phase is triggered by partial phosphorylation of the retinoblastoma protein (Rb) by Cdk4/cyclin D and Cdk6/cyclin D complexes followed by hyper-phosphorylation of Rb by Cdk2/cyclin E in late G1 phase11. Cdk2/cyclin A and Cdk1/cyclin B complexes mediate the orderly progression through S phase and transition from G2 to M phase, respectively11. The Cip/Kip proteins, including p21, p27Kip1 (p27) and p57Kip2 (p57)9, were originally described as paralogous inhibitors of multiple mammalian Cdks. In particular, p21 was described as a universal inhibitor of the Cdk/cyclin repertoire12, including Cdk1, Cdk2, Cdk4 and Cdk6 paired with their respective cyclin partners (e.g., cyclin A, B1, B2, D1 and D3)13,14. Although p21, p27 and p57 exhibit inhibitory activity toward multiple Cyclin/Cdk complexes9, p21 and p27 have also been shown to positively regulate Cdk4 (and Cdk6) by mediating their assembly with D-type cyclins15. Inhibitory interactions between the Cip/Kip proteins and Cdk/cyclin complexes are mediated by a conserved, N-terminal ~61 residue domain termed the kinase inhibitory domain (KID). Subsequent to the discovery that the Cip/Kip family of proteins regulates a multitude of Cdk/cyclin complexes, it was determined that isolated Cip/Kip proteins lacked significant secondary and tertiary structure7,16, and that p21 and p27 folded only upon binding to Cdk/cyclin complexes6,7,16. More than a decade later, the Cip/Kip proteins are considered to be prototypical IDPs5–7 and therefore provide a powerful model system to study relationships between their structural and dynamic features and their biological functions. The crystal structure of the p27 KID bound to Cdk2/cyclin A explained how p27 binds to and inhibits this particular Cdk/cyclin complex17 (Fig. 1a). However, these data alone do not explain the mechanism(s) that mediate promiscuous binding to the full Cdk/cyclin repertoire.
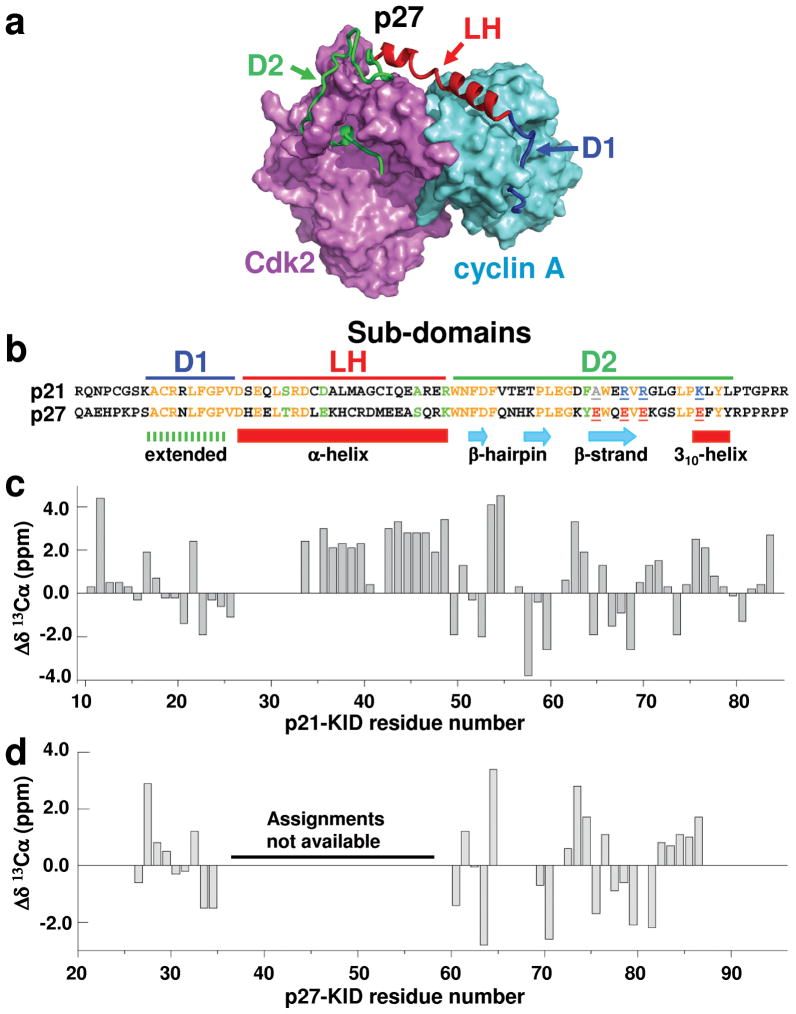
Figure 1.
p21 and p27 adopt similar secondary structure when bound to Cdk2/cyclin A. (a) Structure of p27-KID bound to Cdk2/cyclin A (PDB 1JSU17) showing sub-domains D1 (blue), LH (red) and D2 (green) of p27-KID. Cdk2 and cyclin A are illustrated in magenta and cyan, respectively. (b) Sequence alignment of the kinase inhibitory domains (KIDs) of p21 and p27. Sub-domains are indicated by bars at the top, sequence identities are denoted in orange letters and similarities in green letters. Four non-conserved Glu residues within sub-domain D2 of p27 are colored red and underlined; the corresponding residues in p21 are colored grey (Ala) or blue (Arg and Lys) and also underlined. The secondary structure for p27 observed in the crystal structure p27/Cdk2/cyclin A is indicated at the bottom. Secondary 13Cα chemical shift values (Δδ 13Cα) for residues in (c) p21-KID and (d) p27-KID bound to Cdk2/cyclin A are represented as vertical gray bars. Resonance assignments for seven residues in the LH sub-domain of p21-KID and this entire sub-domain of p27-KID are not available.
In the present study, we investigated relationships between dynamic features of p21 and its function as an inhibitor of multiple Cdk/cyclin complexes using spectroscopic, biochemical and cellular methods. The N-terminal KID of p21 (residues 17–78; p21-KID) and p27 (residues 28–89; p27-KID) can be divided into three sub-domains: D1, LH and D26 (Figs. 1a and b). Based upon sequence homology, structural investigations of isolated p217, and the structure of p27 bound to Cdk2/cyclin A17, it can be generalized that sub-domain D1 of p21 binds to the cyclin subunit of Cdk/cyclin complexes and sub-domain D2 binds to the Cdk subunit. In contrast, sub-domain LH, which adopts a partially α-helical conformation, plays primarily a structural role by tethering sub-domains D1 and D2 together6,17. Interestingly, our multidisciplinary studies revealed that, when p21 is bound to Cdk2/cyclin A, sub-domain LH is not rigid but rather dynamic, allowing it to serve as an adaptable linker between sub-domains D1 and D2. We determined that the natural length of this linker is specifically required for inhibition of multiple Cdk/cyclin complexes by p21. Our findings suggest that the dynamic nature of sub-domain LH allows p21 to adaptively bind to the individual complexes which comprise the Cdk/cyclin repertoire that regulates cell division. Moreover, they provide fundamental physical insights into how the dynamic features of disordered proteins enable binding to multiple targets and perform diverse biological functions.
Results
p21 and p27 bind similarly to Cdk2/cyclin A
Based upon our previous partial resonance assignments18, secondary 13Cα chemical shift values19 (Δδ13Cα; Fig. 1c) were used to analyze the secondary structure of p21-KID bound to Cdk2/cyclin A. Sub-domain D1 of p21-KID, identical at 9/10 positions with respect to sub-domain D1 of p27 (Fig. 1b), exhibits Δδ13Cα values (Fig. 1c) generally consistent with an extended conformation20, as was observed for sub-domain D1 of p27-KID bound to Cdk2/cyclin A in crystals17 (Fig. 1a). In crystals, sub-domain D2 of p27 exhibits a variety of secondary structures, including a β-hairpin (residues 62–69), a β-strand (residues 74–78), and a single turn of 310 helix (residues 86–89) (Fig. 1a, b)17. The Δδ13Cα values observed for sub-domain D2 of p21 in the p21-KID/Cdk2/cyclin A complex in solution are consistent with the latter observations for p27-KID in crystals17. Further, Δδ13Cα values for the 15 residues within sub-domain LH indicate two segments of α-helix (including residues 33–39 and 42–48) separated by Gly 40 and Cys 41 which may break the helix (Fig. 1c). Finally, comparison of Δδ13Cα values for p21-KID and p27-KID bound to Cdk2/cyclin A in solution (Fig. 1c versus d) strongly suggested that the conformations of sub-domains D1 and D2 of the two Cdk inhibitory proteins were very similar.
Sub-domain LH stretches to bind Cdk2/cyclin A
NMR spectroscopy was used to probe the dynamics of p21-KID bound to Cdk2/cyclin A to understand whether flexibility observed in the free state7,21 was fully quenched in the bound state (Fig. 2a). Amide groups within sub-domain D1 exhibited 1H-15N heteronuclear nuclear Overhauser effect (hetNOE) values, which probe dynamics on the picosecond to nanosecond timescale, consistent with rigid secondary structure (hetNOE values near +0.8). Amides in subdomain D2 exhibited a similar dynamic profile, although some segments exhibited hetNOE values near +0.6, indicative of a higher degree of motion. For sub-domain LH, amides at the extreme C-terminus appeared rigid. In contrast, amides in the center of this sub-domain (residues 34–44) exhibited hetNOE values between +0.3 and +0.4, indicative of increased mobility in comparison with those in sub-domains D1 and D2. Further, resonances for seven residues within the N-terminal portion of sub-domain LH were not observed, possibly due to dynamic conformational exchange.
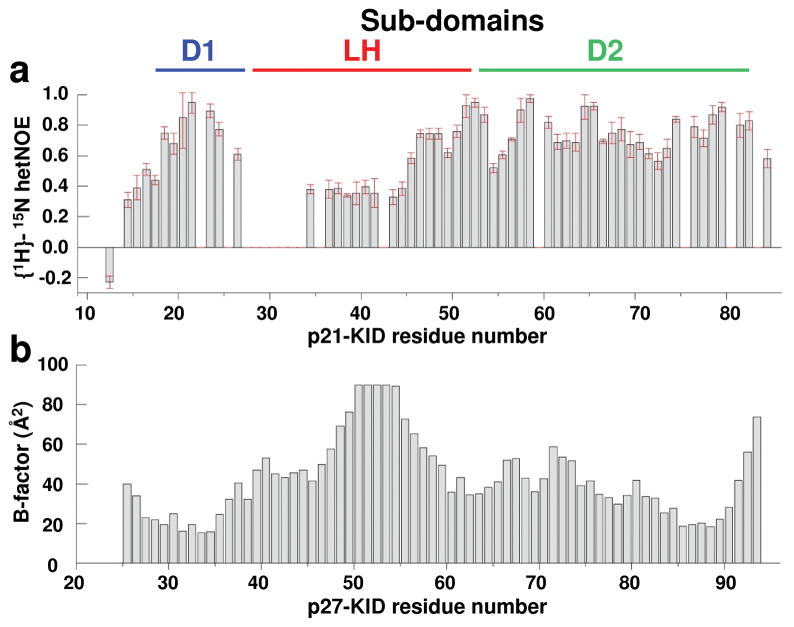
Figure 2.
The LH sub-domains of p21-KID and p27-KID exhibit flexibility and disorder within ternary complexes with Cdk2/cyclin A. (a) {1H}-15N hetNOE values for p21-KID when bound to Cdk2/cyclin A. The error bars represent the standard deviation of the mean based of triplicate measurements. (b) B-factor values for backbone amide N atoms for p27-KID within the ternary complex with Cdk2/cyclin A (PDB 1JSU).
These hetNOE results suggested that many residues within sub-domain LH remain dynamic when p21-KID was bound to Cdk2/cyclin A. This was surprising considering that p21-KID fully inhibits Cdk2/cyclin A with a Ki value of 0.3 nM22, indicative of thermodynamically very favorable interactions between p21-KID and Cdk2/cyclin A. To investigate the origins of flexibility within sub-domain LH, the structure of p21-KID within the p21-KID/Cdk2/cyclin A complex was modeled on the basis of the crystal structure of the corresponding complex containing p27-KID using Swiss-Model23. In this model, the length of p21 sub-domain LH, based on the distance between the Cα atoms of residues 27 and 48, was 36.0 Å. Based on standard parameters (e.g., translation of 1.5 Å/residue24), this distance in an α-helix would be 31.5 Å, not 36 Å. This analysis suggests that the α-helical LH sub-domain of p21-KID, within the ternary complex with Cdk2/cyclin A, is elongated, or stretched, by ~4.5 Å relative to the length of a standard α-helix. This stretching would weaken the hydrogen bonds that normally stabilize α-helices24 and, consequently, allow local flexibility.
The length of sub-domain LH in the p27-KID structure (between residues 38 and 59) is 36.2 Å, also corresponding to a stretched helix. Due to the lack of resonance assignments for this sub-domain within the p27-KID/Cdk2/cyclin A complex, we were unable to probe its dynamic features in solution. However, we analyzed crystallographic B-factors for amide N atoms reported in the coordinate file (1JSU17) (Fig. 2b). B-factor values for most amide groups of p27-KID were <60 Å 2 with the exception of residues 47 to 56 within sub-domain LH and residue 93 at the C-terminus. This indicated that residues within the central region of sub-domain LH of p27-KID exhibited greater flexibility relative to residues in sub-domains D1 and D2, a flexibility pattern which parallels that observed for p21-KID (bound to Cdk2/cyclin A) in solution on the basis of hetNOE values. The similarity of the solution NMR results for p21-KID and the X-ray crystallography results for p27-KID, strongly suggests that sub-domain LH in both molecules is stretched to similar extents in the ternary complexes and that stretching destabilizes helical structure and introduces flexibility. Notably, this conservation of dynamic features within the LH sub-domains of p21 and p27 occurrs despite sequence dissimilarity at 14/22 positions within these stretched helices.
We hypothesized that sub-domain LH is a stretchable linker between sub-domains D1 and D2 to mediate specific and functionally critical interactions with cyclin and Cdk subunits of the Cdk/cyclin repertoire. Further, we propose that the ability of sub-domain LH to stretch is required for p21 to accommodate a range of distances between the Cdk and cyclin subunits of these complexes. The different sub-domains of p27-KID bind sequentially to Cdk2/cyclin A, with sub-domain D1 binding first to cyclin A followed by binding of sub-domain D2 to Cdk26. Subdomain D1 (of p27-KID) “reaches over” to cyclin subunits to block a substrate recruitment site25 that is conserved within the Cdk/cyclin repertoire26. Sub-domain D2 independently mediates kinase inhibition by binding to the N-terminal β-sheet domain of Cdk2 and inserting Tyr 88 into the kinase ATP binding pocket17. Sub-domain LH therefore may function to simply connect subdomains D1 and D2 and may have evolved to accommodate the similar yet subtly different structural features of different Cdk/cyclin complexes. The ability of sub-domain LH to stretch, or structurally adapt, affords a molecular mechanism for the Cdk/cyclin binding diversity observed for p21. We additionally reasoned that the binding promiscuity of p21 would be altered if the length and thus the structural adaptability of sub-domain LH was altered. We tested this hypothesis through spectroscopic, biochemical and cellular studies of p21-KID variants in which sub-domain LH was decreased (p21-KID-LH−3) or increased (p21-KID-LH+3) in length by three residues, corresponding approximately to one helical turn (Fig. 3a).
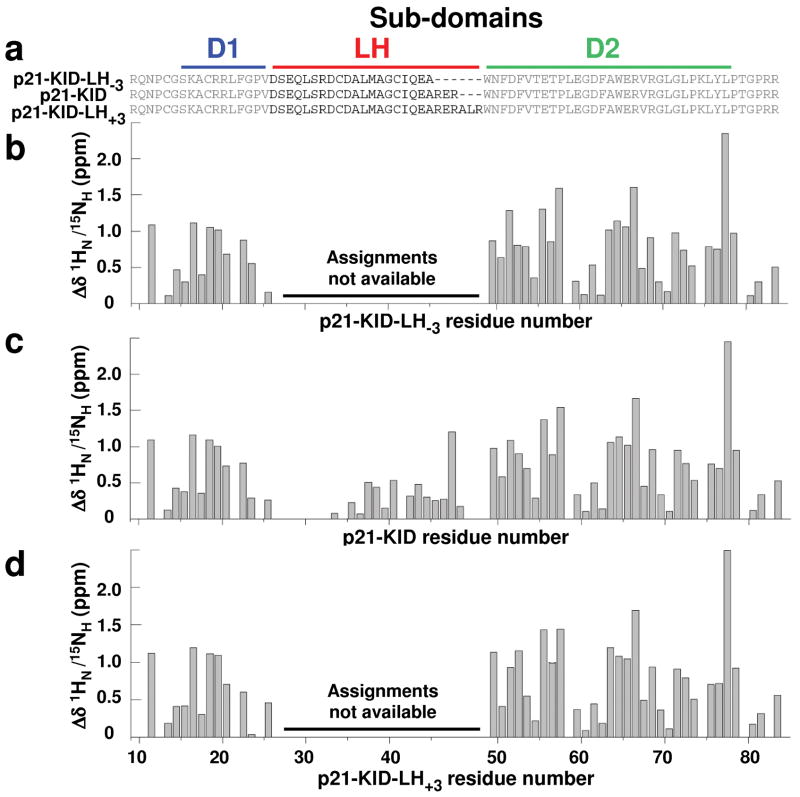
Figure 3.
The Cdk2/cyclin A-bound structures of sub-domains D1 and D2 within the p21 constructs are unaffected by elongation or truncation of sub-domain LH by 3 residues. (a) Sequences of wild-type p21-KID and the variants with a lengthened (p21-KID-LH+3) and shortened (p21-KID-LH−3) LH sub-domain. Differences between experimental composite 1HN/15NH chemical shift values for residues in (b) p21-KID-LH−3, (c) p21-KID, and (d) p21-KID-LH+3 when bound to Cdk2/cyclin A and sequence-adjusted, standard values for residues in random coil peptides (Δδ 1HN/15NH). The magnitude of these Δδ 1HN/15NH values reflects the extent of structural change that occurs when the p21 constructs fold upon binding to Cdk2/cyclin A.
Structural features of LH variants bound to Cdk2/cyclin A
We analyzed the structure of p21-KID-LH−3 and p21-KID-LH+3 bound to Cdk2/cyclin A using NMR spectroscopy (Supplementary Results, Supplementary Fig. 1). The binding-induced chemical shift values for amide groups within sub-domains D1 and D2 for p21-KID, p21-KID-LH−3 and p21-KID-LH+3 (presented as the difference [Δδ1HN /15NH] between experimental 1H-15N chemical shift values [δ1HN /15NH(expt)] and values for random conformations [δ1HN /15NH(rc)]19) were very similar (Fig. 3b–d). Resonance assignments were not made for residues within the altered LH sub-domains of complexes containing p21-KID-LH−3 and p21-KID-LH+3. These results indicate that the LH sub-domains in the different p21-KID constructs structurally adapt so that sub-domains D1 and D2 can interact similarly with cyclin A and Cdk2, respectively, within Cdk2/cyclin A complexes. Based on this analysis, the LH sub-domain of p21-KID-LH+3 may be stretched to the smallest extent relative to a standard α-helix, while that of p21-KID-LH−3 may be stretched to the greatest extent.
The p21-KID constructs variably stabilize Cdk2/cyclin A
While varying the length of sub-domain LH did not affect the structure of sub-domains D1 and D2 when bound to Cdk2/cyclin A, it was possible that these alterations affected the thermodynamics of interactions within this complex. Similar to past observations for p27-KID27, the binding of p21-KID caused the thermal denaturation temperature (Tmapp) of Cdk2/cyclin A to increase from 50.3 °C to 70.5 °C (Table 1 and Supplementary Fig. 2). Interestingly, p21-KID-LH+3 exhibited slightly greater stabilization (Tmapp, 72.9 °C) while p21-KID-LH−3 stabilized Cdk2/cyclin A to a significantly smaller extent (Tmapp, 58.4 °C). These results suggested that the p21-KID-LH+3 ternary complex was slightly more stable than that which contained wild-type p21-KID and that the ternary complex that contained p21-KID-LH−3 was significantly less stable. These results further suggested that the different LH sub-domains were stretched and thus destabilized to different extents when bound to Cdk2/cyclin A. An alternative interpretation was that the LH sub-domain may directly contribute to Cdk2/cyclin A binding. If this is true, altering the length of the LH sub-domain could account for the varied thermal stability of the three ternary complexes. To address this issue, we used isothermal titration calorimetry (ITC) to determine whether the wild-type and variant LH sub-domains directly contributed to the Gibbs free energy of binding (ΔG) to Cdk2/cyclin A. In addition, we analyzed the contributions of the D1 and D2 sub-domains to Cdk2/cyclin A binding. Peptides corresponding to each of the LH sub-domains (wt LH, LH+3 and LH−3) failed to produce significant heat when titrated into Cdk2/cyclin A, indicating that they do not directly contribute to ΔG of binding (Supplementary Fig. 3 and Supplementary Table 1). In contrast, sub-domain D1 exhibited a Kd value of 61 nM and D2 a value of 5.3 μM for binding to Cdk2/cyclin A. Thus, sub-domains D1 and D2 of p21 dominated the thermodynamics of interactions with the Cdk2/cyclin A complex, while the contribution of all LH sub-domain variants were negligible. These results are generally consistent with those obtained previously with sub-domains of p27-KID, wherein D1 bound to cyclin A with a Kd value of 25 nM and D2 bound to Cdk2 with a value of 70 nM6. However, the observation that binding of p21 sub-domain D2 to Cdk2 was weak (Kd, 5.3 μM) in comparison with the relatively tight binding of this sub-domain of p27 (Kd, 70 nM) was surprising. Inspection of the sequences of the two proteins within the D2 sub-domain, however, revealed a possible explanation for the decreased affinity of p21-D2 for Cdk2. First, four Glu residues within p27-D2 are substituted by Ala, two Arg residues and Lys in p21-D2 (Fig. 1b). Second, electrostatic computations showed that the four Glu residues of p27-D2 interact favorably with an electropositive surface of Cdk2 (Supplementary Fig. 4a and b) and that within p21-D2 these interactions are unfavorable (see Supplementary Methods). However, with both p21 and p27, the presence of sub-domains D1 and D2, connected by the LH sub-domain, is associated with high Cdk2 inhibitory potency (IC50 < 5 nM)7,28, despite the relatively weak binding of p21-D2 to Cdk2. Together, the thermal denaturation and ITC results strongly suggested that alteration of the LH sub-domain of p21 had no direct effect on interactions with Cdk2/cyclin A but rather indirectly affected the thermodynamic behavior of the KID constructs through altered LH subdomain stretching.
Table 1.
Values of apparent thermal denaturation temperatures (Tmapp) determined using CD.
| Protein complex | Tmapp (°C) |
|---|---|
| Cdk2/cyclin A | 50.3 ± 0.3 |
| p21-KID/Cdk2/cyclin A | 70.5 ± 0.2 |
| p21-KID-LH−3/Cdk2/cyclin A | 58.4 ± 0.6 |
| p21-KID-LH+3/Cdk2/cyclin A | 72.9 ± 0.4 |
Altering sub-domain LH alters biochemical promiscuity
We hypothesized that, if the structural adaptability of sub-domain LH mediates the binding of p21 to the diverse Cdk/cyclin complexes that regulate cell division, alteration of sub-domain LH should alter binding diversity. To test this hypothesis, we determined the activity of wild-type p21-KID and the LH sub-domain variants in vitro toward a panel of catalytically active Cdk/cyclin complexes, including Cdk1/cyclin B1, Cdk2/cyclin A, Cdk4/cyclin D1, and Cdk6/cyclin D1 (Fig. 4a and Supplementary Fig. 5). p21-KID and p21-KID-LH+3 were essentially equipotent inhibitors of Cdk1/cyclin B1 activity, with IC50 values of 40 nM and 71 nM, respectively. In contrast, the IC50 value for p21-KID-LH−3 (218 nM) was significantly higher, indicating that shortening subdomain LH diminished inhibitory activity toward Cdk1/cyclin B1. Notably, at saturating concentrations (>10 μM) of p21-KID-LH−3, Cdk1 retained 20% activity (Supplementary Fig. 5a). p21-KID and p21-KID-LH+3 were also potent inhibitors of Cdk2/cyclin A kinase activity, with IC50 values of 2.6 and 0.8 nM, respectively, and, as for Cdk1/cyclin B1, p21-KID-LH−3 was a poor inhibitor of Cdk2/cyclin A (IC50, 108 nM). It is interesting that p21-KID-LH+3 was a slightly more potent inhibitor of Cdk2/cyclin A than wild-type p21-KID, suggesting that the length of the wild-type LH sub-domain is non-optimal with regard to inhibition of this particular Cdk/cyclin complex. p21-KID exhibited similar IC50 values toward Cdk4/cyclin D1 and Cdk6/cyclin D1 (7.5 and 11 nM, respectively), while both p21-KID-LH+3 and p21-KID-LH−3 were significantly less potent toward these complexes (Fig. 4a and Supplementary Fig. 5). p21-KID-LH+3 was the more potent Cdk4 and Cdk6 inhibitor between the two variants. These results indicate that shortening sub-domain LH by approximately one helical turn is generally detrimental to p21-dependent Cdk inhibitory activity. In contrast, lengthening this sub-domain by a similar amount either had no effect on Cdk1 or slightly enhanced Cdk2 inhibitory activity, respectively, but diminished inhibitory activity toward cyclin D1 complexes with Cdk4 and Cdk6. It must be emphasized that, while the D1 and D2 sub-domains of p21-KID-LH+3 and p21-KID-LH−3 were shown to bind in a structurally similar manner to Cdk2/cyclin A (Fig. 3), the alterations made within the LH subdomains (which directly influence the ability of this sub-domain to stretch) indirectly influence the thermodynamics of their interactions with different Cdk/cyclin complexes.
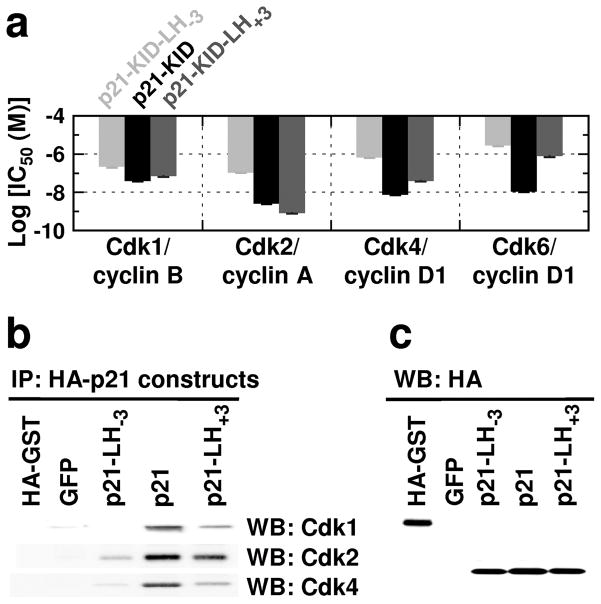
Figure 4.
Elongation or truncation of sub-domain LH within p21 significantly influences cell cycle regulation and interactions with Cdk/cyclin complexes. (a) Summary of log(IC50) values for inhibition of a panel of Cdk/cyclin complexes in vitro by p21-KID (black bars) or the sub-domain LH variants, p21-KID-LH+3 (dark grey bars) and p21-KID-LH−3 (light grey bars). (b) Results of immunoprecipitation of HA-tagged proteins from NIH 3T3 cell lysates followed by SDS-PAGE and immunoblotting using antibodies for Cdk1, Cdk2 and Cdk4. The full-size images of these data are illustrated in Supplementary Fig. 10. (c) Immunoblotting results using an antibody for the HA tag for the NIH 3T3 cell lysates discussed in panel b; from left to right, isolated HA-tagged GFP protein (positive control), and lysates from cells expressing GFP only (GFP, negative control) or GFP and HA-tagged p21-LH−3 (p21-LH−3), HA-tagged wild-type p21 (p21) or HA-tagged p21-LH+3 (p21-LH+3). The full-size image of these data is illustrated in Supplementary Fig. 11.
Altering sub-domain LH alters cell cycle regulation
To further characterize the functional effects of altering the LH sub-domain of p21, we monitored the influence of a series of HA-tagged full-length p21 constructs containing either the wild-type or variant LH sub-domains (p21-LH+3 and p21-LH−3) on the cell division cycle of mouse NIH 3T3 fibroblasts. The p21 constructs were introduced into MSCV-based retroviral vectors co-expressing green fluorescent protein (GFP)29. We used an additional vector that co-expressed GFP and p19Ink4d, a selective inhibitor of Cdk4 and Cdk630, as a positive control for monitoring G1 arrest31. Ectopic over-expression of p21 in human fibroblasts was previously shown to cause G1 arrest32. As expected, mouse fibroblasts expressing wild-type p21 exhibited G1 and G2 arrest (Table 2, Supplementary Fig. 6). The G1/G0 population increased from 25 ± 10 % for cells that expressed GFP only to 50 ± 25 % for cells that expressed both wild-type p21 and GFP, respectively (Table 2, Supplementary Fig. 6). Correspondingly, the S phase population declined from 41 ± 1 % (GFP only) to 5 ± 2 % (p21 and GFP). Furthermore, the G2/M population increased from 34 ± 11 % (GFP only) to 44 ± 27 % (p21 and GFP), indicating modest G2/M arrest. Immunoblotting analysis showed that similar levels of the three HA-tagged p21 constructs were expressed in NIH 3T3 cells (Supplementary Fig. 7). Expression of p19Ink4d caused the expected G1 arrest (Table 2, Supplementary Fig. 6). Expression of either p21-LH−3 or p21-LH+3 (and GFP) arrested cells in G1 phase to significantly smaller extents than wild-type p21 (Table 2, Supplementary Fig. 6). Expression of p21-LH+3 caused modest G1 arrest, with the G1/G0 population increased to 41 ± 9 % (from 25 ± 10 % for GFP only) and the S phase population decreased to 30 ± 5 % (from 41 ± 1 % for GFP only) (Table 2, Supplementary Fig. 6). Expression of p21-LH−3 yielded a cell cycle profile that was most similar to that obtained in cells that expressed GFP only (Table 2, Supplementary Fig. 6). However, p21-LH−3 did appear to slightly accelerate entry into S phase (S phase content of 41 ± 1 % for cells expressing GFP only versus 44 ± 2 % for cells expressing GFP and p21-LH−3; Table 2, Supplementary Fig. 6). These results indicated that wild-type p21 was the most effective cell cycle inhibitor in mouse fibroblast cells at both the G1/S and G2/M transitions and that, despite the ability of p21-KID-LH−3 and p21-KID-LH+3 to bind Cdk2/cyclin A in vitro at high concentrations (Fig. 3), the full-length forms of these LH sub-domain variants were poor inhibitors of cell division in mouse fibroblasts.
Table 2.
Cell cycle analysis of NIH 3T3 cells expressing GFP only or GFP and p19Ink4d, p21-LH−3, p21, or p21-LH+3, as indicated. Representative raw data from a single experiment are illustrated in Supplementary Figure 6.
| Cell cycle inhibitor | Percentage of cells in the different phases of division (%) | ||
|---|---|---|---|
| G1/G0 | S | G2/M | |
| GFP only 1 | 25 ± 10 | 41 ± 1 | 34 ± 11 |
| p19Ink4d 1 | 59 ± 12 | 18 ± 4 | 23 ± 8 |
| p21-LH−32 | 21 ± 8 | 44 ± 2 | 34 ± 10 |
| p21 2 | 50 ± 25 | 5 ± 2 | 44 ± 27 |
| p21-LH+32 | 41 ± 9 | 30 ± 5 | 28 ± 11 |
p21-dependent inhibition of cell cycle progression from G1 to S phase is mediated by inhibition of Cdk4/(and Cdk6)/D-type cyclin complexes, and Cdk2/cyclin E (and cyclin A) complexes12,22. Additionally, p21-dependent arrest in G2 phase is mediated by inhibition of Cdk1/cyclin B112,22,33. The p21 LH sub-domain variants were variably deficient in G1/S and G2 arrest (Table 2, Supplementary Fig. 6). To investigate the biochemical origins of these deficiencies, we determined the extent to which wild-type p21 and the LH sub-domain variants were associated with Cdk/cyclin complexes containing Cdk1, Cdk2 and Cdk4 in lysates from the variously infected NIH 3T3 cells. Immunoblotting analysis of the different forms of HA-tagged p21 after immunoprecipitation with an antibody against the HA showed that wild-type p21 was associated with complexes containing Cdk1, Cdk2 and Cdk4 (Fig. 4b), consistent with binding to and inhibition of the Cdk/cyclin complexes noted above and the G1/S and G2 arrest observed in NIH 3T3 cells. In contrast, compared with wild-type p21, the p21-LH+3 variant exhibited significantly reduced binding to Cdk1 and Cdk4 and slightly reduced binding to Cdk2. These variations reflected decreased affinity of p21-LH+3 for some Cdk/cyclin complexes because the levels of the HA-tagged p21 constructs expressed in NIH 3T3 cells and pulled-down from lysates were virtually the same (Supplementary Fig. 8 and Fig. 4c). Similarly, the levels of Cdk1, Cdk2 and Cdk4 in the variously infected NIH 3T3 cells were comparable (Supplementary Fig. 9). The binding of p21-LH+3 to Cdk2 to almost the same extent as wild-type p21 accounted for the accumulation of cells in G1 phase through inhibition of Cdk2/cyclin E and Cdk2/cyclin A complexes. The larger population of cells in S phase that expressed this variant in comparison with those that expressed wild-type p21, however, may have been due to significantly reduced binding to and inhibition of Cdk4 (and Cdk6)/D-type cyclin complexes. Furthermore, the absence of G2 arrest with p21-LH+3 was likely due to reduced binding to and inhibition of Cdk1/cyclin B activity that mediates entry into mitosis. Cell cycle and Cdk binding results with p21-LH+3 were consistent with the in vitro Cdk inhibition data presented in Fig. 4a. The IC50 value for p21-KID-LH−3 toward Cdk1/cyclin B1 was 5-fold higher than that observed for p21-KID (Fig. 4a). In addition, IC50 values for this LH sub-domain variant toward Cdk2/cyclin A and Cdk4 (and Cdk6)/cyclin D1 complexes were 40-fold, or more, higher than those observed for wild-type p21-KID, reflecting significantly reduced affinity of p21-KID-LH−3 for these Cdk/cyclin complexes. Correspondingly, little Cdk1, Cdk2, or Cdk4 was pulled down by this sub-domain LH variant in NIH 3T3 cells (Fig. 4b). These findings explain why over-expression of p21-LH−3 in cells was not associated with cell cycle arrest (Table 2, Supplementary Fig. 6). Taken together, the results of the HA immunoprecipitation and immunoblotting analyses provide biochemical explanations for the influence of the various p21 constructs on the distribution of NIH 3T3 cells amongst the different phases of the cell division cycle.
Discussion
Many intrinsically disordered proteins have evolved to interact with multiple binding partners and thus can perform diverse biological functions; this phenomenon has been referred to as binding diversity or binding promiscuity7,34. The paralogous Cip/Kip proteins, p21, p27 and p57, exhibit binding diversity through the interactions of their KIDs with the full Cdk/cyclin repertoire. Conserved residues within sub-domains D1 and D2 of p27 contact residues on the surfaces of cyclin A and Cdk2, respectively17, that are conserved in other Cdk/cyclin complexes that regulate cell division6,26. This pattern of sequence conservation suggested that the Cip/Kip proteins adopt similar structures when bound to the complexes which comprise the cell cycle regulatory Cdk/cyclin repertoire, as demonstrated by solution NMR data for p21-KID and p27-KID bound to Cdk2/cyclin A (Fig. 1c, d). However, beyond the surface regions of the cyclins and Cdks that contact conserved regions of p27, sequence conservation declines6. This structural variability accounts for the functional diversity of Cdk/cyclin complexes that phosphorylate different sites on the same or different substrates at different times during cell division35. Considering this coordinate structural and functional diversity, it is remarkable that the Cip/Kip proteins regulate the full Cdk/cyclin repertoire. While the sequences of sub-domains D1 and D2 are conserved, that of sub-domain LH is poorly conserved between the three human paralogs6. This suggests that residues within this sub-domain do not directly contact conserved features of Cdk/cyclin complexes, but rather that sub-domain LH modulates the functions of the other two sub-domains which are the primary mediators Cdk/cyclin inhibition6. Our data strongly suggest that the helical sub-domain LH structurally adapts, by stretching and pivoting, to enable subdomains D1 and D2 to bind conserved features of Cdk/cyclin complexes, allowing the full repertoire to be inhibited. Structural adaptation can readily be accommodated as the Cip/Kip proteins sequentially fold upon binding their Cdk/cyclin targets.
Support for this mechanistic model was developed by studying the effects of lengthening or shortening sub-domain LH on the structural, dynamic and functional properties of p21. First, while the altered LH sub-domains were sufficiently adaptable to allow sub-domains D1 and D2 of p21 to adopt similar structures when bound to the cyclin and Cdk subunits of the Cdk2/cyclin A complex (Fig. 3), lengthening or shortening sub-domain LH by approximately one turn of α-helix significantly altered in vitro Cdk inhibitory function (Fig. 4a). Thus, alteration of the structural adaptability of the linker between sub-domains D1 and D2 significantly altered promiscuous binding of p21 to a variety of Cdk/cyclin complexes. Alteration of the LH subdomain also modulated binding promiscuity in cells, with the effects on cell division essentially exactly as predicted by our biochemical findings (Table 2, Supplementary Fig. 6, Fig. 4b, c). The strong correlation between results from the in vitro Cdk/cyclin inhibition assays, cell cycle analyses and cellular Cdk co-IP assays supports our hypothesis that structural adaptability of sub-domain LH is requisite for promiscuous binding to and inhibition of multiple Cdk/cyclin complexes.
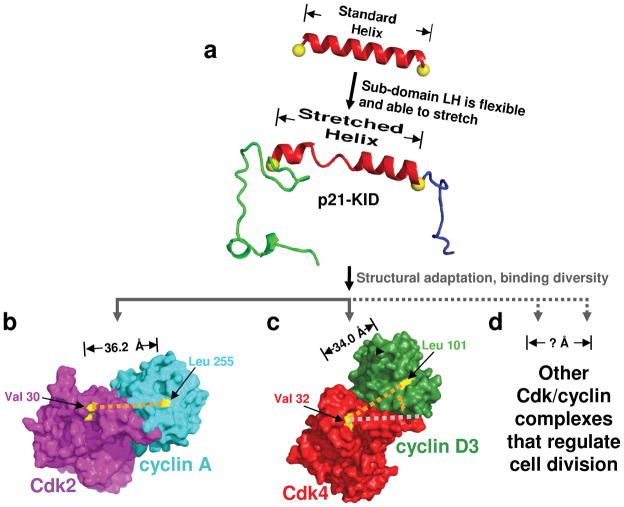
The structural adaptation model for binding promiscuity (Fig. 5a) is further supported by structural data for a diverse panel of Cdk/cyclin complexes. In the crystal structure of p27-KID bound to Cdk2/cyclin A, His 38 and Trp 60, at opposite ends of sub-domain LH, are in close proximity to Val 30 and Leu 255 of Cdk2 and cyclin A, respectively. The latter two residues are conserved as identities in the Cdk/cyclin complexes that regulate cell division6 and thus constitute conserved features of the p27 binding surface of these complexes. In the Cdk2/cyclin A complex, the distance between the Cα atoms of these two conserved residues is 36.2 Å17 (Fig. 5b), and this distance is 36.5 Å in the structure of Cdk2/cyclin B136 and 35.8 Å in that for Cdk2/cyclin E137 (Table 2). Thus, p27 (and the paralogs, p21 and p57) can bind to these three Cdk2/cyclin complexes such that the distances between sub-domains D1 and D2, as defined by the end-to-end length of sub-domain LH, are very similar. In the p21-KID/Cdk2/cyclin A (studied here) and p27-KID/Cdk2/cyclin A (studied by Russo, et al.17) complexes, this requires that sub-domain LH stretch beyond the length of a standard α-helix (pictured for p21-KID in Fig. 5a). In contrast, the distance between the same two conserved residues in the two available structures of Cdk4, Cdk4/cyclin D138 and Cdk4/cyclin D339, is 34.0 Å (Fig. 5c). Thus, contraction of the sub-domain LH helix to near-standard α-helical dimensions (33 Å for an α-helix comprised of 23 residues) would position sub-domains D1 and D2 of p21 and p27 to fold onto the surfaces of cyclin D1 and Cdk4, respectively, in a manner comparable to p27 binding the surface of Cdk2/cyclin A. However, sub-domain LH would be forced to pivot to accommodate the different orientation of cyclin D1 relative to Cdk4 in comparison with the relative orientation of these two subunits in the Cdk2/cyclin A complex (Fig. 5b and c). Due to their intrinsic flexibility and disordered nature in isolation, the different sub-domains of Cip/Kip proteins are structurally independent34; therefore, the subtly different topology of the Cdk4/cyclin D1 surface, relative to that of Cdk2/cyclin A, can readily be accommodated through sequential folding upon binding6. We note, however, that in the crystal structures of Cdk4/cyclin D138 and Cdk4/cyclin D339, Cdk4 appears to adopt an inactive conformation despite phosphorylation on Thr 172 (equivalent to Thr 160 in Cdk2). Importantly, however, the Cdk4/cyclin D complexes used for crystallization were shown to be biochemically active38,39. Therefore, crystallization may have trapped an inactive conformer and adaptive folding upon binding of p21 to Cdk4/cyclin D may occur in the context of as yet uncharacterized, active conformers. The ability of the LH sub-domain to structurally adapt upon binding may mediate the assembly function of p21 and p27 toward Cdk4/D-type cyclin complexes15, as suggested by thermal denaturation data for complexes containing p21-KID, or the LH+3 variant, and Cdk2/cyclin A (Supplementary Fig. 2). However, similar data are not available for the related Cdk4/D-type cyclin complexes; therefore, we are unable to confirm this assembly model. Furthermore, structural data are not available for other Cdk/cyclin complexes that are regulated by the Cip/Kip proteins (e.g., Cdk1 paired with either cyclin E or cyclin A that, together with the respective Cdk2 complexes, regulate progression from G1 to S phase, and Cdk1/cyclin B, which regulates entry into and passage through mitosis). However, we anticipate that the distances between and relative orientation of the conserved binding surfaces for sub-domains D1 and D2 within the cyclin and Cdk subunits of these complexes will vary, requiring sub-domain LH to adapt and pivot during the sequential binding and folding process (Fig. 5d). Thus, this analysis of high-resolution structural data for a subset of the Cdk/cyclin complexes targeted by the Cip/Kip proteins in cells supports our hypothesis that the process of adaptive folding upon binding enables this protein family to inhibit the diverse Cdk/cyclin repertoire that regulates mammalian cell division.
Figure 5.
Adaptive folding mediates promiscuous binding of p21 to the diverse Cdk/cyclin complexes that regulate cell division. Sub-domain LH of p21 (a) and the other Cip/Kip proteins can stretch to accommodate the varied orientations of the Cdk and cyclin subunits of Cdk/cyclin complexes that regulate cell division, including (b) Cdk2/cyclin A and (c) Cdk4/cyclin D3, for which high-resolution structural data is available (PDB files 1JST17 and 3G3339, respectively), as well as (d) other, uncharacterized Cdk/cyclin complexes that regulate cell division. In (a), the structure of p21-KID bound to Cdk2/cyclin A was modeled using that of p27-KID bound to Cdk2/cyclin A (1JSU17). In (b) the surface of Cdk2 is illustrated in magenta and that of cyclin A in cyan. The yellow patches and orange dotted line illustrate the locations of Val 30 in Cdk2 and Leu 255 in cyclin A, which are located at the interfaces, respectively, with Trp 60 and His 38 at the C- and N-termini of sub-domain LH of p27. In (c) the surface of Cdk4 is illustrated in red and that of cyclin D3 in green. The yellow patches and the orange dotted line illustrate the locations of Val 32 in Cdk4 and Leu 101 in cyclin D3, which are homologous to, respectively, the two illustrated in (b). The gray dotted line in (c) corresponds to that drawn in orange in (b).
Intrinsically disordered proteins generally lack secondary and tertiary structure and exist in isolation as dynamic conformational ensembles40. The association of these properties with diverse functions has been discussed since 19967; however, how IDPs perform their diverse functions is not understood in mechanistic terms. Our studies show how conserved features of the Cip/Kip proteins (sub-domains D1 and D2) mediate specific folding upon binding to conserved molecular features of the Cdk/cyclin repertoire. However, the structures of these complexes have diverged so as to phosphorylate different, specific substrates at different times during the division cycle and thus represent a diverse set of molecular targets for the Cip/Kip proteins. Through the lack of pre-existing tertiary structure, these IDPs can adaptively fold into relatively similar inhibitory conformations through the ability of sub-domain LH to stretch and pivot, as needed, to adapt to the unique molecular surfaces presented by the Cdk/cyclin repertoire. Interestingly, prior to binding Cdk/cyclin complexes, the regions of p27-KID which are most highly conserved within the Cip/Kip family (sub-domains D1 and D2) are highly dynamic while the poorly conserved LH sub-domain exhibits nascent helicity and partially restricted dynamics6,41. The partial helicity of sub-domain LH may position sub-domain D2 near the Cdk subunit of Cdk/cyclin complexes after sub-domain D1 initiates binding through interactions with the surface of the cyclin subunit. Importantly, we integrated results from several disciplines, including structural biophysics, biochemistry and cell biology to reveal the functional mechanism through which the Cip/Kip proteins regulate cell division. Further, we emphasize that knowledge of the structural and dynamic features of IDPs, both in their free and bound forms, is required to understand how these prevalent proteins perform their diverse biological functions. Insights into the location of functionally important regions of the thousands of IDPs that are currently poorly characterized may be gained through sequence analysis to identify conserved regions as well as disordered and partially structured regions. These bioinformatics studies must be coupled with investigations into the structural features of IDPs prior to binding their targets and others to identify their biological targets. Finally, high-resolution structural and dynamics data for IDPs bound to their biological targets is invaluable in deciphering functional mechanisms. Of course, models for the functional mechanisms of IDPs must ultimately be validated in biological assays. It is likely that the adaptive, folding upon binding mechanism that mediates the diverse functionality of the Cip/Kip protein family will be recapitulated by other IDPs. Wide application of the experimental strategy applied herein will provide broad insights into the relationships between the structural and dynamic properties of IDPs and their diverse biological functions.
Methods
Protein expression and purification
Full length human Cdk2, T160 phosphorylated Cdk2, and truncated human cyclin A (residues 173–432 of human cyclin A) were expressed in E. coli and purified using established procedures28. Cdk4 and cyclin D1 were cloned into a pBacPAK8 vector (Clonetech) as a single chain fusion protein with a PreScission protease (GE Healthcare) cleavage site between the two proteins and used to infect Sf9 cells for fusion protein expression. After purification by Ni-affinity chromatography, the fusion protein was cleaved by PreScission protease and the complex was further purified using size exclusion chromatography (Superdex 200, GE Healthcare). Baculoviruses viruses expressing GST-Cdk6 and 6xHis-cyclin D1 were graciously provided by Ludger Hengst (University of Innsbruck, Innsbruck, Austria). cDNA for Cdk1 and cyclin B1 were generated by PCR using synthetic oligonucleotides and sub-cloned into the pBacPAK8 vector fused to the C-terminus of GST. Both GST-Cdk6/6xHis-cyclin D1 and GST-Cdk1/GST-cyclin B1 complexes were expressed by co-infection of Sf9 cells and purified by GST-affinity chromatography.
p21-KID (residues 9–84 of human p21 with a C-terminal 6xHis tag)7,18 and p27-KID (residues 22–104 of human p27 with a thrombin-cleavable N-terminal 6xHis tag)6 were expressed in E. coli BL21(DE3) and purified using established procedures6,7. Isotope labeled proteins were expressed in E. coli BL21(DE3) grown in 3-(N-morpholino) propane sulfonic acid (MOPS)-based minimal medium42 enriched with isotope-labeled compounds. 15NH4Cl, glycerol, and 2H2O were used to express 2H/15N-labeled p21-KID; 15NH4Cl, 13C-acetate, and 2H2O were used to express 2H/13C/15N-labeled p21-KID and 15NH4Cl, 13C-glucose, and 2H2O were used to express 2H/13C/15N-labeled p27-KID. Expression plasmids for p21-KID-LH−3 and p21-KID-LH+3 were prepared by PCR using synthetic oligonucleotides. Three residues (ALR) were inserted between the end of sub-domain LH and the beginning of sub-domain D2 (between residues 48 and 49 of p21) to create p21-KID-LH+3. These residues were selected due to their compatibility with α-helical secondary structure43 and their structural features (e.g., Ala is compatible with its predicted location at the interface with Cdk2 and Leu and Arg are compatible with their predicted solvent exposure). The last three residues in sub-domain LH (residues 46 to 48 of p21) were deleted to create p21-KID-LH−3. Expression, 2H/15N-labeling and purification of p21-KID-LH+3 and p21-KID-LH−3 were performed as described above for wild-type p21-KID7,18 using 15 NH4Cl, glycerol, and 2H2O.
Samples of ternary complexes containing p21-KID, p21-KID -LH+3 or p21-KID -LH−3 and Cdk2/cyclin A for CD or NMR studies were prepared by mixing a 1.2-fold molar excess of each p21-KID species with Cdk2/cyclin A followed by purification using size exclusion chromatography (Superdex 200, GE Healthcare) in HEPES buffer (20 mM HEPES, pH 7.5, 300 mM NaCl, 5 mM DTT) and exchanged into CD or NMR buffers using ultrafiltration (Centricon units, Amicon).
Plasmids containing the C-terminal domain of Rb (RbC) were graciously provided by Brenda Schulman (St. Jude Children’s Research Hospital, Memphis, Tennessee, USA) and Peter Adams (Fox Chase Cancer Center, Philadelphia, Pennsylvania, USA). The coding region corresponding to Rb residues 773–928 was sub-cloned into a vector expressing N-terminal 6xHis and solubility enhancement tags (His-SET)44. The His-SET- RbC fusion protein was expressed in E. coli BL21(DE3) using standard procedures and purified using Ni-affinity (Chelating-Sepharose, GE Healthcare) and size exclusion chromatography (Superdex 75, GE Healthcare).
NMR spectroscopy
The NMR buffer for all studies was 20 mM potassium phosphate, pH 6.5, 50 mM arginine, 8% v/v 2H2O, 5 mM DTT and 0.02% w/v sodium azide. All NMR experiments were performed at 35 °C using a Bruker Avance 800 MHz spectrometer equipped with cryogenically-cooled triple resonance z-gradient probe. Backbone and 13Cβ resonance assignments for p21-KID bound to Cdk2/cyclin A were previously reported18. Secondary 13Cα chemical shift values (Δδ 13Cα) and composite 1H/15N chemical shift values (Δδ 1H/15N) for p21-KID constructs were calculated by subtracting sequence-dependent random coil values compiled by Schwarzinger, et al.45, from the experimental values. 2D 1H-15N TROSY and TROSY-based {1H}-15N heteronuclear nuclear Overhauser effect (hetNOE)46 for complexes of the p21-KID constructs with Cdk2/cyclin A were recorded using pulse sequences provided by Bruker Biospin. Spectra were processed using NMRPipe software47 and analyzed using Felix software (Accelerys). For all spectra, the 1H dimension was referenced to external TSP and the 13C and 15N dimensions were referenced indirectly using the appropriate ratios of gyromagnetic ratios48.
Thermal denaturation monitored by CD spectropolarimetry
CD measurements were performed using an AVIV model 62A DS circular dichroism spectropolarimeter using a 1 cm quartz cell. For thermal denaturation experiments, ellipticity at 222 nm was measured at 1° C intervals in the temperature range from 25 to 93 °C at a heating rate of 1 °C min−1. Samples containing 1.5 μM protein in 5 mM sodium phosphate buffer, pH 7.5, and 100 mM NaCl were incubated for 1 min. at each temperature prior to measurement. Thermal denaturation curves were analyzed as previously described49. Thermal denaturation of p21-KID/Cdk2/cyclin A complexes is irreversible due to precipitation of Cdk2 and cyclin A, therefore apparent thermal denaturation temperatures (Tmapp) are reported.
In vitro Cdk kinase activity assays
Kinase assay buffer contained 20 mM HEPES, pH 7.3, 25 mM sodium glycerolphosphate, 15 mM MgCl2, 16 mM EGTA, 0.5 mM Na3VO4, and 10 mM DTT. The in vitro Cdk1, Cdk2, Cdk4 and Cdk6 kinase activity assays were performed using established procedures28, as follows: a Cdk/cyclin complex (2 nM Cdk1/cyclin B1; 80 pM Cdk2/cyclin A; 3 nM Cdk4/cyclin D1; or 5 nM Cdk6/cyclin D1), substrate (1.0 μM RbC) and different concentrations of the p21-KID constructs (p21-KID, p21-KID-LH+3, or p21-KID-LH−3) were incubated at 4 °C for 2 hours. After equilibration, 6 μCi [γ-32P]-ATP and 40 μM non-radioactive ATP were added and the reactions were incubated for 35 minutes at 30 °C. Reactions were terminated by addition of SDS loading buffer and the [32P]-labeled products were resolved using 10% SDS-PAGE followed by analysis using phosphorimaging (Typhoon 9200; Molecular Dynamics, Inc.). IC50 values were determined after fitting normalized percentage of kinase activity versus log ([inhibitor]) employing the variable slope model using Prism software (GraphPad Software, Inc.).
Cell culture, virus infection, cell cycle analysis, immunoblotting and immunoprecipitation analyses
The methodological details of the cellular experiments are given under Supplementary Methods.
Supplementary Material
Table 3.
Distances between conserved residues in the Cdk and cyclin subunits of Cdk/cyclin complexes that, in the structure of p27-KID/Cdk2/cyclin A, interact with the C-terminus of subdomain D2 and the N-terminus of sub-domain D1, respectively, of p276,17.
| Cdk/cyclin complex | Distance (Å) 1 | PDB ID |
|---|---|---|
| Cdk2/cyclin A | 36.2 | 1JST17 |
| Cdk2/cyclin B1 | 36.5 | 2JGZ36 |
| Cdk2/cyclin E1 | 35.8 | 1W9837 |
| Cdk4/cyclin D1 | 34.0 | 2W9638 |
| Cdk4/cyclin D3 | 34.0 | 3G3339 |
Distances were measured between the Cα atoms of a conserved Val residue in the Cdks (Val 30 in Cdk2 and Val 32 in Cdk4) and a conserved Leu residue in the cyclins [Leu 255 in cyclin A, Leu 190 (isoform 1) in cyclin E1, Leu 246 in cyclin B, and Leu 101 in cyclins D1 and D3]. These residues are at the interface with p27 based on the crystal structure of p27-KID/Cdk2/cyclin A6,17.
Acknowledgments
The authors gratefully acknowledge Steven I. Reed (Scripps Research Institute) for providing a vector to express a cyclin D1-Cdk4 fusion protein, Ludger Hengst (Innsbruck Medical University) for providing baculoviruses that co-express Cdk6 and cyclin D1, Brenda Schulman (St. Jude Children’s Research Hospital) and Peter D. Adams (Fox Chase Cancer Center) for providing Rb constructs, Charles J. Sherr (St. Jude Children’s Research Hospital) for providing anti-sera for Cdk2 and Cdk4 and for stimulating discussions, Marie Assem (St. Jude Children’s Research Hospital) for technical assistance with cell cycle assays, Cheon-Gil Park (St. Jude Children’s Research Hospital) for preparation of Cdk/cyclin complexes for kinase assays, Nickolas Pytel (St. Jude Children’s Research Hospital) for preparation of p21-KID protein samples, Elaine Tuomanen (St. Jude Children’s Research Hospital) for assistance with kinase assays, and Richard Ashmun (St. Jude Children’s Research Hospital) for FACS analysis. The authors acknowledge support from NIH core grant P30CA21765 (St. Jude Children’s Research Hospital), 5R01CA082491 (to R.W.K.), R01CA71907 (to M. F. R.), the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital, and an NSF CAREER Award (NSF MCB 0952514 to J. C.).
Footnotes
Contributions
R.W.K. and M.R. designed the research; Y.W., J.F., L.O., S.O., R.M. and J.C. performed the research; Y.W., J.F., L.O., S.O., J.C., M.R. and R.W.K. analyzed data; L.X. and J.S. provided critical technical assistance; and Y.W., J.F., L.O., S.O., J.C., M.R. and R.W.K. wrote the manuscript.
Competing Financial Interests Statement
The authors have no competing financial interests related to the topic of this manuscript.
References
- 1.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–30. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Martin AC, et al. Protein folds and functions. Structure. 1998;6:875–84. doi: 10.1016/s0969-2126(98)00089-6. [DOI] [PubMed] [Google Scholar]
- 3.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 4.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–71. [PubMed] [Google Scholar]
- 5.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 6.Lacy ER, et al. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat Struct Mol Biol. 2004;11:358–364. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- 7.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21(waf1/cip1/sdi1) in the free and Cdk2-bound state: Conformational disorder mediates binding diversity. Proc Natl Acad Sci USA. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tompa P, Szasz C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem Sci. 2005;30:484–9. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;15:1149–63. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 10.Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 11.Weinbeg RA. The Biology of Cancer. Garland Science; London: 2006. [Google Scholar]
- 12.Xiong Y, et al. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 13.Harper JW, et al. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Xiong Y, Beach D. Proliferating cell nuclear antigen and p21 are components of multiple cell cycle kinase complexes. Molecular Biology of the Cell. 1993;4:897–906. doi: 10.1091/mbc.4.9.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng M, et al. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–83. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galea CA, Wang Y, Sivakolundu SG, Kriwacki RW. Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits. Biochemistry. 2008;47:7598–609. doi: 10.1021/bi8006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Filippov I, Richter C, Luo R, Kriwacki RW. Solution NMR studies of an intrinsically unstructured protein within a dilute, 75 kDa eukaryotic protein assembly; probing the practical limits for efficiently assigning polypeptide backbone resonances. Chembiochem. 2005;6:2242–6. doi: 10.1002/cbic.200500260. [DOI] [PubMed] [Google Scholar]
- 19.Schwarzinger S, et al. Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc. 2001;123:2970–8. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 20.Wishart DS, Sykes BD. Chemical shifts as a tool for structure determination. Meth Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 21.Kriwacki RW, Wu J, Siuzdak G, Wright PE. Probing Protein/Protein Interactions with Mass Spectrometry and Isotopic Labeling: Analysis of the p21/Cdk2 Complex. J Amer Chem Soc. 1996;118:5320–5321. [Google Scholar]
- 22.Harper JW, et al. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 24.Creighton TE, editor. Protein Structure, A Practical Approach. IRL Press; New York: 1989. [Google Scholar]
- 25.Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci U S A. 1998;95:10453–8. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lacy ER, et al. Molecular Basis for the Specificity of p27 Toward Cyclin-dependent Kinases that Regulate Cell Division. J Mol Biol. 2005;349:764–73. doi: 10.1016/j.jmb.2005.04.019. Epub 2005 Apr 26. [DOI] [PubMed] [Google Scholar]
- 27.Bowman P, Galea CA, Lacy E, Kriwacki RW. Thermodynamic characterization of interactions between p27(Kip1) and activated and non-activated Cdk2: intrinsically unstructured proteins as thermodynamic tethers. Biochim Biophys Acta. 2006;1764:182–9. doi: 10.1016/j.bbapap.2005.12.016. Epub 2006 Jan 11. [DOI] [PubMed] [Google Scholar]
- 28.Grimmler M, et al. Cdk-Inhibitory Activity and Stability of p27(Kip1) Are Directly Regulated by Oncogenic Tyrosine Kinases. Cell. 2007;128:269–80. doi: 10.1016/j.cell.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 29.Zindy F, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherr CJ, Roberts JM. Cdk inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 31.Hirai H, Roussel MF, Kato JY, Ashmun RA, Sherr CJ. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol Cell Biol. 1995;15:2672–81. doi: 10.1128/mcb.15.5.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 33.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 34.Galea CA, et al. Role of intrinsic flexibility in signal transduction mediated by the cell cycle regulator, p27 Kip1. J Mol Biol. 2008;376:827–38. doi: 10.1016/j.jmb.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 36.Brown NR, et al. Cyclin B and cyclin A confer different substrate recognition properties on CDK2. Cell Cycle. 2007;6:1350–9. doi: 10.4161/cc.6.11.4278. [DOI] [PubMed] [Google Scholar]
- 37.Honda R, et al. The structure of cyclin E1/CDK2: implications for CDK2 activation and CDK2-independent roles. EMBO J. 2005;24:452–63. doi: 10.1038/sj.emboj.7600554. Epub 2005 Jan 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Day PJ, et al. Crystal structure of human CDK4 in complex with a D-type cyclin. Proc Natl Acad Sci U S A. 2009;106:4166–70. doi: 10.1073/pnas.0809645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takaki T, et al. The structure of CDK4/cyclin D3 has implications for models of CDK activation. Proc Natl Acad Sci U S A. 2009;106:4171–6. doi: 10.1073/pnas.0809674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Sivakolundu SG, Bashford D, Kriwacki RW. Disordered p27 (Kip1) Exhibits Intrinsic Structure Resembling the Cdk2/Cyclin A-bound Conformation. J Mol Biol. 2005;353:1118–1128. doi: 10.1016/j.jmb.2005.08.074. Epub 2005 Sep 20. [DOI] [PubMed] [Google Scholar]
- 42.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bact. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Creighton TE. Proteins: Structures and Molecular Properties. W. H. Freeman & Co; New York, NY: 1993. [Google Scholar]
- 44.Zhou P, Lugovskoy AA, Wagner G. A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. J Biomol NMR. 2001;20:11–14. doi: 10.1023/a:1011258906244. [DOI] [PubMed] [Google Scholar]
- 45.Schwarzinger S, et al. Sequence-dependent correction of random coil NMR chemical shifts. J Amer Chem Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 46.Zhu G, Xia Y, Nicholson LK, Sze KH. Protein dynamics measurements by TROSY-based NMR experiments. J Magn Reson. 2000;143:423–6. doi: 10.1006/jmre.2000.2022. [DOI] [PubMed] [Google Scholar]
- 47.Delaglio F, et al. NMR Pipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 48.Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton NJ. Protein NMR spectroscopy: Principle and Practice. Academic Press; New York: 2007. [Google Scholar]
- 49.Bowman P, Galea CA, Lacy E, Kriwacki RW. Thermodynamic characterization of interactions between p27 (Kip1) and activated and non-activated Cdk2: intrinsically unstructured proteins as thermodynamic tethers. Biochim Biophys Acta. 2006;1764:182–9. doi: 10.1016/j.bbapap.2005.12.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.