Abstract
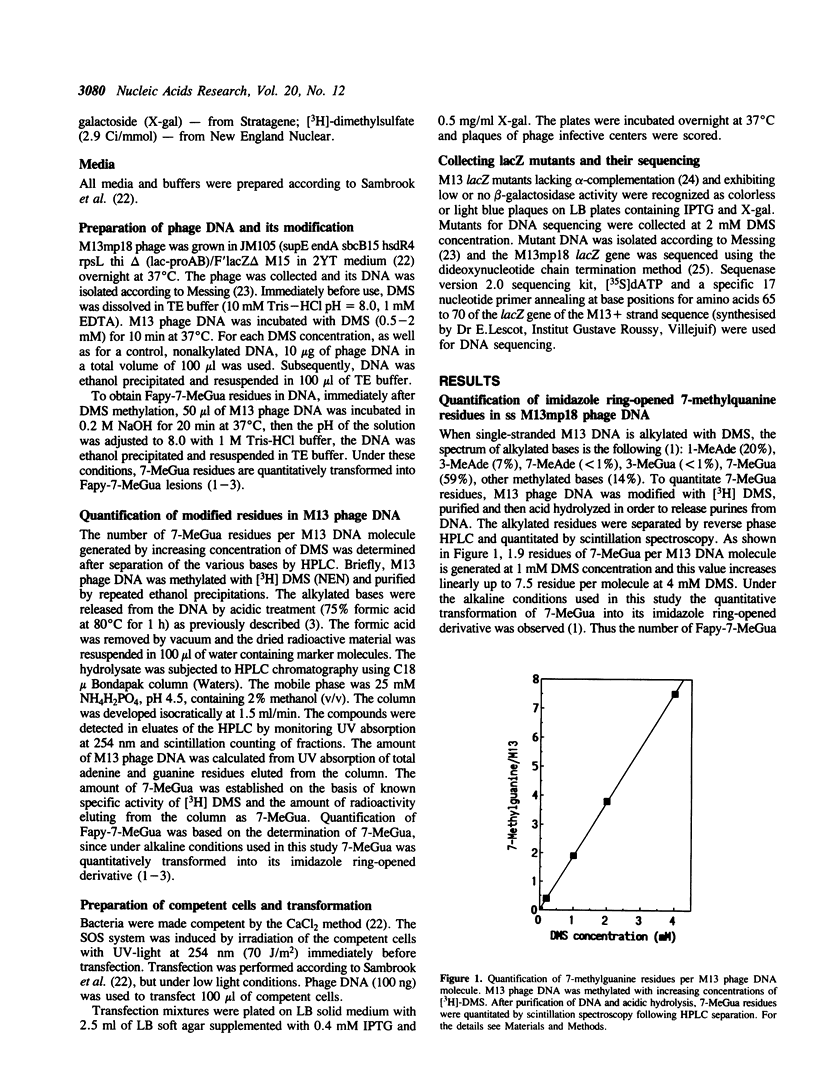
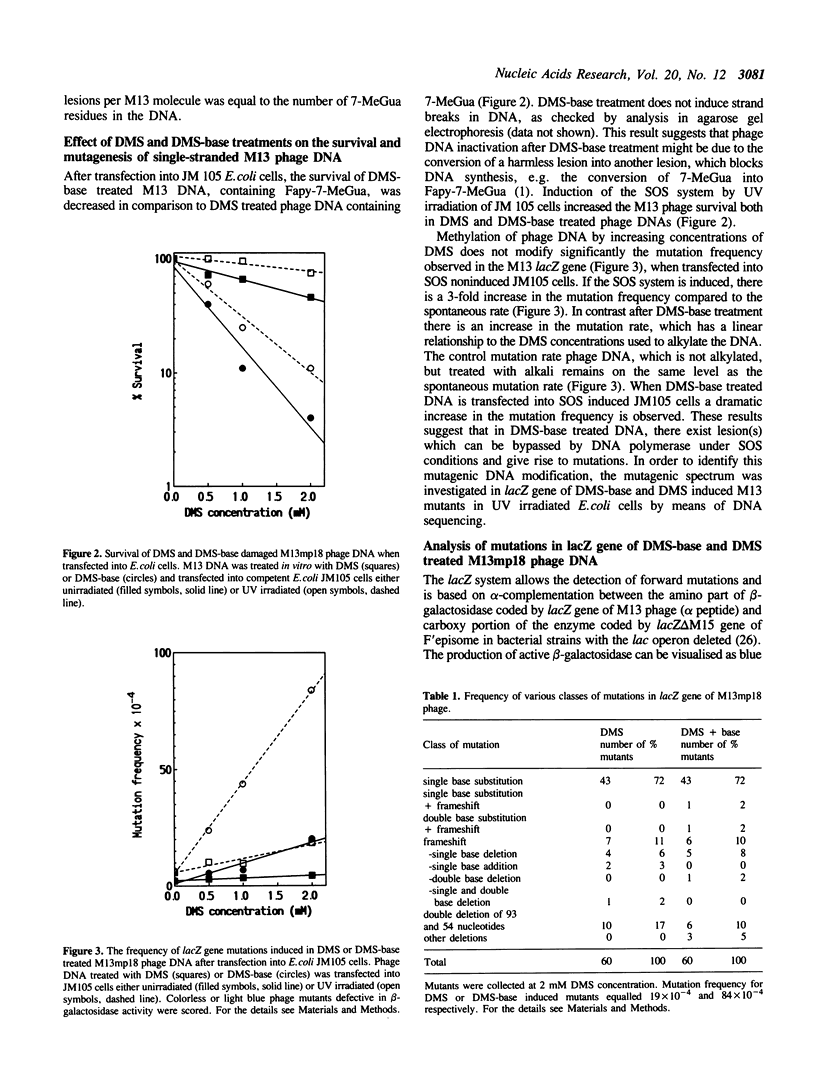
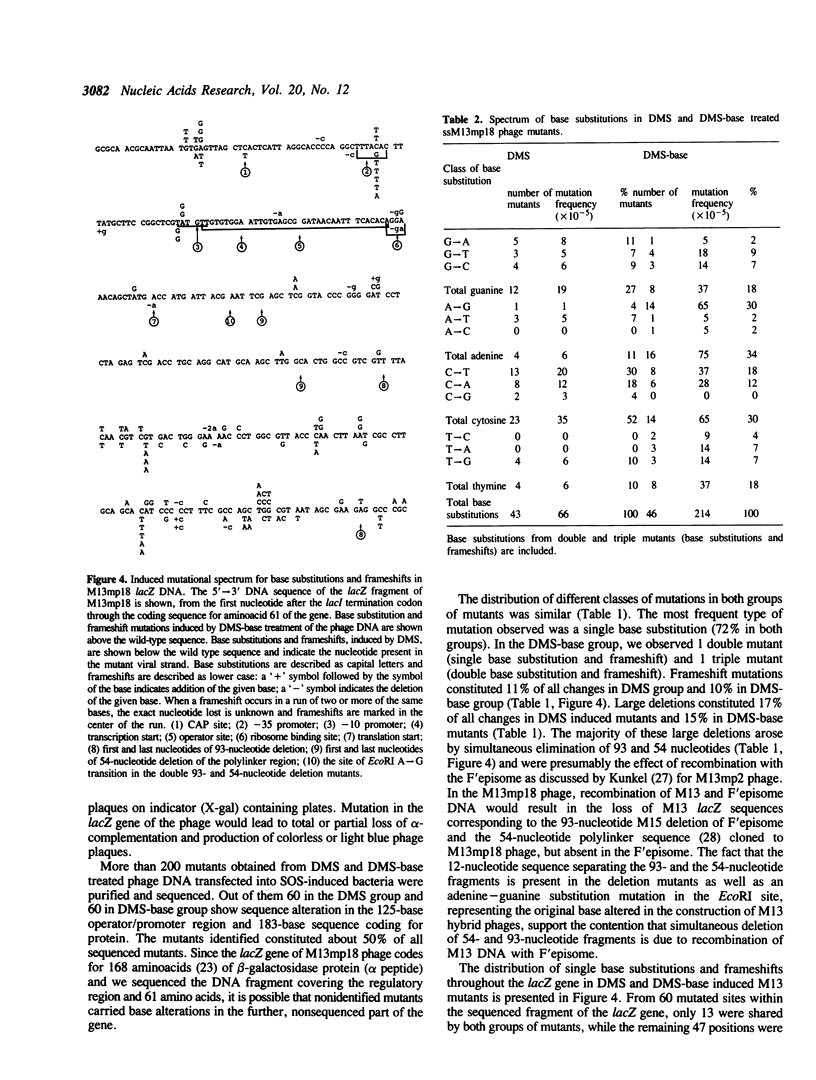
Guanine residues methylated at the N-7 position (7-MeGua) are susceptible to cleavage of the imidazole ring yielding 2,6-diamino-4-hydroxy-5N-methyl-formamidopyrimidine (Fapy-7-MeGua). The presence of Fapy-7-MeGua in DNA template causes stops in DNA synthesis in vitro by E. coli DNA polymerase I. The biological consequences of Fapy-7-MeGua lesions for survival and mutagenesis were investigated using single-stranded M13mp18 phage DNA. Fapy-7-MeGua lesions were generated in vitro in phage DNA by dimethylsulfate (DMS) methylation and subsequent ring opening of 7-MeGua by treatment with NaOH (DMS-base). The presence of Fapy-7-MeGua residues in M13 phage DNA correlated with a significant decrease in transfection efficiency and an increase in mutation frequency in the lacZ gene, when transfected into SOS-induced JM105 E.coli cells. Sequencing analysis revealed unexpectedly, that mutation rate at guanine sites was only slightly increased, suggesting that Fapy-7-MeGua was not responsible for the overall increase in the mutagenic frequency of DMS-base treated DNA. In contrast, mutation frequency at adenine sites yielding A----G transitions was the most frequent event, 60-fold increased over DMS induced mutations. These results show that treatment with alkali of methylated single-stranded DNA generates a mutagenic adenine derivative, which mispairs with cytosine in SOS induced bacteria. The results also imply that the Fapy-7-MeGua in E. coli cells is primarily a lethal lesion.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aruoma O. I., Halliwell B., Dizdaroglu M. Iron ion-dependent modification of bases in DNA by the superoxide radical-generating system hypoxanthine/xanthine oxidase. J Biol Chem. 1989 Aug 5;264(22):13024–13028. [PubMed] [Google Scholar]
- Beranek D. T., Weis C. C., Evans F. E., Chetsanga C. J., Kadlubar F. F. Identification of N5-methyl-N5-formyl-2,5,6-triamino-4-hydroxypyrimidine as a major adduct in rat liver DNA after treatment with the carcinogens, N,N-dimethylnitrosamine or 1,2-dimethylhydrazine. Biochem Biophys Res Commun. 1983 Jan 27;110(2):625–631. doi: 10.1016/0006-291x(83)91195-6. [DOI] [PubMed] [Google Scholar]
- Beranek D. T., Weis C. C., Evans F. E., Chetsanga C. J., Kadlubar F. F. Identification of N5-methyl-N5-formyl-2,5,6-triamino-4-hydroxypyrimidine as a major adduct in rat liver DNA after treatment with the carcinogens, N,N-dimethylnitrosamine or 1,2-dimethylhydrazine. Biochem Biophys Res Commun. 1983 Jan 27;110(2):625–631. doi: 10.1016/0006-291x(83)91195-6. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Belleney J., Roques B. P., Laval J. Two rotameric forms of open ring 7-methylguanine are present in alkylated polynucleotides. Nucleic Acids Res. 1984 Jul 11;12(13):5429–5439. doi: 10.1093/nar/12.13.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Bichara M., Fuchs R. P., Laval J. Excision of the imidazole ring-opened form of N-2-aminofluorene-C(8)-guanine adduct in poly(dG-dC) by Escherichia coli formamidopyrimidine-DNA glycosylase. Carcinogenesis. 1989 Oct;10(10):1905–1909. doi: 10.1093/carcin/10.10.1905. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Gajewski E., Laval J., Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992 Jan 14;31(1):106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Laval J. Coding properties of poly(deoxycytidylic acid) templates containing uracil or apyrimidinic sites: in vitro modulation of mutagenesis by deoxyribonucleic acid repair enzymes. Biochemistry. 1982 Dec 21;21(26):6746–6751. doi: 10.1021/bi00269a020. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Laval J. Mutagenesis by alkylating agents: coding properties for DNA polymerase of poly (dC) template containing 3-methylcytosine. Biochimie. 1982 Aug-Sep;64(8-9):637–641. doi: 10.1016/s0300-9084(82)80103-x. [DOI] [PubMed] [Google Scholar]
- Boiteux S., O'Connor T. R., Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987 Oct;6(10):3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., O'Connor T. R., Lederer F., Gouyette A., Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J Biol Chem. 1990 Mar 5;265(7):3916–3922. [PubMed] [Google Scholar]
- Breimer L. H. Enzymatic excision from gamma-irradiated polydeoxyribonucleotides of adenine residues whose imidazole rings have been ruptured. Nucleic Acids Res. 1984 Aug 24;12(16):6359–6367. doi: 10.1093/nar/12.16.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri I., Essigmann J. M. 3-Methyladenine mutagenesis under conditions of SOS induction in Escherichia coli. Carcinogenesis. 1991 Dec;12(12):2283–2289. doi: 10.1093/carcin/12.12.2283. [DOI] [PubMed] [Google Scholar]
- Chetsanga C. J., Frenette G. P. Excision of aflatoxin B1-imidazole ring opened guanine adducts from DNA by formamidopyrimidine-DNA glycosylase. Carcinogenesis. 1983 Aug;4(8):997–1000. doi: 10.1093/carcin/4.8.997. [DOI] [PubMed] [Google Scholar]
- Chetsanga C. J., Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979 Aug 10;6(11):3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan B. K., Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982 Aug;151(2):750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett E. R., Mehta P. J. Solvolysis of adenine nucleosides. II. Effects of sugars and adenine substituents on alkaline solvolyses. J Am Chem Soc. 1972 Nov 29;94(24):8542–8547. doi: 10.1021/ja00779a041. [DOI] [PubMed] [Google Scholar]
- Kadlubar F. F., Beranek D. T., Weis C. C., Evans F. E., Cox R., Irving C. C. Characterization of the purine ring-opened 7-methylguanine and its persistence in rat bladder epithelial DNA after treatment with the carcinogen N-methylnitrosourea. Carcinogenesis. 1984 May;5(5):587–592. doi: 10.1093/carcin/5.5.587. [DOI] [PubMed] [Google Scholar]
- Kallama S., Hemminki K. Stabilities of 7-alkylguanosines and 7-deoxyguanosines formed by phosphoramide mustard and nitrogen mustard. Chem Biol Interact. 1986 Jan;57(1):85–96. doi: 10.1016/0009-2797(86)90051-7. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Mutational specificity of depurination. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1494–1498. doi: 10.1073/pnas.81.5.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAWLEY P. D., BROOKES P. FURTHER STUDIES ON THE ALKYLATION OF NUCLEIC ACIDS AND THEIR CONSTITUENT NUCLEOTIDES. Biochem J. 1963 Oct;89:127–138. doi: 10.1042/bj0890127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley K. E., Villarejo M. R., Fowler A. V., Zamenhof P. J., Zabin I. Molecular basis of beta-galactosidase alpha-complementation. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1254–1257. doi: 10.1073/pnas.72.4.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval J., Boiteux S., O'Connor T. R. Physiological properties and repair of apurinic/apyrimidinic sites and imidazole ring-opened guanines in DNA. Mutat Res. 1990 Nov-Dec;233(1-2):73–79. doi: 10.1016/0027-5107(90)90152-t. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974 Jul 30;13(16):3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Malins D. C., Haimanot R. Major alterations in the nucleotide structure of DNA in cancer of the female breast. Cancer Res. 1991 Oct 1;51(19):5430–5432. [PubMed] [Google Scholar]
- Malins D. C., Ostrander G. K., Haimanot R., Williams P. A novel DNA lesion in neoplastic livers of feral fish: 2,6-diamino-4-hydroxy-5-formamidopyrimidine. Carcinogenesis. 1990 Jun;11(6):1045–1047. doi: 10.1093/carcin/11.6.1045. [DOI] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- O'Connor T. R., Boiteux S., Laval J. Ring-opened 7-methylguanine residues in DNA are a block to in vitro DNA synthesis. Nucleic Acids Res. 1988 Jul 11;16(13):5879–5894. doi: 10.1093/nar/16.13.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffhill R. Differences in the promutagenic nature of 3-methylcytosine as revealed by DNA and RNA polymerising enzymes. Carcinogenesis. 1984 May;5(5):691–693. doi: 10.1093/carcin/5.5.691. [DOI] [PubMed] [Google Scholar]
- Saffhill R., Margison G. P., O'Connor P. J. Mechanisms of carcinogenesis induced by alkylating agents. Biochim Biophys Acta. 1985 Dec 17;823(2):111–145. doi: 10.1016/0304-419x(85)90009-5. [DOI] [PubMed] [Google Scholar]
- Sambamurti K., Callahan J., Luo X., Perkins C. P., Jacobsen J. S., Humayun M. Z. Mechanisms of mutagenesis by a bulky DNA lesion at the guanine N7 position. Genetics. 1988 Dec;120(4):863–873. doi: 10.1093/genetics/120.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Kunkel T. A., Loeb L. A. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc Natl Acad Sci U S A. 1983 Jan;80(2):487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B., Chavez F., Goodman M. F., Essigmann J. M., Dosanjh M. K. Effect of 3' flanking neighbors on kinetics of pairing of dCTP or dTTP opposite O6-methylguanine in a defined primed oligonucleotide when Escherichia coli DNA polymerase I is used. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8271–8274. doi: 10.1073/pnas.86.21.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B., Kuśmierek J. T., Fraenkel-Conrat H. In vitro discrimination of replicases acting on carcinogen-modified polynucleotide templates. Proc Natl Acad Sci U S A. 1983 Feb;80(4):969–972. doi: 10.1073/pnas.80.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchou J., Kasai H., Shibutani S., Chung M. H., Laval J., Grollman A. P., Nishimura S. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman J. S., McCarthy B. J. Alkali deamination of cytosine residues in DNA. Biochim Biophys Acta. 1973 Feb 4;294(1):396–404. doi: 10.1016/0005-2787(73)90094-4. [DOI] [PubMed] [Google Scholar]
- Zielenska M., Horsfall M. J., Glickman B. W. The dissimilar mutational consequences of SN1 and SN2 DNA alkylation pathways: clues from the mutational specificity of dimethylsulphate in the lacI gene of Escherichia coli. Mutagenesis. 1989 May;4(3):230–234. doi: 10.1093/mutage/4.3.230. [DOI] [PubMed] [Google Scholar]



