Abstract
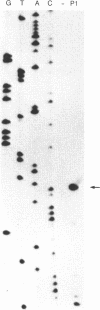
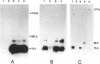
The c4 repressors of the temperate bacteriophages P1 and P7 inhibit antirepressor synthesis and are essential for establishment and maintenance of lysogeny. Using in vivo complementation tests we have previously shown that c4 is an antisense RNA acting on a target, ant mRNA, which is transcribed from the same promoter. Here we identify the c4 repressor molecule of P1 as a 77 +/- 1 base RNA by mapping its termini and show that the c4 RNA in P7 lysogens has the same or a similar size. P1 c4 RNA is encoded in a region shown to be sufficient for c4 complementation. It covers exactly the 74 bases previously suggested to fold into a stem-loop secondary structure essential for c4 function. Furthermore, we demonstrate that the 5' end of c4 RNA is generated by processing. Thus, c4 is the first example of an antisense RNA to be processed. A possible mechanism of processing is discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Altman S. Ribonuclease P: an enzyme with a catalytic RNA subunit. Adv Enzymol Relat Areas Mol Biol. 1989;62:1–36. doi: 10.1002/9780470123089.ch1. [DOI] [PubMed] [Google Scholar]
- Appleyard R K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954 Jul;39(4):440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumstark B. R., Scott J. R. The c4 gene of phage P1. Virology. 1987 Feb;156(2):197–203. doi: 10.1016/0042-6822(87)90398-9. [DOI] [PubMed] [Google Scholar]
- Chesney R. H., Scott J. R. Superinfection immunity and prophage repression in phage P1. II. Mapping of the immunity-difference and ampicillin-resistance loci of P1 and phi amp. Virology. 1975 Oct;67(2):375–384. doi: 10.1016/0042-6822(75)90439-0. [DOI] [PubMed] [Google Scholar]
- Citron M., Schuster H. The c4 repressors of bacteriophages P1 and P7 are antisense RNAs. Cell. 1990 Aug 10;62(3):591–598. doi: 10.1016/0092-8674(90)90023-8. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Hansen E. B. Structure and regulation of the lytic replicon of phage P1. J Mol Biol. 1989 May 5;207(1):135–149. doi: 10.1016/0022-2836(89)90445-2. [DOI] [PubMed] [Google Scholar]
- Heisig A., Riedel H. D., Dobrinski B., Lurz R., Schuster H. Organization of the immunity region immI of bacteriophage P1 and synthesis of the P1 antirepressor. J Mol Biol. 1989 Oct 20;209(4):525–538. doi: 10.1016/0022-2836(89)90591-3. [DOI] [PubMed] [Google Scholar]
- KONDO E., MITSUHASHI S. DRUG RESISTANCE OF ENTERIC BACTERIA. IV. ACTIVE TRANSDUCING BACTERIOPHAGE P1 CM PRODUCED BY THE COMBINATION OF R FACTOR WITH BACTERIOPHAGE P1. J Bacteriol. 1964 Nov;88:1266–1276. doi: 10.1128/jb.88.5.1266-1276.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck-Miller K. A., Altman S. Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J Mol Biol. 1991 Sep 5;221(1):1–5. doi: 10.1016/0022-2836(91)80194-y. [DOI] [PubMed] [Google Scholar]
- Scott J. R., Kropf M., Mendelson L. Clear plaque mutants of phage P7. Virology. 1977 Jan;76(1):39–46. doi: 10.1016/0042-6822(77)90279-3. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Biological regulation by antisense RNA in prokaryotes. Annu Rev Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- Smith H. W. Ampicillin resistance in Escherichia coli by phage infection. Nat New Biol. 1972 Aug 16;238(85):205–206. doi: 10.1038/newbio238205a0. [DOI] [PubMed] [Google Scholar]
- Sternberg N. A characterization of bacteriophage P1 DNA fragments cloned in a lambda vector. Virology. 1979 Jul 15;96(1):129–142. doi: 10.1016/0042-6822(79)90179-x. [DOI] [PubMed] [Google Scholar]
- Takayama K. M., Inouye M. Antisense RNA. Crit Rev Biochem Mol Biol. 1990;25(3):155–184. doi: 10.3109/10409239009090608. [DOI] [PubMed] [Google Scholar]
- Wandersman C., Yarmolinsky M. Bipartite control of immunity conferred by the related heteroimmune plasmid prophages, P1 and P7 (formerly phi Amp). Virology. 1977 Mar;77(1):386–400. doi: 10.1016/0042-6822(77)90435-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]