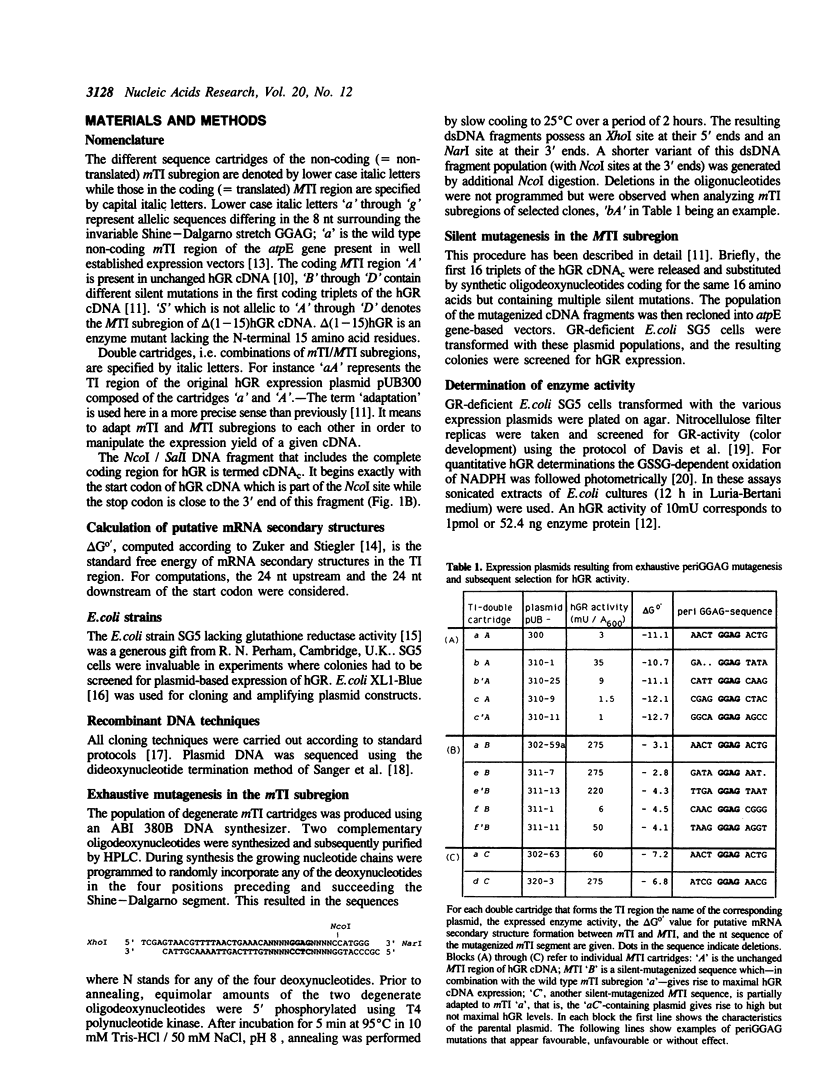
Abstract
Adaptation of eucaryotic cDNA to heterologous expression was studied by mutating the translation initiation (TI) region upstream (mTI) and downstream (MTI) of the start codon. In the mTI subregion the 8 bases flanking the invariant Shine-Dalgarno motif GG-AG were mutagenized exhaustively, while the MTI subregion was subjected to random silent mutations at the wobble positions. The quality of a given TI sequence was judged on the basis of expressed enzyme activity. Low-yield and high-yield mutants of both TI subregions were selected and recombined systematically. The analysis of these double cartridges gave the following results: 1. As a rule, an unfavourable MTI subregion can be compensated for by mutations in the mTI subregion and vice versa. 2. The compatibility between mTI and MTI subregion is explainable at least in part by a low interaction tendency; a delta G(o)'-value of -10.7 kcal/mol appears to be a physical threshold for heterologous cDNA expression. 3. On the basis of periShine-Dalgarno mutations, the expression yield for different cDNA sequences could be increased by 1 to 2 orders of magnitude. One of these sequences encoded delta(1-15)human glutathione reductase, a mutant lacking the flexible N-terminal extension of the protein. In conclusion, to study and overcome TI region-based expression problems it is worthwhile to start out with a versatile vector containing exhaustive mutations in the periShine-Dalgarno sequences; as a rule the coding MTI subregion can be kept unchanged.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boni I. V., Isaeva D. M., Musychenko M. L., Tzareva N. V. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991 Jan 11;19(1):155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley M., Bücheler U. S., Walsh C. T. Redox enzyme engineering: conversion of human glutathione reductase into a trypanothione reductase. Biochemistry. 1991 Jun 25;30(25):6124–6127. doi: 10.1021/bi00239a006. [DOI] [PubMed] [Google Scholar]
- Bücheler U. S., Werner D., Schirmer R. H. Random silent mutagenesis in the initial triplets of the coding region: a technique for adapting human glutathione reductase-encoding cDNA to expression in Escherichia coli. Gene. 1990 Dec 15;96(2):271–276. doi: 10.1016/0378-1119(90)90263-q. [DOI] [PubMed] [Google Scholar]
- Dalbøge H., Carlsen S., Jensen E. B., Christensen T., Dahl H. H. Expression of recombinant growth hormone in Escherichia coli: effect of the region between the Shine-Dalgarno sequence and the ATG initiation codon. DNA. 1988 Jul-Aug;7(6):399–405. doi: 10.1089/dna.1.1988.7.399. [DOI] [PubMed] [Google Scholar]
- Davis N. K., Greer S., Jones-Mortimer M. C., Perham R. N. Isolation and mapping of glutathione reductase-negative mutants of Escherichia coli K12. J Gen Microbiol. 1982 Jul;128(7):1631–1634. doi: 10.1099/00221287-128-7-1631. [DOI] [PubMed] [Google Scholar]
- Faxén M., Plumbridge J., Isaksson L. A. Codon choice and potential complementarity between mRNA downstream of the initiation codon and bases 1471-1480 in 16S ribosomal RNA affects expression of glnS. Nucleic Acids Res. 1991 Oct 11;19(19):5247–5251. doi: 10.1093/nar/19.19.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer S., Perham R. N. Glutathione reductase from Escherichia coli: cloning and sequence analysis of the gene and relationship to other flavoprotein disulfide oxidoreductases. Biochemistry. 1986 May 6;25(9):2736–2742. doi: 10.1021/bi00357a069. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Gold L. Influence of mRNA determinants on translation initiation in Escherichia coli. J Mol Biol. 1991 Mar 5;218(1):83–97. doi: 10.1016/0022-2836(91)90875-7. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Glutathione-dependent synthesis of deoxyribonucleotides. Characterization of the enzymatic mechanism of Escherichia coli glutaredoxin. J Biol Chem. 1979 May 10;254(9):3672–3678. [PubMed] [Google Scholar]
- Iserentant D., Fiers W. Secondary structure of mRNA and efficiency of translation initiation. Gene. 1980 Apr;9(1-2):1–12. doi: 10.1016/0378-1119(80)90163-8. [DOI] [PubMed] [Google Scholar]
- Karplus P. A., Schulz G. E. Refined structure of glutathione reductase at 1.54 A resolution. J Mol Biol. 1987 Jun 5;195(3):701–729. doi: 10.1016/0022-2836(87)90191-4. [DOI] [PubMed] [Google Scholar]
- Lohrer H., Krauth-Siegel R. L. Purification and characterization of lipoamide dehydrogenase from Trypanosoma cruzi. Eur J Biochem. 1990 Dec 27;194(3):863–869. doi: 10.1111/j.1432-1033.1990.tb19480.x. [DOI] [PubMed] [Google Scholar]
- Petersen G. B., Stockwell P. A., Hill D. F. Messenger RNA recognition in Escherichia coli: a possible second site of interaction with 16S ribosomal RNA. EMBO J. 1988 Dec 1;7(12):3957–3962. doi: 10.1002/j.1460-2075.1988.tb03282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke-Appel J., Jünke N., Stade K., Brimacombe R. The path of mRNA through the Escherichia coli ribosome; site-directed cross-linking of mRNA analogues carrying a photo-reactive label at various points 3' to the decoding site. EMBO J. 1991 Aug;10(8):2195–2202. doi: 10.1002/j.1460-2075.1991.tb07755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder B., Blöcker H., Frank R., McCarthy J. E. Inducible expression vectors incorporating the Escherichia coli atpE translational initiation region. Gene. 1987;52(2-3):279–283. doi: 10.1016/0378-1119(87)90054-0. [DOI] [PubMed] [Google Scholar]
- Schulz G. E., Schirmer R. H., Sachsenheimer W., Pai E. F. The structure of the flavoenzyme glutathione reductase. Nature. 1978 May 11;273(5658):120–124. doi: 10.1038/273120a0. [DOI] [PubMed] [Google Scholar]
- Sprengart M. L., Fatscher H. P., Fuchs E. The initiation of translation in E. coli: apparent base pairing between the 16srRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990 Apr 11;18(7):1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutic M., Lu X. A., Schirmer R. H., Werner D. Cloning and sequencing of mammalian glutathione reductase cDNA. Eur J Biochem. 1990 Mar 30;188(3):523–528. doi: 10.1111/j.1432-1033.1990.tb15431.x. [DOI] [PubMed] [Google Scholar]
- Untucht-Grau R., Schirmer R. H., Schirmer I., Krauth-Siegel R. L. Glutathione reductase from human erythrocytes: amino-acid sequence of the structurally known FAD-binding domain. Eur J Biochem. 1981 Nov;120(2):407–419. doi: 10.1111/j.1432-1033.1981.tb05718.x. [DOI] [PubMed] [Google Scholar]
- Worthington D. J., Rosemeyer M. A. Glutathione reductase from human erythrocytes. Catalytic properties and aggregation. Eur J Biochem. 1976 Aug 1;67(1):231–238. doi: 10.1111/j.1432-1033.1976.tb10654.x. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]



