Abstract
The precise mechanisms leading to the emergence of low-level glycopeptide resistance in Staphylococcus aureus are poorly understood. In this study, we used whole genome deep sequencing to detect differences between two isogenic strains: a parental strain and a stable derivative selected stepwise for survival on 4 µg/ml teicoplanin, but which grows at higher drug concentrations (MIC 8 µg/ml). We uncovered only three single nucleotide changes in the selected strain. Nonsense mutations occurred in stp1, encoding a serine/threonine phosphatase, and in yjbH, encoding a post-transcriptional negative regulator of the redox/thiol stress sensor and global transcriptional regulator, Spx. A missense mutation (G45R) occurred in the histidine kinase sensor of cell wall stress, VraS. Using genetic methods, all single, pairwise combinations, and a fully reconstructed triple mutant were evaluated for their contribution to low-level glycopeptide resistance. We found a synergistic cooperation between dual phospho-signalling systems and a subtle contribution from YjbH, suggesting the activation of oxidative stress defences via Spx. To our knowledge, this is the first genetic demonstration of multiple sensor and stress pathways contributing simultaneously to glycopeptide resistance development. The multifactorial nature of glycopeptide resistance in this strain suggests a complex reprogramming of cell physiology to survive in the face of drug challenge.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) infections is a major cause of morbidity and mortality worldwide, including in the health care setting. The mortality rate associated with invasive MRSA infections is significant and in the United States MRSA infections are now quoted as the leading cause of death by an infectious agent [1]. MRSA infections are particularly difficult to treat because of the development of antibiotic resistance and limited therapeutic options. Glycopeptide antibiotics (vancomycin and teicoplanin) are still the preferred drugs for treatment of serious hospital or community-acquired MRSA infections, despite reports of increasing numbers of glycopeptide-resistant MRSA isolates [2], [3].
Glycopeptide resistance in Staphylococcus have emerged by two mechanisms. Highly glycopeptide-resistant S. aureus strains (VRSA; MIC≥16 µg/ml) acquired the exogeneous multigene VanA complex carried on transposon Tn1546 from Enterococcus faecalis by horizontal gene transfer. Fortunately, these events are infrequent and only few examples are known worldwide [4], [5]. The second mechanism of resistance, termed endogenous or low-level (MICs with >2 µg/ml to <16 µg/ml), arises because spontaneous mutation(s) are thought to confer a selective survival advantage. Endogeneous resistance is thought to occur stepwise: emergence of resistance to low-antibiotic levels must be first acquired to allow growth in progressively higher antibiotic concentrations [6], [7]. The exact molecular mechanism(s) leading to endogeneous resistance to teicoplanin, or vancomycin, is/are unknown. A common resistance pathway has being suggested since in general reduced susceptibility to vancomycin strains also display reduced teicoplanin sensitivity. However, teicoplanin resistance can be acquired without alteration in vancomycin susceptibility [3], [6].
Endogenous resistance is more often observed and clinical studies have linked glycopeptide clinical failure with progressive selection of bacterial isolates showing increasing glycopeptide resistance levels. In some reported cases, as little as a two-fold change in MIC altered clinical outcome [8], [9]. Such concerns have resulted in the recent re-evaluation of clinical breakpoints for glycopeptides [10] (www.srga.org/). In light of these concerns, understanding the molecular changes permitting survival of S. aureus during drug challenge is of paramount importance.
Glycopeptides are non-penetrating cell wall acting antibiotics whose site of action lies outside the cell membrane, implying that changes in physical-chemical barriers, detection, signalling and response mechanisms could conceivably promote resistance. Several phenotypic responses leading to resistance are observed in some, but not all cases and include thicker cell wall, reduced autolysis and increased cell wall crosslinking [11], [12], [13], [14], [15], [16], [17]. These changes are thought to be correlated with a differential expression of genes involved in cell wall metabolism. In several studies, point mutations occurring in only one gene (vraRS, graRS or tcaA) have been described and confirmed by genetic analysis to contribute to glycopeptide resistance [15], [18], [19]. No studies concerning the functional links between these pathways have been provided, however. Only one study has uncovered multiple mutations by sequence analysis when comparing Mu50 (VISA) and a revertant Mu50Ω (VSSA) that were conclusively proven to contribute to low-level glycopeptide resistance [20].
In this study, we identified genomic changes using whole genome deep sequencing of a teicoplanin-susceptible and its teicoplanin-resistant derivative. Point mutations occurred in three genes and complete genetic analysis proved their causal link to glycopeptide resistance. We demonstrate multiple pathways contributing to glycopeptide resistance emergence including two types of phosphosignalling (histidine and serine/threonine) combined with a global redox/thiol stress sensor.
Results
Genomic sequencing of isogenic ISP794 and AR376 strains
To identify genomic changes selected during teicoplanin exposure, whole genome deep sequencing of both ISP794 and AR376 was performed. AR376 is an in vitro derived stable teicoplanin-resistant mutant from laboratory strain ISP794 [21].
Since ISP794 and AR376 are derivatives of the sequenced NCTC8325 strain (NCBI Accession No. NC_007795), we used this as a reference to evaluate genome coverage and facilitate contig assembly. Using Illumina-Solexa technology, we obtained 5,659,670 and 3,609,916 of 35-bp paired-end reads for ISP794 and AR376 strains, respectively, mapping the reference genome. The raw coverage depth was 35 and 23 times for ISP794 and AR376, respectively. Mapping covered 2,819,664 and 2,819,622 bases in ISP794 and AR376 strains, resulting in 99.8% and 99.6% of NCTC8325 genome coverage (2,821,361 bp), respectively. Most of the reference genome was covered sufficiently (minimum coverage of 3 read bases per reference base) to allow single nucleotide polymorphism (SNP) detection. The paired end information was used to detect with high confidence insertions and deletions (InDel).
SNP and InDel differences detected between ISP794 and AR376 strains
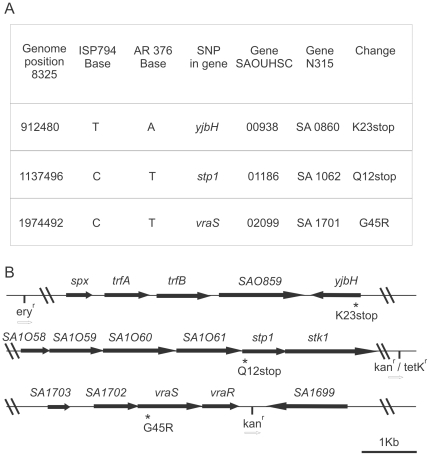
Computer analysis of interstrain differences between the assembled ISP794 and AR376 genomes first revealed 9 potential SNPs and 11 potential InDels. Each difference was subsequently re-examined by genomic DNA PCR amplification using appropriate primers and sequencing. Six potential SNPs and all InDel differences were rejected by this analysis. As shown in Figure 1, only 3 SNP differences apparently distinguished AR376 from its parental strain ISP794.
Figure 1. Genomic differences detected by whole genome sequencing.
(A) Chromosomal location of detected SNPs and predicted protein changes. (B) Physical map of each locus containing yjbH(K23stop), stp1(Q12stop) and vraS(G45R). For genetic analysis, each mutation was tagged by site-specific insertion of a nearby selectable marker to facilitate strain constructions using bacteriophage-mediated transduction.
SNPs were detected in three different open reading frames corresponding to reference sequence tag SAOUHSC 00938, SAOUHSC 01186 and SAOUHSC 02099 genes (equivalent to N315 ordered sequence tags SA0860, SA1062 and SA1701, respectively). Similarity searches with SAOUHSC 00938 indicated the presence in Bacillus subtilis of a gene showing partial similarity and annotated as yjbH (SwissProt accession BG13137). In B. subtilis YjbH is an adaptor protein, which, together with ClpXP protease, regulates the degradation of the global transcriptional regulator Spx [22]. In S. aureus, YjbH is a hypothetical protein showing 34% identity and 73% amino acid sequence similarity with YjbH of B. subtilis. The yjbH t/a SNP occurred at nucleotide position 67 generating a nonsense mutation at amino acid 23 (K23stop). The SAOUHSC 01186 gene encodes Stp1, a serine/threonine phosphatase [23]. The stp1 c/t SNP occurred at nucleotide position 34 generating a nonsense mutation at amino acid 12 (Q12stop). Finally, the SAOUHSC 02099 gene encodes the histidine kinase sensor VraS. The vraS c/t SNP occurred at nucleotide position 133 generating a missense mutation at amino acid 45 by substituting arginine for glycine (G45R) (Figure 1). The amino acid G45 in VraS is located in a region predicted to lie between two N-terminal transmembrane domains suggesting a possible role in extracellular signal sensing.
Genetic analysis of each SNP and its contribution to teicoplanin-resistance
To determine whether each individual SNP detected in AR376 contributed to reduced teicoplanin susceptibility, we first re-engineered each SNP change by site-specific mutation or phage mediated transduction backcross in ISP794 (see Table 1). As judged by broth macrodilution MICs and confirmed by spot population analysis profiles (spot PAP assays), vraS(G45R), or stp1(Q12stop), led to reduced teicoplanin susceptibility (MIC = 2 µg/ml) compared to the parent ISP794 (MIC = 1 µg/ml) (Table 2). Spot PAP assays revealed detectably enhanced growth of both single mutants on MHA supplemented with increasing concentrations of teicoplanin, while no growth was observed with parent ISP794. Identical results were obtained when comparing a second independent isolate of each single mutant (Figure 2A). In contrast to vraS(G45R) and stp1(Q12stop) mutants, a small increase of growth was observed for yjbH(K23stop) while no growth was observed for the parent ISP794. Growth was observed when 1×105 and 1×104 colony forming units were applied on MHA supplemented with 0.5 µg/ml of teicoplanin (Figure 2A). However, MIC analysis showed no difference between ISP794 and its isogenic yjbH(K23stop) mutant (MIC = 1 µg/ml) (Table 2).
Table 1. Strains and plasmids used in this study.
| Strain/plasmid | Revelant genotype | Characteristics | Source/reference |
| S. aureus | |||
| RN4220 | 8325-4; r- m+, restriction defective laboratory strain | [55] | |
| ISP794 | 8325 pig 131 | [56] | |
| AR376 | ISP4-2-1 | [21] | |
| AR774 | vraS (G45R) | ISP794, vraS (G45R) kanr nearby | This study |
| AR758 | kanr nearby vraS | ISP794, vraS kanr nearby | [32] |
| AR756 | vraS::ermB | ISP794, vraS::ermB | [32] |
| AR802 | kanr nearby stp1(Q12stop) | AR376, stp1(Q12stop) kanr nearby | This study |
| AR860 | kanr nearby stp1 | ISP794, stp1kanr nearby | This study |
| AR826 | stp1 (Q12stop) | ISP794, stp1(Q12stop) kanr nearby | This study |
| AR853 | tetKr nearby stp1(Q12stop) | AR376, stp1(Q12stop) tetKr nearby | This study |
| AR858 | vraS (G45R), stp1 (Q12stop) | ISP794, vraS (G45R) kanr nearby, stp1(Q12stop) tetKr nearby | This study |
| AR864 | vraS (G45R), yjbH (K23stop) | AR376, stp1 kanr nearby | This study |
| AR854 | stp1 (Q12stop), yjbH (K23stop) | AR376, vraS kanr nearby | This study |
| AR1077 | eryr nearby yjbH (K23stop) | AR376, yjbH (K23stop) eryr nearby | This study |
| AR1079 | yjbH (K23stop) | ISP794, yjbH(K23stop) eryr nearby | This study |
| AR1082 | vraS (G45R), yjbH (K23stop), stp1 (Q12stop) | ISP794, vraS (G45R) kanr nearby, yjbH (K23stop) eryr nearby, stp1(Q12stop) tetKr nearby | This study |
| AR964 | AR774/pAM1483 | This study | |
| AR1001 | AR826/pAR992 | This study | |
| AR1085 | AR1082/pAR973 | This study | |
| SaΔclpP | 8325-4 derived strain | clpP deleted strain | [57] |
| Δspx | 8325-4 derived strain | spx deleted strain | [27] |
| spx+ | 8325-4 Δspx, Pspx-spx::geh | spx deleted strain and chromosomally complemented with the intact copy of spx inserted into geh locus. | [27] |
| Plasmids | |||
| pTYB12 | N-terminal fusion IMPACT intein and chitin binding domain plasmid | New England (Biolabs) | |
| pMK4 | E.coli-S.aureus shuttle vector, ampr and camr | [58] | |
| pBT2 | E.coli-S.aureus thermosensitive-shuttle vector, ampr and camr | [59] | |
| pBluescript II KS+ | routine multicopy E.coli cloning vector ampr | Stratagene | |
| pAR749 | pBT2, vraS-G45R kanr nearby ts shuttle vector | This study | |
| pAR712 | pBT2, vraS-kanr-SA1699 intergenic ts shuttle vector | [32] | |
| pAR784 | pBT2, stp1-kanr nearby ts shuttle vector | This study | |
| pAR787 | pBT2, stp1-tetr nearby ts shuttle vector | This study | |
| pAR1063 | pBT2, yjbH-eryr nearby ts shuttle vector | This study | |
| pAM1483 | pMK4- 3.3 kb entire vraR operon and upstream promoter region Kpn-Pst | 32 | |
| pAR992 | pMK4- containing Not1-Kpn pHU promoter region, Kpn-Pst1 Stp1 gene | This study | |
| pAR973 | pMK4- containing Not1-Kpn pGlyS promoter region, Kpn-Pst1 yjbH gene | This study | |
| pAM1101 | pTYB12-SpxA (Nde-Pst) | This study |
Table 2. Glycopeptide susceptibility profiles of ISP794 and its derivatives.
| MIC* µg/ml | ||
| Strain | Teicoplanin | Vancomycin |
| ISP794 | 1 | 2 |
| AR376 | 8 | 4 |
| vraS * | 2 | 2 |
| stp1 * | 2 | 2–4 |
| yjbH * | 1 | 2 |
| vraS * /stp1 * | 4 | 4 |
| vraS * /yjbH * | 1 | 2 |
| stp1 * /yjbH * | 1 | 2 |
| vraS * /stp1 * /yjbH * | 8 | 4 |
*MIC, Modal minimum inhibitory concentration measured by broth macrodilution. VraS*, stp1* and yjbH * correspond to vraS (G45R), stp1 (Q12stop) and yjbH (K23stop) mutations, respectively.
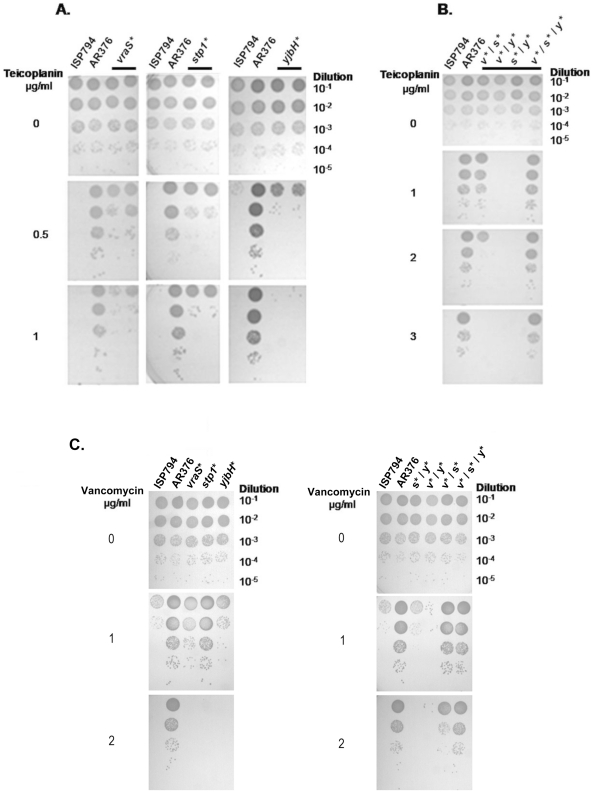
Figure 2. Stepwise genetic reconstitution of all SNP permutations and analysis of their contribution to glycopeptide resistance.
(A and B) Teicoplanin spot plating population analysis (Spot PAP) on teicoplanin of ISP794 and each of its strain derivatives. Spot serial dilutions are indicated at the right margin. The first spot 10 µl corresponds to 1×105 colony forming units (CFU). Results for two independent isolates are shown for each single mutant (solid bar). For convenience, genes are marked with an asterisk such that vraS*, stp1* and yjbH* correspond to vraS(G45R), stp1(Q12stop) and yjbH(K23stop) mutations, respectively. (C) Vancomycin spot plating population analysis (Spot PAP) of ISP794 and each of its strain derivatives. Conditions used were as in panels A, B.
Complementation of either vraS(G45R) or stp1(Q12stop) mutants by a multicopy plasmid carrying the corresponding wild-type genes, led to restored sensitivity to teicoplanin (Figure S1). As yjbH(K23stop) mutation has a subtle effect on teicoplanin resistance, complementation of the yjbH mutation was performed by introducing a multicopy plasmid carrying yjbH wild-type gene into the triple mutant (see below), a condition where its role could be readily assessed. We observed restored sensitivity of the triple mutant to levels comparable with the double mutant vraS(G45R)/stp1(Q12stop) mutant (Figure S1).
Collectively, we conclude that single vraS (G45R), stp1 (Q12stop) and, to a lesser extent, yjbH(K23stop) mutations in parent ISP794 decreased teicoplanin susceptibility, however, none of the single mutations alone could account for the parental AR376 MIC levels. We conclude that some combination of these mutants must therefore contribute to the observed teicoplanin-resistant phenotype of AR376.
Effect of mutant combinations on teicoplanin resistance
We next analysed the drug resistance phenotypes of each of the three possible pairwise mutants: stp1/yjbH, stp1/vraS, and yjbH/vraS. As shown in Figure 2B and Table 2, a further decrease of teicoplanin susceptibility was observed only with double mutant vraS(G45R)-stp1(Q12stop). Spot PAP assays revealed growth of the double vraS(G45R)-stp1(Q12stop) mutant on MHA supplemented with 2 µg/ml teicoplanin and showing a macrodilution MIC of 4 µg/ml. Notably, the double mutant displayed enhanced growth compared to either of the single mutants alone (Figure 2A and B, compare growth and log10 –fold scale, teicoplanin 1 µg/ml panels, for the single stp1 or vraS mutants versus the stp1/vraS double mutant). In contrast, and as detected by spot PAP assay and macrodilution MIC, the other two pairwise mutants did not show any decrease in teicoplanin susceptibility (Figure 2B and Table 2). Interestingly, the decrease in teicoplanin susceptibility observed in vraS(G45R), or stp1(Q12stop) single mutants, was abolished by the yjbH(K23stop) mutation (Figure 2A, B and Table 2). These results suggest that the loss of yjbH exerts a negative effect on the pathways leading to drug sensitivity changes engendered by vraS(G45R) or stp1(Q12stop) mutations.
Although the double mutant vraS(G45R)-stp1(Q12stop) decreased teicoplanin susceptibility, it still did not fully recapitulate the AR376 MIC levels. The effect on teicoplanin susceptibility of the fully reconstructed triple mutant was therefore tested. Genetic analysis of the re-engineered triple mutant revealed complete restoration of the teicoplanin resistance phenotype indistinguishable from AR376. Spot PAP assays revealed identical growth of both the triple mutant vraS(G45R) - stp1(Q12stop) - yjbH(K23stop) and AR376 on MHA supplemented with increasing concentrations of teicoplanin (Figure 2B) and identical macrodilution MIC levels were also observed (MIC = 8 µg/ml) (Table 2).
Effect of single mutations and their combinations on vancomycin susceptibility
The impact of all mutations was also examined for their contribution to changes in vancomycin susceptibility, despite the fact that the in-vitro derived teicoplanin-resistant AR376 strain showed only a 2-fold marginal increase in vancomycin MIC compared to its susceptible counterpart ISP794 (Table 2). Changes in vancomycin MICs were not as dramatic as the changes observed in teicoplanin MICs for the various mutant combinations (Table 2). Nevertheless, as shown in Figure 2C, similar results with teicoplanin susceptibilities were observed using the more sensitive log10-scaled growth assay. Spot PAP assays revealed detectable enhanced growth of both single vraS(G45R) or stp1(Q12stop) mutants on MHA supplemented with 1 µg/ml of vancomycin compared to the susceptible strain ISP794. In contrast no difference was observed with yjbH(K23stop) mutant. At 2 µg/ml vancomycin concentration only AR376 showed growth and comparable colony formation as observed with control agar plates with no drug. Further analysis of all pairwise and triple mutants revealed a detectable decrease in vancomycin susceptibility only for the double vraS(G45R)-stp1(Q12stop) and reconstructed triple mutants. As previously noted above and in results presented in Figure 2B, we also observed that the yjbH(K23stop) mutation, when paired with either vraS(G45R) or stp1(K12stop) exerted a negative effect by reversing drug sensitivity changes arising from these single mutations.
Taken together, we conclude that all three SNPs were collectively responsible for the decreased glycopeptide susceptibility of AR376 strain. Furthermore, vraS(G45R) and stp1(Q12stop) mutations acted synergistically to reduce glycopeptide susceptibility while yjbH(K23Stop) contributed significantly to low level glycopeptide resistance in this strain only when paired with the dual phospho-signalling mutants. These results also reveal that although AR376 was originally selected for reduced susceptibility to teicoplanin, the genetic changes also impact vancomycin susceptibility, albeit to a lesser degree in this strain background. The sensitivity of the spot PAP assay to reveal these subtle changes underscores its powerful application to unravelling the genetics of endogenous glycopeptide resistance.
Analysis of cell wall thickness by electron microscopy
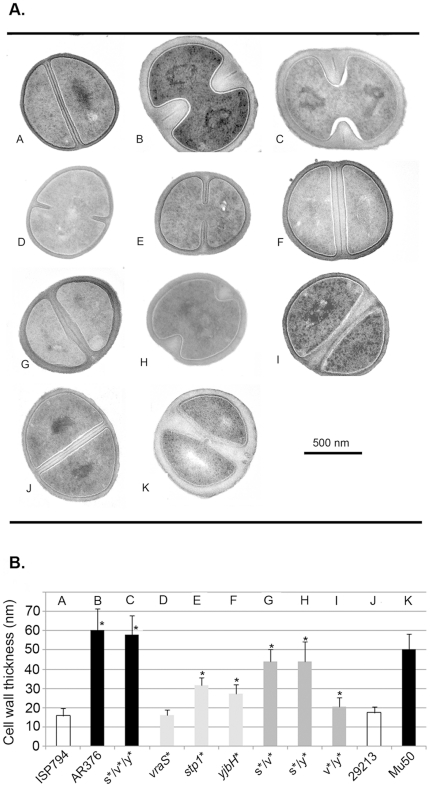
Since the VraRS two-component sensor system regulates a response to cell wall stress and controls, in part, the expression of genes involved cell wall metabolism [24], and further, since stk1/stp1 modulate cell wall metabolism [25], we analysed the cell wall thickness of all strains used in this study by transmission electron microscopy (Figure 3). As reference control strains, we first analysed the cell wall thickness of the glycopeptide resistant strain Mu50, which displays a significantly thicker cell wall (50.4±8 nm) than the glycopeptide susceptible control strain ATCC29213 (17.35±2.9 nm) (Figure 3A and 3B; [11]). Analysis of AR376 showed a significantly (p<0.001) thicker cell wall compared to its susceptible counterpart ISP794 and furthermore, this enhanced thickness was comparable to, and in fact greater than, Mu50. Both of the stp1(Q12stop) and yjbH(K23stop) single mutations significantly increased (p<0.001) cell wall thickness compared to the susceptible parental strains ISP794, while in contrast, no significant difference was observed between vraS(G45R) mutant and ISP794. Analysis of all pairwise mutants showed a significantly greater (p<0.001) increase in cell wall thickness compared to ISP794; however, no double mutation attained AR376 cell wall thickness levels. Indeed, the observed mean thickness for both the stp1/vraS and stp1/yjbH mutations were significantly less (p<0.001) than AR376. Only the fully reconstituted triple mutant AR1082 showed comparable cell wall thickness as AR376 strain. It is worthwhile noting that the yjbH(K23stop) mutation enhanced cell wall thickness when paired with either stp1(Q12stop) or vraSG45R in these assays in contrast to the negative effect of yjbH(K23stop) on teicoplanin MIC and spot PAP assay growth (on either teicoplain or vancomycin) when paired with these mutants as described above. Apart from these changes in cell wall thickness, we did not observe overt changes in cell division such as aberrant septum formation or other peculiar morphology for any mutation examined.
Figure 3. Cell wall thickness analysis.
(A) Transmission electron microscopy showing one representative image of each bacterial strain used in this study, after growth in the absence of antibiotic to post-exponential phase on TSB media. Magnification ×37,000. Scale bar automatically inserted by the microscope imaging system is shown. (B) Quantification of cell wall thickness (nm) of each bacterial strain. Reported values correspond to the mean ± SD (n = at least 40) of each bacterial strain. Asterisk (*) represents results significantly different by student's two-tailed t-test (p<0.05) from ISP794.
Collectively, these results show that all mutations contribute to significant alterations in cell wall thickness in our strain background; however, the observed changes in cell wall thickness do not precisely correlate with the observed changes in glycopeptide susceptibility. We conclude that changes in glycopeptide sensitivities in this strain background must arise primarily from physiological changes other than pronounced changes in cell wall thickness.
YjbH negatively regulates Spx levels in S. aureus
A role for YjbH has not been previously described in S. aureus; however, in B. subtilis, YjbH acts as a post-transcriptional negative regulator of the global oxidative/thiol stress regulator Spx [26]. YjbH binds Spx and functions as an adaptor protein directing ClpXP-dependent degradation of Spx. Disruption of yjbH or clpP in B. subtilis is known to result in enhanced levels of Spx since its proteolytic turnover is greatly attenuated. A rise in Spx levels leads to substantial changes in gene expression profiles.
To address the hypothesis that S. aureus YjbH acts as a negative regulator of Spx analogous to its B. subtilis counterpart, we first examined Spx protein levels by western blot. As shown in Figure 4, rabbit polyclonal anti-Spx antibody recognized purified S. aureus Spx migrating with the expected molecular weight of 15 kDa. No Spx was detected in a negative control S. aureus 8325-4 derived strain extract containing an internal disruption of spx, whereas Spx was strongly detected in cell extract lacking clpP as previously described [27]. Spx was also detected in an extract from AR376, harbouring the triple vraS(G45R)-stp1(Q12stop)-yjbH(K23stop) mutant combination. Importantly, comparison of extracts from ISP794 and its isogenic derivative AR1079 containing the yjbH(K23Stop) mutation, showed that Spx was undetectable in the presence of wild type yjbH, but clearly detectable in the presence of the yjbH(K23Stop). We conclude from these results that the loss of yjbH results in significant stabilization of Spx protein in S. aureus and that YjbH probably functions similarly to its B. subtilis counterpart by aiding Spx proteolytic turnover. Consequently, the triple mutant AR376 harbors changes in two distinct phosphosignalling pathways and possesses enhanced Spx protein levels arising from the loss of its negative regulator yjbH.
Figure 4. Western blot showing the effect of yjbH(K23stop) mutation on Spx protein levels.
Total soluble protein extracts (50 µg) from S. aureus strains were loaded per well in an SDS 15% acrylamide gel. Spx protein (15 kDa) was detected using rabbit-polyclonal anti-Spx antibodies. Purified-Spx protein (600 ng) (lane 1), Δspx strain (lane 2), ISP794 containing wild-type yjbH gene (lane 3), yjbH* correspond to yjbH(K23stop) mutant (lane 4), SaΔclpP strain (lane 5) and AR376 (lane 6). The position of protein markers are shown in the right margin.
Influence of Spx and teicoplanin on transcription of msrA1 encoding methionine sulfoxide reductase in S. aureus
Several lines of evidence link oxidative stress and S. aureus responses to glycopeptide antibiotics. First, recent work suggests that the mode of killing by several classes of antibiotic, including glycopeptides, involves the endogenous production of reactive oxygen radicals [28]. Secondly, transcriptional profiling revealed that an oxidative stress defence gene, msrA1, encoding methionine sulfoxide reductase was among those genes most strongly upregulated in response to various cell wall active antibiotics, including glycopeptides, in S. aureus [29], [30]. As msrA1 transcription is known to be positively regulated by Spx in B. subtilis [31], we hypothesized that a possible functional consequence of the loss of yjbH in AR376, would be enhanced transcription of Spx-dependent genes such as msrA1 (N315 ordered sequence tag SA1257). To explore this possibility, we tested whether msrA1 was indeed subject to Spx-dependent transcription regulation in S. aureus.
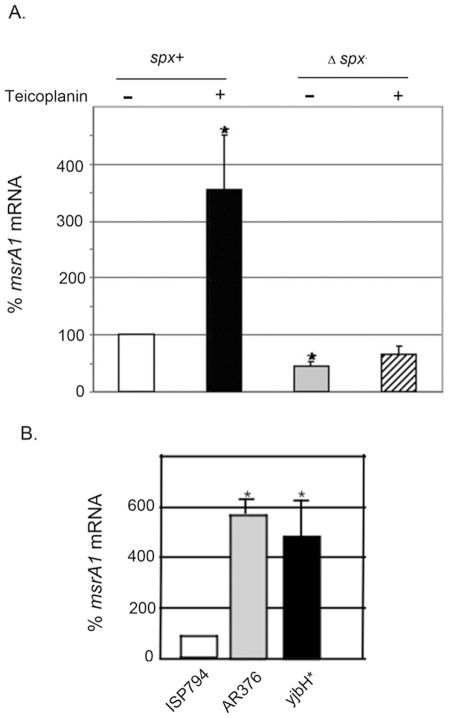
The results (Figure 5A) show that in a Δspx strain, teicoplanin exposure resulted in no significant induction of msrA1 compared to uninduced control, whereas in the Δspx strain complemented with a chromosomally integrated copy of wild type spx, msrA1 induction was significantly induced more than 3-fold (p<0.05) following exposure to teicoplanin. Indeed, in the absence of spx, even basal msrA1 transcript levels were significantly lower (p<0.05) compared with msrA1 transcript levels in the spx complemented strain.
Figure 5. Effect of spx on msrA1 transcription.
(A) Steady-state levels of msrA1 transcript of wild type strain (spx +) compared to mutant Δspx strain, determined by qRT-PCR. Values represent the mean ± SEM of four independent experiments performed in triplicate. Data are depicted setting spx + values in the absence of teicoplanin as 100%. Asterisk (*) represents results significantly different by student's two-tailed t-test (p<0.05) from spx + without teicoplanin. (B) Steady-state levels of msrA1 transcript of ISP794 containing wild-type yjbH gene, AR376 and yjbH* containing both yjbH(K23stop) mutation, determined by qRT-PCR. Values represent the mean ± SEM of three independent experiments performed in triplicate. Data are depicted setting ISP794 values as 100%. Asterisk (*) represents results significantly different by student's two-tailed t-test (p<0.05) from ISP794.
Since the yjbH(K23stop) mutation results in significant stabilization of Spx protein, we also analysed whether the steady state msrA1 transcription in the absence of drug induction was affected in either AR376 or the yjbH(K23stop) single mutant. Figure 5B shows that basal msrA1 transcript levels were indeed significantly higher (p<0.05) in both AR376 and yjbH(K23stop) mutant compared to the parental strain ISP794. We conclude from these results that msrA1 is subject to transcriptional regulation dependent upon spx in S. aureus.
We also recently described the discovery of trfA (teicoplanin resistance factor A) as a gene whose loss restores glycopeptide sensitivity in AR376 [21]. We have found that trfA transcript levels are also significantly increased by the loss of yjbH and that the trfA promoter is also positively regulated by Spx (A. Renzoni, manuscript in preparation). Taken together, these findings strongly suggest that stabilization of Spx through disruption of yjbH results in important functional consequences within the cell since Spx positively regulates the expression of at least two genes, msrA1 and trfA, known to contribute to glycopeptide resistance.
Effect of pre-existing stp1(Q12stop), yjbH(K23stop), vraS(G45R) or spx mutations on the emergence teicoplanin resistance in S. aureus
Since AR376 was originally obtained by stepwise cultivation on agar plates containing low levels of teicoplanin we wished to determine the extent to which each individual mutation identified in our study contributed to a change in a detectable frequency of emergence of teicoplanin resistant colonies using our standard assay conditions [21], [32]. In light of our findings that disruption of yjbH altered Spx levels, we also tested the effect of spx deletion in the same assay. The results are shown in Table 3.
Table 3. Effect of the single point mutations and spx on the emergence of teicoplanin resistance.
| No. bacteria under non-selective conditions T0 | No. bacteria under selective conditions T2 | Frequency of emergence1 | |||||
| Strain | Relevant genotype | Mean (n = 7) | SEM | Mean (n = 7) | SEM | T2/T0 | % |
| ISP794 | ISP794, rsbU − | 7.82×10+7 | 2.01×10+7 | 255 | 79 | 3.26×10−6 | 1003 |
| AR1079 | ISP794, yjbH(K23stop) eryr nearby | 6.98×10+7 | 1.95×10+7 | 895 | 201 | 1.28×10−5 | 394 |
| AR774 | ISP794, vraS(G45R) kan r nearby | 1.25×10+7 | 2.24×10+6 | >10002 | 8.00×10−5 | >1000 | |
| AR826 | ISP794, stp1(Q12stop) kanr nearby | 2.80×10+7 | 5.73×10+6 | >10002 | 3.50×10−5 | >1000 | |
Frequency of emergence expressed as the ratio of colony forming units (CFU) under selective (Teicoplanin 2 µg/ml) and non-selective conditions (No teicoplanin). Colony forming units were counted at 48 h, 37°C.
Viable counts on agar containing 2 µg/ml of teicoplanin were too high to accurately measure using these conditions. More than 1000 CFU were estimated in each experiment.
The effect of rsbU + on glycopeptide emergence is discussed in Galbusera et al [(30)]. ISP794 emergence frequency was set equal to 100% for normalization and comparison.
The emergence frequency was set equal to 100% for normalization and comparison with its derivative Δspx.
Compared to ISP794, strains harboring any of the three mutations, stp1(Q12Stop), yjbH(K23Stop) or vraS(G45R) - each backcrossed by bacteriophage mediated transduction into ISP794 and never having been previously exposed to glycopeptides, resulted in strongly enhanced frequency (from 4- to 10-fold) of emergence of low-level teicoplanin resistant colonies on MH agar plates. In contrast, loss of spx was associated with at least a 10-fold reduction in the frequency of emergence of teicoplanin resistant colonies compared to the identical strain background harboring a second wild-type copy of spx. We conclude that loss of spx reduced the emergence of teicoplanin resistant colonies while mutation in any of the three mutations discovered by genomic sequence analysis of AR376 enhanced the emergence of teicoplanin resistance.
Discussion
In the present study, we show by deep sequencing and genetic analysis that a laboratory selected low level teicoplanin intermediate resistant S. aureus strain has only three point mutations which distinguished it from its parental susceptible strain. These changes occur in two distinct (histidine kinase and serine/threonine kinase) phospho-signalling systems and in a gene contributing to a global oxidative/thiol stress defence response pathway. Moreover, we present evidence revealing a synergistic interaction between the dual phospho signalling pathways. The contribution of the third mutation in yjbH is subtle, and its properties suggest that it most probably exerts its effect following the first step acquisition of low level glycopeptide resistance mediated by the combined effects of stp/vraS mutations.
S. aureus glycopeptide resistance has been often correlated with morphological changes such as increased cell wall thickness, peptidoglycan crosslinking or decreased autolysis. Both vraS and stp1 are known to affect cell wall metabolism, although precisely how is unknown [23], [24], [33], [34], [35].
The VraRS two-component system (TCS) responds to cell-wall active antibiotics [24], [33]. Under cell wall stress, expression of vraRS and several cell wall biosynthetic genes are strongly upregulated. Disruption of the histidine kinase vraS gene blocks this transcriptional response and reduces ß-lactam or glycopeptide resistance levels [32], [33], [36] Missense mutations in vraS altering the susceptibility to glycopeptides are commonly observed [17], [20], [37], [38].
How vraS(G45R) contributes to enhanced glycopeptide resistance is unknown. Amino acid 45 is located in the short region predicted to span two putative transmembrane segments and suggests a role in signal detection. The G45R mutation could also conceivably affect interaction with other regulatory proteins. For example, B. subtilis LiaF, a membrane protein, is thought to negatively regulate LiaS, a putative S. aureus VraS ortholog. B. subtilis LiaF is syngenic to S. aureus SA1702, and mutations in SA1702 have also been reported to affect glycopeptide resistance [17], [38]. Indeed, a recent report suggests that S. aureus SA1702/YvqF and VraS directly interact lending support to the notion that SA1702/YvqF may modulate VraS signalling and detection of cell wall stress [39].
The vraS(G45R) mutation does not detectably increase VraR operon mRNA levels compared to its isogenic parent strain, or any gene (sgtB, prsA, htrA1, murZ) thought to be regulated by VraR that we tested by qRT-PCR and our experimental conditions. Nevertheless, this mutation by itself does result in pronounced changes in emergence of glycopeptide resistance frequencies. This result is curious, since in most reported cases, glycopeptide resistant bacteria show increased expression of vraR operon transcription compared to susceptible parental strains [24], [37], [40]. Nevertheless, one report, consistent with our findings, showed that increased transcription of vraS or vraRS operon is not essential for acquisition of low-level glycopeptide resistance [41]. A closer study of vraR operon regulation and VraR phosphorylation mechanisms is warranted to resolve precisely how VraS missense mutations disrupt signalling.
In S. aureus, Stp1 plays a role in phospho-signalling, together with Stk1 [23], [34], [35], [42], [43]. Stp1 (N315-SA1062), encodes a manganese-dependent serine/threonine phosphatase, capable of dephosphorylating Stk1 [23], [34]. Stk1 is thought to be a membrane protein which possesses three extracellular PASTA domains (penicillin-binding protein and serine/threonine kinase associated domain) [44]. Stk1 or Stp1 have been shown to have a role in S. aureus virulence, cell wall metabolism and antibiotic resistance (for review see [25]). The effects of a single stp1 mutant, however, have not been tested for glycopeptide resistance [23], [34], [35], [42], [43]. To our knowledge, our work is the first study to report mutation in stp1 alone directly affecting glycopeptide resistance levels.
It is not known how stk1/stp1-mediated signalling contributes to the detection of antibiotic encounter and orchestration of cellular responses. One plausible hypothesis is that Stk1 senses some feature(s) of the cell wall assembly process via its PASTA domains and phosphorylates multiple downstream factors that coordinate proper assembly and quality control [44]. In the event of encounter with cell wall active drugs such as glycopeptides, Stk1 could conceivably signal damage and initiate a cascade of signalling events that permit survival in the face of drug stress. The loss of function of stp1 would have the predictable consequence of enhancing the duration of substrates phosphorylated by Stk1. Stk1 and Stp1 are only beginning to be understood in S. aureus, however. To date, several Stp1 client substrates have been described including the global accessory regulator SarA, MgrA, and the nucleoid organizing protein HU [43], [45]. MgrA is known to be a major cell-wall autolytic regulator which modulates antibiotic resistance, including glycopeptides [45], providing a possible mechanistic link between signal systems and altered cell wall assembly.
Changes in cell wall thickening resulting from stp1 disruption appear to be strain dependent and divergent results have been reported for strains Newman and N315, for example [23], [34], [43]. The reason for these discrepancies is unknown and additional studies will be needed to resolve this important issue. The stp1(Q12stop) mutation present in our strain background nearly doubled cell wall thickness compared to ISP794. A recent metabolome study using 8325, a strain closely related to ISP794, detected striking coordinate increases in peptidoglycan intermediates depending upon the sequential action of MurA to MurF enzymes in a strain lacking stp1 compared to its isogenic parent [46]. These results strongly suggest that stk1/stp are indeed intimately involved in the regulation of cell wall biosynthesis.
A striking feature of our study is the enhanced glycopeptide resistance of the double stp1(Q12stop)/vraS(G45R) mutation compared to the resistance levels of the single mutants alone. This observation suggests the intriguing possibility that these signalling systems collaborate to modulate genes necessary to elicit drug resistance. Understanding the molecular mechanism of endogenous glycopeptide resistance arising from this dual signalling synergy is being vigorous pursued.
The yjbH(K23stop) mutation displays both positive and negative effects upon glycopeptide resistance. By itself, the yjbH(K23stop) mutation displays little contribution to glycopeptide resistance, whereas when combined as a double mutant with either the vraS(G45R) or stp1 null mutation, yjbH(K23stop) mutation reduces resistance. These findings suggest that mutation of yjbH exerts a negative influence on endogenous glycopeptide resistance achieved with either phosphosignalling pathway mutant alone. In contrast, when yjbH mutation is combined with the double mutant stp1(Q12stop)/vraS(G45R), a two-fold increase in modal teicoplanin MIC was observed (4 to 8 µg/ml) and was accompanied by increased colony counts indistinguishable from the parental triple mutant sequenced strain AR376. In this instance, yjbH clearly acts positively. The negative effects due to yjbH(K23stop) were also observed with vancomycin although its positive contribution to reduced susceptibility to this drug was less apparent in both spot PAP assays and modal MIC values. Future research will resolve in detail how YjbH and Spx contribute to glycopeptide resistance.
The drug resistance profiles of the three mutations detected in our study suggest that their temporal appearance was hierarchical. If either signalling mutation had occurred first, followed by yjbH, then the results we provide upon genetic reconstruction suggest little or no possibility for growth of the yjbH(K23stop)/stp1 or yjbH(K23stop)/vraS(G45R) mutation on teicoplain 2 µg/ml and thus little chance for continued selection. In contrast, it is reasonable to assume that the dual signalling mutants emerged first, followed by the acquisition of the yjbH mutation. This genetic order has the effect of pushing the teicoplanin MIC from 4 µg/ml to 8 µg/ml. It is thus tempting to speculate that the effect of yjbH, either by itself, or more likely through Spx, is manifest only when a threshold pre-existing level of glycopeptide resistance is attained.
We have shown that loss of yjbH leads to stabilization of Spx protein levels and provide evidence that Spx modulates the expression of at least two genes (msrA1 and trfA) involved in glycopeptide resistance. Spx is necessary for the upregulation of msrA1, encoding methionine sulfoxide reductase, an enzyme required to combat oxidative stress. msrA1 (SA1257) is transcribed as a four-gene operon (SA1254-SA1257) and induced by several cell wall active antibiotics [24], [29], [30], [33]. In addition, activating oxidative stress defences is a plausible strategy to combat antibiotic stress since recent studies reveal that several antibiotics, including glycopeptides, kill cells by promoting the production of endogenous reactive oxygen species [28], [47], [48].
Teicoplanin resistance factor A, encoded by trfA, is also regulated by YjbH and Spx (A. Renzoni, manuscript in preparation). TrfA is important for glycopeptide resistance in several strain backgrounds including AR376 used in this and other studies, although its precise function has yet to be established [21]. Studies are underway to dissect which precise pathways are affected by the trfA mutation.
The three mutations uncovered in our study each contribute to enhanced cell wall thickness, although vraS(G45R) evokes only minimal change by itself and acts in conjunction with either of the other mutations. These findings could partly explain the reduced glycopeptide sensitivites observed in some, but not all of our strains, since in many instances no change in glycopeptide MICs were detected despite strong changes in cell wall thickness. To date, the majority of clinical glycopeptide resistant strains exhibit cell wall thickening [3], [11]; however, reduced susceptibility to glycopeptides can be achieved without alterations in cell wall thickness [49], [50]. Other metabolic changes must be affected by the mutations uncovered in our study which modulate low level glycopeptide resistance. While herein we present a detailed analysis suggesting an intricate interplay among signalling pathways in one particular strain, it is certainly possible that additional genes and pathways contributing to the acquisition of endogeneous glycopeptides resistance will be uncovered.
Materials and Methods
Bacterial strains and culture conditions
Bacterial strains used in this study are listed in Table 1. The NCTC8325 strain ISP794 (MIC = 1 µg/ml) and its teicoplanin-derivative AR376 (MIC = 8 µg/ml) are MLST type ST8 (3-3-1-1-4-4-3) and were described previously [21]. All S. aureus and E. coli strains were grown in Mueller-Hinton broth (MHB) and Luria-Bertani medium, respectively. When required, media were supplemented with the following antibiotics: 15 µg/ml chloramphenicol, 5 µg/ml erythromycin, 40 µg/ml kanamycin, and 3 µg/ml tetracycline.
Genome sequencing
Genomic DNA (gDNA) was prepared from an overnight culture grown in MHB at 37°C as previously described [21]. Solexa technology was used to sequence the genomic DNA of both ISP794 and AR376 strains on an Illumina Genome Analyzer GAII (Illumina; Fasteris, SA, Geneva, Switzerland). The quality-controlled filtered reads, followed by alignment to the reference sequence NCTC8325 strain, were used to generate a consensus sequence for both ISP794 and AR376 strains, using the IUPAC ambiguity code. The generated consensus ISP794 and AR376 sequences were then compared to detect SNPs and InDel differences. All detected SNPs and InDels were subsequently verified or rejected by PCR of relevant genomic DNA and capillary sequencing. Additional details are available upon request.
Construction and analysis of genetic point mutants
To test the contribution of each individual mutation to glycopeptide resistance, or reconstruct all possible combinations, we used genetic linkage analysis by conveniently tagging each mutation with a nearby resistance marker (Figure 1B), using pBT2 thermosensitive plasmid [21]. All plasmids were first electroporated into non-restricting strain RN4220 prior to electroporation into ISP794 and its derivative strains. After growth with applied marker selection at a non-permissive temperature for pBT2 replication, double cross-over events were screened using antibiotic markers and all linked mutants were confirmed by PCR and sequencing. We also verified in every case that the insertion of the antibiotic-resistant marker alone did not alter the susceptibility phenotype to either teicoplanin or vancomycin. The intergenic regions chosen were devoid of known small regulatory RNAs. While we cannot exclude that the markers used confer some changes to the cell which we do not detect, they are importantly neutral with respect to all assays used in this study to measure glycopeptide susceptibility. All antibiotic-linked SNP mutations were subsequently backcrossed to ISP794 using bacteriophage Φ80α-mediated generalized transduction. In some cases, allelic exchange using nearby markers linked to the wild type locus were used to remove one or several point mutations and restoration of wild type sequence. Non-cell wall active antibiotic selectable markers kanamycin, tetracycline, and erythromycin were obtained as previously described [21], [32]. Details of the construction of each single, pairwise, or reconstituted triple mutation are indicated below. In no case did we observe that single, double mutants, or the reconstructed triple mutant displayed altered growth rates or detectable fitness changes compared to ISP794.
Generation of point mutant vraS(G45R) in ISP794
VraS(G45R) point mutation linked to a kanamycin-resistant marker was constructed in ISP794 as follows: a chromosomal fragment containing a nucleotide change in position 133 (C to T) of vraS open-reading frame (ORF) to generate an amino acid change from glycine to arginine, was amplified from ISP794 using primers described in Table S1. A restriction site (Bgl2) was used to ligate a kanamycin resistance marker at position 1972957 (NCTC 8325 genome sequence coordinates).
The resulting plasmid, pAR749, was electroporated into vraS::ermB mutant (described in [32] selecting for kanamycin resistance. Double cross-over events were screened on agar containing 40 µg/ml kanamycin and then replica streaked on 15 µg/ml chloramphenicol and 5 µg/ml erythromycin plates to screen for chloramphenicol and erythromycin-sensitive but kanamycin-resistant colonies. Clone AR774a was selected and used to backcross vraS(G45R) linked to kanamycin resistant marker to ISP794 using Φ80α lysates. A single mutant vraS(G45R) clone, AR774, was retained for further study.
Generation of stp1(Q12stop) and yjbH(K23stop) single mutants in ISP794
A similar strategy was used to generate stp1(Q12stop) mutant in ISP794 strain. We first pre-marked strain AR376 by targeted insertion of a kanamycin resistance marker nearby the stp1 gene (chromosomal locations 1142731 and 1142744 from NCTC 8325 genome sequence coordinates), using primer pairs described in Table S1. A restriction site (Bgl2) was used to insert a kanamycin resistant marker as described above. The resulting plasmid, pAR784, was electroporated into AR376, selecting with 40 µg/ml kanamycin and then replica streaked on 15 µg/ml chloramphenicol plates to screen for chloramphenicol-sensitive but kanamycin-resistant colonies. Clone AR802 was chosen and used to backcross the stp1(Q12stop) linked to kanamycin resistant marker to ISP794 using bacteriophage generalized transduction with Φ80α lysates. A single mutant stp1(Q12stop) AR826 was retained for further study.
To generate the yjbH(K23stop) mutant, strain AR376 was pre-marked by targeted insertion of an erythromycin resistance gene nearby yjbH gene in position 905525 (NCTC 8325 genome sequence coordinates) using plasmid pAR1063 generated with primers described in Table S1. One colony was selected and designated AR1077. Transfer of yjbH(K23stop) mutation into ISP794 strain was next performed by bacteriophage transduction using Φ80α lysates of AR1077 selecting for erythromycin-resistance marker. A single mutant yjbH(K23stop), AR1079, was fully verified by PCR and sequence analysis and retained for further study.
Generation of pairwise and triple mutants
The double mutant vraS(G45R)-stp1(Q12stop) was constructed as follows: we first pre-marked strain AR376 nearby stp1 gene (AR853) using plasmid pAR787 as described above, but instead inserting a tetracycline resistance marker at the same chromosomal location as we had inserted the kanamycin marker. Transfer of the stp1(Q12stop) mutation to vraS(G45R) mutant (AR774) was next performed by bacteriophage transduction using Φ80α lysates of AR853 selecting for the tetracycline-resistance marker. The selected kanamycin and tetracycline-resistant transductant (AR858) was analysed for acquisition of both vraS(G45R) and stp1(Q12stop) mutations by sequencing. The double mutant vraS(G45R)-yjbH(K23stop) was constructed as follows: we pre-marked strain ISP794 nearby stp1 gene (AR860) using plasmid pAR784 as described above, but instead inserting a kanamycin resistance marker. Transfer of the stp1 wild-type gene to AR376 was next performed by bacteriophage transduction using Φ80α lysates of AR860 selecting for the kanamycin-resistance marker. The selected kanamycin-resistant transductant (AR864) was analysed for acquisition of wild-type stp1 gene by allelic exchange and the presence of both vraS(G45R) and yjbH(K23stop) mutations by sequencing. To construct the double mutant stp1(Q12stop)-yjbH(K23stop), we pre-marked strain ISP794 with a kanamycin resistance marker nearby wild-type vraS gene (AR758) using plasmid pAR712 as previously described [32]. Transfer of wild-type vraS gene to AR376 was next performed by bacteriophage transduction using Φ80α lysates of AR758 selecting for the kanamycin-resistance marker. The selected kanamycin-resistant transductant (AR854) was analysed for acquisition of wild-type vraS gene and the presence of both stp1(Q12stop) and yjbH(K23stop) mutations by sequencing.
Finally, to reconstruct the vraS(G45R)-stp1(Q12stop)-yjbH(K23stop) triple mutant, we transfered the yjbH(K23stop) mutation from AR376 by bacteriophage transduction using Φ80α lysates of AR1077 to the double mutant vraS(G45R)-stp1(K23stop) (AR858) selecting for the erythromycin-resistance marker. The selected erythromycin, kanamycin and tetracycline-resistant transductant (AR1082) was analysed for acquisition of vraS(G45R), stp1(Q12stop) and yjbH(K23stop) mutations by sequencing.
Plasmids for complementation
Plasmid pMK4 was used to express the entire wild-type vraR operon under control of its own promoter and stp1 or yjbH under the control of the heterologous p HU or p GlyS promoters (Table 1). Briefly, pMK4 plasmid containing Not1-Kpn p GlyS or p HU promoters were obtained as described [51], [52]. PCR fragments containing vraR operon, or promoterless stp1 or yjbH were cloned into Kpn-Pst1 restriction sites using primers described in Table S1. All constructions were sequenced verified. Plasmids constructed in E. coli were first introduced into strain RN4220 prior to transformation of the corresponding mutant strains. Plasmids for negative controls in these experiments were either empty pMK4, or pMK4 harboring a heterologous gene, namely GFPuv4, driven by the same p HU or p GlyS promoters.
Minimal inhibitory concentration (MIC) and population analysis
Broth macrodilution MIC assays were performed in triplicate as previously described according to CLSI (Clinical and Laboratory Standards Institute) standard procedures [21]. Results were reported as the modal MIC values. The spot PAP method was used to assess population analysis, as previously described [21], [32]. Bacterial cultures were adjusted to 0.5 MacFarland standard (1.5×108 bacteria/ml), corresponding to an OD600 = 0.1. Serial log10 dilutions (resulting in a range 10−1 to 10−5) were prepared, then aliquots of each dilution (10 µl) were spotted on freshly prepared MH agar (MHA) containing different concentrations of teicoplanin or vancomycin. Viable colonies were examined after 48 h at 37°C. The results reported were consistant across at least five independent assays. It is important to emphasize that the MIC values reported throughout our studies were determined using broth macrodilution; this method may give higher values than other susceptibility testing methods. Indeed, a recent study conducted in our laboratory indicates that microdilution methods tend to underestimate glycopeptide resistance [53].
Purification of recombinant Spx protein and antibody production
The open reading frame of the spx gene (SA0856 N315) was PCR amplified with primers indicated in Table S1 and cloned in pBluescriptII KS+. A sequenced verified spx fragment was next subcloned into E. coli expression vector pTYB12 (New England Biolabs) using Nde1-Pst1 sites, generating plasmid pAM1101. N-terminal intein-recombinant Spx protein was then purified using chitin affinity column (IMPACT system, New England Biolabs) as follows: E. coli strain ER2566 (New England Biolabs) containing pTYB12-Spx protein was grown in Luria-Bertani media containing carbenicillin at 100 µg/ml until an OD600 of 0.7, induced with 0.4 mM isopropyl–β-d-1-thiogalactopyranoside (IPTG) for an additional 2 h30 min at 22°C with vigorous shaking. Bacteria were harvested by low speed centrifugation, resuspended in buffer containing 0.1% Tween 20, followed by affinity chromatography and intein-mediated proteolytic cleavage of the affinity tag with 50 mM DTT. Recombiant Spx protein was eluted with 20 mM Tris-HCl pH 7.5, 500 mM NaCl and 1 mM EDTA, according to the manufacturer's recommendations. Thiol-induced intein cleavage resulted in the N-terminal attachment of three additional amino acids AlaGlyHis. Protein concentrations were determined using Bradford reagent (BioRad) and a bovine serum albumin standard. Recombinant Spx protein was stored at 4°C and used to generate polyclonal antibodies using the Rabbit Express™ extended protocol (Pickcell Laboratories, Netherlands).
Western blot analysis
Total protein extracts were obtained as follows: 10 ml of an overnight culture of S. aureus strains in MHB were harvested. After centrifugation, cell pellets were washed and resuspended in 500 uL TES (10 mM Tris pH 7.5, 1 mM EDTA and 0.1% SDS). Bacterial cells were disrupted by adding 500 µl of acid washed glass beads (100–200 micron, Sigma) and using FastPrep cell disrupter (MP Biomedicals). The cell debris were separated from soluble protein extracts by centrifugation for 10 min at 4°C at 17000 rpm (JA-20 rotor Beckman) followed by a second centrifugation step to remove additional insoluble and aggregated proteins (30 min, 4°C, 17000 rpm). Supernatant was removed and proteins were concentrated on Amicon spin columns (4 kDa cut-off, Milian, Geneva, Switzerland). Protein concentrations were determined by Bradford assay (Bio-Rad). Aliquots of total proteins (50 µg) were loaded on 15% SDS-PAGE gels and blot transferred onto a polyvinylidene difluoride membranes (PVDF, Bio-Rad). After blocking using 5% (w/v) low fat milk in phosphate buffered saline, membranes were probed with a 1∶1500 dilution of Spx antibody followed by incubation with a secondary HRP-conjugated goat anti-rabbit antibody (1∶10,000 BioRad). Chemiluminescence was detected using the Western Pico Super Signal reagent (Pierce).
Assays for emergence of teicoplanin resistance
Emergence of glycopeptide resistance was performed as previously described [21], [32]. Overnight bacterial cultures were adjusted to McFarland 2.0 in 0.9% NaCl and ca. 3×108 CFU/ml were plated on Muller Hinton agar (BD BBL 211438) containing the indicated freshly prepared teicoplanin concentration and incubated for 48 h at 37°C. To determine the frequency of emergence, serial dilutions of each culture were also plated on MHB plates without drug. Results are expressed as the mean ± SEM of seven independent experiments for ISP794, AR774, AR826, AR1079 and five independent experiments for spx+ and Δspx. A subset of colonies was retested by replica plating on selective agar plates to estimate the percentage of false positives (persisters cells which score as susceptible upon replating on selective medium) arising in each experiment. False positive rate of CFUs detected in the emergence assay were found to be 5–20% depending upon the experiment. The raw data are reported without correction and thus calculated emergence frequencies represent an upper limit. ISP794 used in this study is defective in rsbU. The effects of this mutation, its restoration, and the role of sigB in the emergence of teicoplanin resistance in this strain background have been thoroughly addressed in our previous study [32].
Total RNA extraction
Overnight cultures of 8325-4spx-c and 8325-4Δspx were diluted and grown at 37°C for 4 h in MHB without shaking. When indicated, sub-inhibitory concentrations of teicoplanin (0.5 µg/ml) were added and incubated for an additional hour. Bacteria were harvested and RNA extraction was performed as previously described [21]. The absence of contaminating DNA was verified by PCR using qRT-PCR probes in the absence of reverse transcription.
Quantitative real-time qRT-PCR
mRNA levels were determined by quantitative RT-PCR (qRT-PCR) using the one-step reverse transcriptase qPCR Master Mix Kit (Eurogentec, Seraing, Belgium) as described [54]. Primers and probes were designed using PrimerExpress software (version 1.5; Applied Biosystems), and obtained from Eurogentec. MsrA1 primers (from SA1257) and probe are: msrA1-334F 5′GGAAGTAACCTCTGGATCAAACGT, MsrA1-415R 5′CCCTACTTATGAACAGGTATGTACGAAT and msrA1-359T 5′ATTTGTACTGCTTCGACATGGCCGGTT. 16S primers and probe are described [54]. Reverse transcription and PCR were performed using primers and probes at a concentration of 0.2 and 0.1 µM, respectively. The msrA1 mRNA levels were normalized on the basis of their 16S rRNA levels, which were assayed in each round of qRT-PCR as internal controls as described [54].
Transmission electron microscopy
S. aureus strains were grown on TSB at 37°C under shaking conditions until post-exponential phase. One milliter of post-exponential phase bacteria was pelleted by low speed centrifugation, washed with PBS and fixed for 30 min in PBS containing 4% (v/v) glutaraldehyde. Fixed cells were further treated with 2% osmium tetroxide in buffer and immersed in a solution of 0,25% uranyl acetate to enhance membrane contrast. The pellets were dehydrated in increasing concentrations of ethanol followed by pure propylene oxide, then embedded in Epon resin. Thin sections for electron microscopy were stained with uranyl acetate and lead citrate and observed in a Technai 20 electron microscope (FEI Company, Eindhoven, The Netherlands). Digital images were captured at 37,000×. Cell wall thickness was determined using equatorial sections and four independent measurements for each image from random placement of a cardinal point grid. The reported values represent the mean of at least 40 independent images for each strain.
Supporting Information
Complementation of either yjbH (K23stop), stp1 (Q12stop) or vraS (G45R) mutants by multicopy plasmid carrying wild-type genes. (A) Spot plating population analysis of each single mutant and its corresponding complemented strain on teicoplanin. V* and s* complementation was tested on teicoplanin containing 0.5 µg/ml. Spot serial dilutions are indicated at the right margin. As yjbH(23stop) mutation has a subtle effect on teicoplanin resistance, complementation of the yjbH mutation was performed by introducing a multicopy plasmid carrying wild-type yjbH gene into the triple strain mutant, a condition where its role could be unambiguously assessed. Y* complementation was tested on teicoplanin containing 2 µg/ml. For convenience genes marked with an asterisk such as v*, s* and y* correspond to vraS(G45R), stp1(Q12stop) or yjbH(K23stop) mutations, respectively.
(TIF)
Primers used in this study.
(DOC)
Acknowledgments
We thank Dorte Frees and Hanne Ingmer (Royal Veterinary and Agricultural University, Denmark) for kindly supplying S. aureus spx and clp mutant strains, Laurent Farinelli, Loic Baerlocher and the bioinformatic staff of Fasteris S.A. (Geneva) for their attention to our custom sequencing needs, and Michelangelo Foti and the staff of the University of Geneva Medical School ultrastructure core facility.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Swiss National Science Foundation grant 3100A0-120428 (WLK) and 310030-125109 (DL) and an F. Hoffmann-La Roche MD-PhD training grant (DOA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.DeLeo FR, Chambers HF. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest. 2009;119:2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maclayton DO, Hall RG., 2nd Pharmacologic treatment options for nosocomial pneumonia involving methicillin-resistant Staphylococcus aureus. Ann Pharmacother. 2007;41:235–244. doi: 10.1345/aph.1H414. [DOI] [PubMed] [Google Scholar]
- 3.Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev. 2010;23:99–139. doi: 10.1128/CMR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perichon B, Courvalin P. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:4580–4587. doi: 10.1128/AAC.00346-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu W, Murray PR, Huskins WC, Jernigan JA, McDonald LC, et al. Dissemination of an Enterococcus Inc18-Like vanA Plasmid, Associated with Vancomycin-Resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2010 doi: 10.1128/AAC.00185-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiramatsu K. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect Dis. 2001;1:147–155. doi: 10.1016/S1473-3099(01)00091-3. [DOI] [PubMed] [Google Scholar]
- 7.Berger-Bachi B, McCallum N. State of the knowledge of bacterial resistance. Injury. 2006;37(Suppl 2):S20–25. doi: 10.1016/j.injury.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Tenover FC, Sinner SW, Segal RE, Huang V, Alexandre SS, et al. Characterisation of a Staphylococcus aureus strain with progressive loss of susceptibility to vancomycin and daptomycin during therapy. Int J Antimicrob Agents. 2009;33:564–568. doi: 10.1016/j.ijantimicag.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lodise TP, Graves J, Evans A, Graffunder E, Helmecke M, et al. Relationship between vancomycin MIC and failure among patients with methicillin-resistant Staphylococcus aureus bacteremia treated with vancomycin. Antimicrob Agents Chemother. 2008;52:3315–3320. doi: 10.1128/AAC.00113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tenover FC, Moellering RC., Jr The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin Infect Dis. 2007;44:1208–1215. doi: 10.1086/513203. [DOI] [PubMed] [Google Scholar]
- 11.Cui L, Ma X, Sato K, Okuma K, Tenover FC, et al. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J Clin Microbiol. 2003;41:5–14. doi: 10.1128/JCM.41.1.5-14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui L, Lian JQ, Neoh HM, Reyes E, Hiramatsu K. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2005;49:3404–3413. doi: 10.1128/AAC.49.8.3404-3413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howden BP, Johnson PD, Ward PB, Stinear TP, Davies JK. Isolates with low-level vancomycin resistance associated with persistent methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob Agents Chemother. 2006;50:3039–3047. doi: 10.1128/AAC.00422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sieradzki K, Tomasz A. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus. J Bacteriol. 2003;185:7103–7110. doi: 10.1128/JB.185.24.7103-7110.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maki H, McCallum N, Bischoff M, Wada A, Berger-Bachi B. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2004;48:1953–1959. doi: 10.1128/AAC.48.6.1953-1959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neoh HM, Cui L, Yuzawa H, Takeuchi F, Matsuo M, et al. Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob Agents Chemother. 2008;52:45–53. doi: 10.1128/AAC.00534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mwangi MM, Wu SW, Zhou Y, Sieradzki K, de Lencastre H, et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci U S A. 2007;104:9451–9456. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda M, Kuwahara-Arai K, Hiramatsu K. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem Biophys Res Commun. 2000;269:485–490. doi: 10.1006/bbrc.2000.2277. [DOI] [PubMed] [Google Scholar]
- 19.Meehl M, Herbert S, Gotz F, Cheung A. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51:2679–2689. doi: 10.1128/AAC.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui L, Neoh HM, Shoji M, Hiramatsu K. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:1231–1234. doi: 10.1128/AAC.01173-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renzoni A, Kelley WL, Barras C, Monod A, Huggler E, et al. Identification by genomic and genetic analysis of two new genes playing a key role in intermediate glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:903–911. doi: 10.1128/AAC.01287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garg SK, Kommineni S, Henslee L, Zhang Y, Zuber P. The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J Bacteriol. 2009;191:1268–1277. doi: 10.1128/JB.01289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beltramini AM, Mukhopadhyay CD, Pancholi V. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect Immun. 2009;77:1406–1416. doi: 10.1128/IAI.01499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, et al. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol. 2003;49:807–821. doi: 10.1046/j.1365-2958.2003.03599.x. [DOI] [PubMed] [Google Scholar]
- 25.Ohlsen K, Donat S. The impact of serine/threonine phosphorylation in Staphylococcus aureus. Int J Med Microbiol. 2010;300:137–141. doi: 10.1016/j.ijmm.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 26.Nakano S, Nakano MM, Zhang Y, Leelakriangsak M, Zuber P. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc Natl Acad Sci U S A. 2003;100:4233–4238. doi: 10.1073/pnas.0637648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pamp SJ, Frees D, Engelmann S, Hecker M, Ingmer H. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J Bacteriol. 2006;188:4861–4870. doi: 10.1128/JB.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 29.Utaida S, Dunman PM, Macapagal D, Murphy E, Projan SJ, et al. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology. 2003;149:2719–2732. doi: 10.1099/mic.0.26426-0. [DOI] [PubMed] [Google Scholar]
- 30.McCallum N, Spehar G, Bischoff M, Berger-Bachi B. Strain dependence of the cell wall-damage induced stimulon in Staphylococcus aureus. Biochim Biophys Acta. 2006;1760:1475–1481. doi: 10.1016/j.bbagen.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 31.You C, Sekowska A, Francetic O, Martin-Verstraete I, Wang Y, et al. Spx mediates oxidative stress regulation of the methionine sulfoxide reductases operon in Bacillus subtilis. BMC Microbiol. 2008;8:128. doi: 10.1186/1471-2180-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galbusera E, Renzoni A, Andrey DO, Monod A, Barras C, et al. Site-specific mutation of Staphylococcus aureus VraS reveals a crucial role for the VraR-VraS sensor in the emergence of glycopeptide resistance. Antimicrob Agents Chemother. 2010 doi: 10.1128/AAC.00720-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardete S, Wu SW, Gill S, Tomasz A. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:3424–3434. doi: 10.1128/AAC.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Debarbouille M, Dramsi S, Dussurget O, Nahori MA, Vaganay E, et al. Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J Bacteriol. 2009;191:4070–4081. doi: 10.1128/JB.01813-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donat S, Streker K, Schirmeister T, Rakette S, Stehle T, et al. Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J Bacteriol. 2009;191:4056–4069. doi: 10.1128/JB.00117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin S, Daum RS, Boyle-Vavra S. VraSR two-component regulatory system and its role in induction of pbp2 and vraSR expression by cell wall antimicrobials in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:336–343. doi: 10.1128/AAC.50.1.336-343.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katayama Y, Murakami-Kuroda H, Cui L, Hiramatsu K. Selection of heterogeneous vancomycin-intermediate Staphylococcus aureus by imipenem. Antimicrob Agents Chemother. 2009;53:3190–3196. doi: 10.1128/AAC.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kato Y, Suzuki T, Ida T, Maebashi K. Genetic changes associated with glycopeptide resistance in Staphylococcus aureus: predominance of amino acid substitutions in YvqF/VraSR. J Antimicrob Chemother. 2010;65:37–45. doi: 10.1093/jac/dkp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCallum N, Meier PS, Heusser R, Berger-Bachi B. Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:1391–1402. doi: 10.1128/AAC.01213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renzoni A, Barras C, Francois P, Charbonnier Y, Huggler E, et al. Transcriptomic and functional analysis of an autolysis-deficient, teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:3048–3061. doi: 10.1128/AAC.00113-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howden BP, Smith DJ, Mansell A, Johnson PD, Ward PB, et al. Different bacterial gene expression patterns and attenuated host immune responses are associated with the evolution of low-level vancomycin resistance during persistent methicillin-resistant Staphylococcus aureus bacteraemia. BMC Microbiol. 2008;8:39. doi: 10.1186/1471-2180-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamber S, Schwartzman J, Cheung AL. Role of PknB kinase in antibiotic resistance and virulence in community-acquired methicillin-resistant Staphylococcus aureus strain USA300. Infect Immun. 2010;78:3637–3646. doi: 10.1128/IAI.00296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burnside K, Lembo A, de Los Reyes M, Iliuk A, Binhtran NT, et al. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS One. 2010;5:e11071. doi: 10.1371/journal.pone.0011071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paracuellos P, Ballandras A, Robert X, Kahn R, Herve M, et al. The extended conformation of the 2.9-A crystal structure of the three-PASTA domain of a Ser/Thr kinase from the human pathogen Staphylococcus aureus. J Mol Biol. 2010;404:847–858. doi: 10.1016/j.jmb.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Chen PR, Nishida S, Poor CB, Cheng A, Bae T, et al. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol Microbiol. 2009;71:198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liebeke M, Meyer H, Donat S, Ohlsen K, Lalk M. A metabolomic view of Staphylococcus aureus and its ser/thr kinase and phosphatase deletion mutants: involvement in cell wall biosynthesis. Chem Biol. 2010;17:820–830. doi: 10.1016/j.chembiol.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 47.Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dwyer DJ, Kohanski MA, Collins JJ. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol. 2009;12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyle-Vavra S, Carey RB, Daum RS. Development of vancomycin and lysostaphin resistance in a methicillin-resistant Staphylococcus aureus isolate. J Antimicrob Chemother. 2001;48:617–625. doi: 10.1093/jac/48.5.617. [DOI] [PubMed] [Google Scholar]
- 50.Pfeltz RF, Singh VK, Schmidt JL, Batten MA, Baranyk CS, et al. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob Agents Chemother. 2000;44:294–303. doi: 10.1128/aac.44.2.294-303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrey DO, Renzoni A, Monod A, Lew DP, Cheung AL, et al. Control of the Staphylococcus aureus toxic shock tst promoter by the global regulator SarA. J Bacteriol. 2010;192:6077–6085. doi: 10.1128/JB.00146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tu Quoc PH, Genevaux P, Pajunen M, Savilahti H, Georgopoulos C, et al. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect Immun. 2007;75:1079–1088. doi: 10.1128/IAI.01143-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaudaux P, Huggler E, Bernard L, Ferry T, Renzoni A, et al. Underestimation of vancomycin and teicoplanin MICs by broth microdilution leads to underdetection of glycopeptide-intermediate isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 2010;54:3861–3870. doi: 10.1128/AAC.00269-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaudaux P, Francois P, Bisognano C, Kelley WL, Lew DP, et al. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect Immun. 2002;70:5428–5437. doi: 10.1128/IAI.70.10.5428-5437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 56.Stahl ML, Pattee PA. Confirmation of protoplast fusion-derived linkages in Staphylococcus aureus by transformation with protoplast DNA. J Bacteriol. 1983;154:406–412. doi: 10.1128/jb.154.1.406-412.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frees D, Qazi SN, Hill PJ, Ingmer H. Alternative roles of ClpX and ClpP in Staphylococcuss aureus stress tolerance and virulence. Mol Microbiol. 2003;48:1565–1578. doi: 10.1046/j.1365-2958.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- 58.Sullivan MA, Yasbin RE, Young FE. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- 59.Bruckner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complementation of either yjbH (K23stop), stp1 (Q12stop) or vraS (G45R) mutants by multicopy plasmid carrying wild-type genes. (A) Spot plating population analysis of each single mutant and its corresponding complemented strain on teicoplanin. V* and s* complementation was tested on teicoplanin containing 0.5 µg/ml. Spot serial dilutions are indicated at the right margin. As yjbH(23stop) mutation has a subtle effect on teicoplanin resistance, complementation of the yjbH mutation was performed by introducing a multicopy plasmid carrying wild-type yjbH gene into the triple strain mutant, a condition where its role could be unambiguously assessed. Y* complementation was tested on teicoplanin containing 2 µg/ml. For convenience genes marked with an asterisk such as v*, s* and y* correspond to vraS(G45R), stp1(Q12stop) or yjbH(K23stop) mutations, respectively.
(TIF)
Primers used in this study.
(DOC)