Abstract
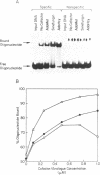
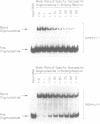
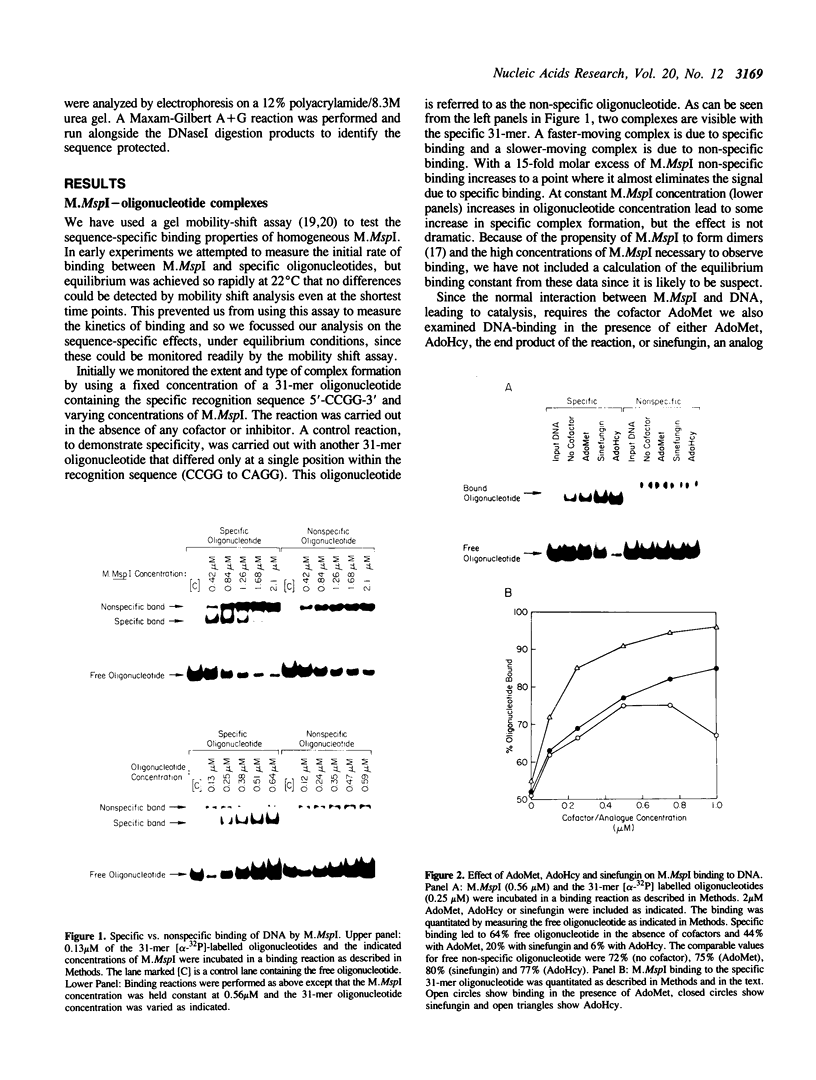
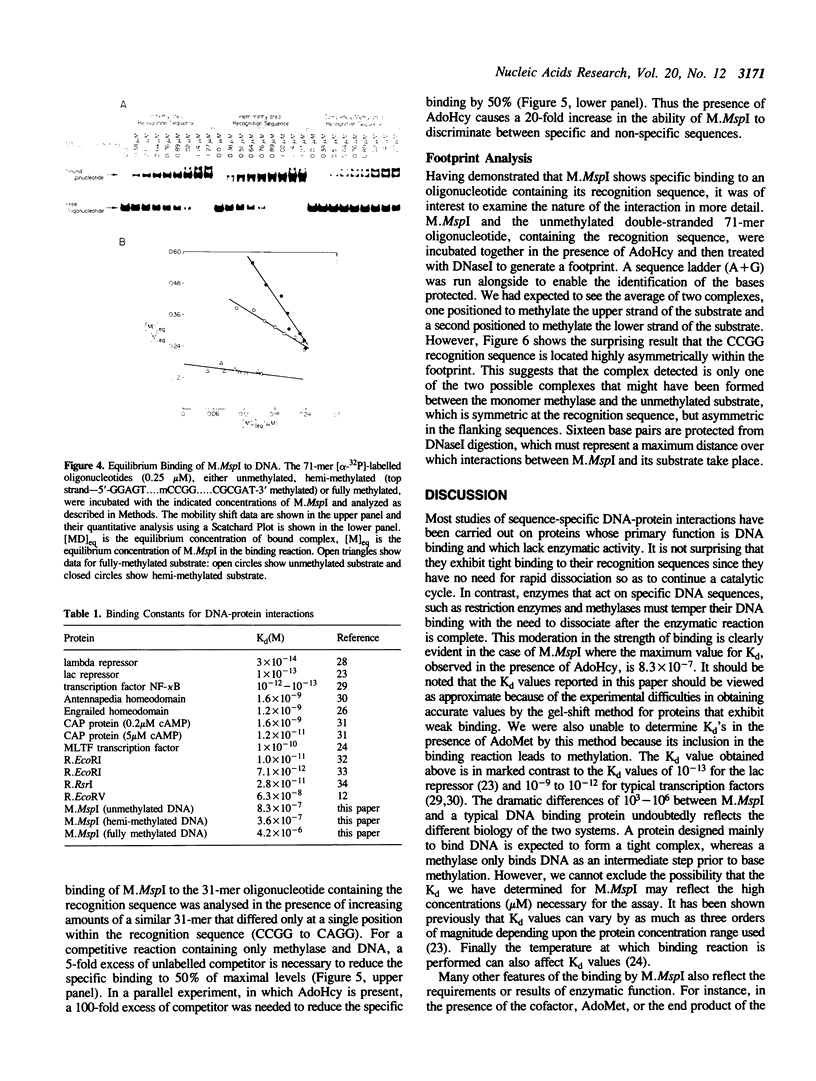
The MspI methyltransferase (M.MspI) recognizes the sequence CCGG and catalyzes the formation of 5-methylcytosine at the fist C-residue. We have investigated the sequence-specific DNA-binding properties of M.MspI under equilibrium conditions, using gel-mobility shift assays and DNasel footprinting. M.MspI binds to DNA in a sequence-specific manner either alone or in the presence of the normal methyl donor S-adenosyl-L-methionine as well as the analogues, sinefungin and S-adenosyl-L-homocysteine. In the presence of S-adenosyl-L-homocysteine, M.MspI shows the highest binding affinity to DNA containing a hemimethylated recognition sequence (Kd = 3.6 x 10(-7) M), but binds less well to unmethylated DNA (Kd = 8.3 x 10(-7) M). Surprisingly it shows specific, although poor, binding to fully methylated DNA (Kd = 4.2 x 10(-6) M). M.MspI binds approximately 5-fold more tightly to DNA containing its recognition sequence, CCGG, than to nonspecific sequences in the absence of cofactors. In the presence of S-adenosyl-L-methionine, S-adenosyl-L-homocysteine or sinefungin the discrimination between specific and non-specific sequences increases up to 100-fold. DNasel footprinting studies indicate that 16 base pairs of DNA are covered by M.MspI, with the recognition sequence CCGG located asymmetrically within the footprint.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Affolter M., Percival-Smith A., Müller M., Leupin W., Gehring W. J. DNA binding properties of the purified Antennapedia homeodomain. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4093–4097. doi: 10.1073/pnas.87.11.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiken C. R., Fisher E. W., Gumport R. I. The specific binding, bending, and unwinding of DNA by RsrI endonuclease, an isoschizomer of EcoRI endonuclease. J Biol Chem. 1991 Oct 5;266(28):19063–19069. [PubMed] [Google Scholar]
- Becker M. M., Lesser D., Kurpiewski M., Baranger A., Jen-Jacobson L. "Ultraviolet footprinting" accurately maps sequence-specific contacts and DNA kinking in the EcoRI endonuclease-DNA complex. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6247–6251. doi: 10.1073/pnas.85.17.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett S. P., Halford S. E. Recognition of DNA by type II restriction enzymes. Curr Top Cell Regul. 1989;30:57–104. doi: 10.1016/b978-0-12-152830-0.50005-0. [DOI] [PubMed] [Google Scholar]
- Butkus V., Petrauskiene L., Maneliene Z., Klimasauskas S., Laucys V., Janulaitis A. Cleavage of methylated CCCGGG sequences containing either N4-methylcytosine or 5-methylcytosine with MspI, HpaII, SmaI, XmaI and Cfr9I restriction endonucleases. Nucleic Acids Res. 1987 Sep 11;15(17):7091–7102. doi: 10.1093/nar/15.17.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh L. A., Carthew R. W., Sharp P. A. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol Cell Biol. 1986 Dec;6(12):4723–4733. doi: 10.1128/mcb.6.12.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R. DNAase I footprinting of restriction enzymes. Biochem Biophys Res Commun. 1988 Sep 15;155(2):779–785. doi: 10.1016/s0006-291x(88)80563-1. [DOI] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. Equilibrium studies of the cyclic AMP receptor protein-DNA interaction. J Mol Biol. 1984 Jan 25;172(3):241–262. doi: 10.1016/s0022-2836(84)80025-x. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S. Binding of the EcoRII methylase to azacytosine-containing DNA. Nucleic Acids Res. 1986 Jun 11;14(11):4543–4556. doi: 10.1093/nar/14.11.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Jentsch S., Freund M. Restriction and modification in Bacillus subtilis: two DNA methyltransferases with BsuRI specificity. II. Catalytic properties, substrate specificity, and mode of action. J Biol Chem. 1981 Sep 10;256(17):9346–9351. [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Jen-Jacobson L., Kurpiewski M., Lesser D., Grable J., Boyer H. W., Rosenberg J. M., Greene P. J. Coordinate ion pair formation between EcoRI endonuclease and DNA. J Biol Chem. 1983 Dec 10;258(23):14638–14646. [PubMed] [Google Scholar]
- King K., Benkovic S. J., Modrich P. Glu-111 is required for activation of the DNA cleavage center of EcoRI endonuclease. J Biol Chem. 1989 Jul 15;264(20):11807–11815. [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Nelson J. L., Roberts R. J. The sequence specificity domain of cytosine-C5 methylases. Nucleic Acids Res. 1991 Nov 25;19(22):6183–6190. doi: 10.1093/nar/19.22.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauster R., Trautner T. A., Noyer-Weidner M. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J Mol Biol. 1989 Mar 20;206(2):305–312. doi: 10.1016/0022-2836(89)90480-4. [DOI] [PubMed] [Google Scholar]
- Lin P. M., Lee C. H., Roberts R. J. Cloning and characterization of the genes encoding the MspI restriction modification system. Nucleic Acids Res. 1989 Apr 25;17(8):3001–3011. doi: 10.1093/nar/17.8.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman T. M. Kinetics of protein-nucleic acid interactions: use of salt effects to probe mechanisms of interaction. CRC Crit Rev Biochem. 1986;19(3):191–245. doi: 10.3109/10409238609084656. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renbaum P., Abrahamove D., Fainsod A., Wilson G. G., Rottem S., Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res. 1990 Mar 11;18(5):1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Macelis D. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2077–2109. doi: 10.1093/nar/19.suppl.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin R. A., Modrich P. EcoRI methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1977 Oct 25;252(20):7265–7272. [PubMed] [Google Scholar]
- Taylor J. D., Badcoe I. G., Clarke A. R., Halford S. E. EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry. 1991 Sep 10;30(36):8743–8753. doi: 10.1021/bi00100a005. [DOI] [PubMed] [Google Scholar]
- Terry B. J., Jack W. E., Rubin R. A., Modrich P. Thermodynamic parameters governing interaction of EcoRI endonuclease with specific and nonspecific DNA sequences. J Biol Chem. 1983 Aug 25;258(16):9820–9825. [PubMed] [Google Scholar]
- Vovis G. F., Horiuchi K., Zinder N. D. Kinetics of methylation of DNA by a restriction endonuclease from Escherichia coli B. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3810–3813. doi: 10.1073/pnas.71.10.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- Wright D. J., King K., Modrich P. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J Biol Chem. 1989 Jul 15;264(20):11816–11821. [PubMed] [Google Scholar]
- Zabel U., Schreck R., Baeuerle P. A. DNA binding of purified transcription factor NF-kappa B. Affinity, specificity, Zn2+ dependence, and differential half-site recognition. J Biol Chem. 1991 Jan 5;266(1):252–260. [PubMed] [Google Scholar]