Abstract
Genomic DNA in the eukaryotic nucleus is hierarchically packaged by histones into chromatin to fit inside the nucleus. The dynamics of higher-order chromatin compaction play a critical role in transcription and other biological processes inherent to DNA. Many factors, including histone variants, histone modifications, DNA methylation and the binding of non-histone architectural proteins regulate the structure of chromatin. Although the structure of nucleosomes, the fundamental repeating unit of chromatin, is clear, there is still much discussion on the higher-order levels of chromatin structure. In this review, we focus on the recent progress in elucidating the structure of the 30-nm chromatin fiber. We also discuss the structural plasticity/dynamics and epigenetic inheritance of higher-order chromatin and the roles of chromatin higher-order organization in eukaryotic gene regulation.
Genomic DNA in the eukaryotic nucleus is hierarchically packaged by histones into chromatin. The fundamental repeating unit of chromatin is the nucleosome that comprises 146 base pairs of DNA wrapped in 1.7 superhelical turns around an octamer of histone proteins [1]. The nucleosomal array, a “beads-on-a-string” fiber with a diameter of 11-nm, represents the first level of chromatin organization [1]. The binding of the linker histone (H1 or H5) organizes the nucleosome arrays into a more condensed 30-nm chromatin fiber which is typically posited as being the second structural level of DNA organization [2]. While folding of chromatin fiber to the next higher layer is much more obscure, it has been clear that the plasticity of and the dynamics of higher-order chromatin compaction are key regulators of transcription and other biological processes inherent to DNA. In this review, we focus on the recent progress in elucidating the structure of the 30-nm chromatin fiber. We also discuss the structural plasticity/dynamics and epigenetic inheritance of higher-order chromatin and the roles of chromatin higher-order organization in eukaryotic gene regulation.
Nucleosome dynamics and gene regulation
During DNA replication and gene transcription, DNA needs to be accessed by DNA binding factors at appropriate regions and at appropriate times, however the compaction of DNA into chromatin makes it refractory to such factors. DNA methylation, core histone posttranslational modifications and histone variants have been shown to affect the intrinsic properties of nucleosomes, and thus have important functions in regulating nucleosome dynamics [3]. Using single molecular fluorescence resonance energy transfer (FRET), DNA methylation was found to result in a closed and rigid nucleosomal structure, changing the dynamics of mono-nucleosomes [4•]. FRET analyses of mononucleosomes in vitro show that the histone variant H2A.Z stabilizes the histone octamer within the nucleosome and prevents eviction of the histone H2A.Z-H2B dimer [5,6]. Interestingly, Felsenfeld and colleagues have shown that histone H3.3 can modulate the effect of H2A.Z on nucleosome stability in vivo. Nucleosomes containing histones H3.3 and H2A.Z exhibit a much more unstable structure and higher turnover rates compared to those containing H3.1 and H2A.Z histones or H3.3 and canonical H2A histones [7]. However, biochemical and biophysical analyses in vitro demonstrated that recombinant histone variants H2A.Z and H3.3, (either one or in combination), display only subtle effects on the compaction and stability of the nucleosome particle [8]. This result suggests that the posttranslational modifications of H2A.Z and/or H3.3 may be responsible for H2A.Z-mediated decreases in nucleosome stability as observed in vivo. Histone H2A.Bbd and macroH2A represent two unusual histone variants. Both H2A.Bbd and macroH2A display intrinsic and extrinsic effects on nucleosome stability and dynamics, thereby impacting gene regulation through their unusual structural and functional properties [9–13].
30-nm chromatin fiber: solenoid vs Zig-Zag
The X-ray crystal structure of the nucleosome core particles has been resolved at high resolution, showing the precise path of the 146 bp of DNA and the localization of individual core histones [1]. Despite considerable efforts during the last three decades however, the arrangement of chromatin in higher-order structures remains largely uncharacterized. Early electron microscopy (EM) studies of native chromatin fibers have led to the proposal of two models for the 30-nm fiber: (i) the one-start helix/solenoid model, in which adjacent nucleosomes are connected by linker DNA bent between them to follow the superhelical path, with about 6 to 8 nucleosomes per turn [14], and (ii) the two-start helix model, in which adjacent nucleosomes are connected by straight linker DNA, based on the Zig-Zag arrangement of nucleosomes observed by EM in the case of chromatin in low ionic strength buffers [15,16].
The study of chromatin isolated from nuclei has an advantage in that it presumably represents the “native” state. However, native chromatin is very heterogeneous, reflective of its different DNA sequences, different histone compositions and their modifications, and irregular nucleosome spacing [17]. In an attempt to reduce the effect of these variables, scientists have recently developed an in vitro well-defined reconstituted nucleosomal array system that incorporates a strong “widom 601 nucleosome positioning” sequence [18,19]. Using this system, Richmond and colleagues proposed a two-start crossed-linker model for 30-nm chromatin fiber based on EM imaging and the crystal structure of a tetranucleosome core array lacking the linker histone [18,20]; (Figure 1). Their proposed idealized model is a twisted ribbon with a diameter of about 25 nm and a compaction density of 5–6 nucleosomes per 11 nm. However, the structure was determined for a tetranucleosome array that is too short to form a solenoid structure. Furthermore, their study employed a very short nucleosome repeat length (167 bp) and a high concentration of divalent cation Mg2+ (120 mM), which is uncommon in nature.
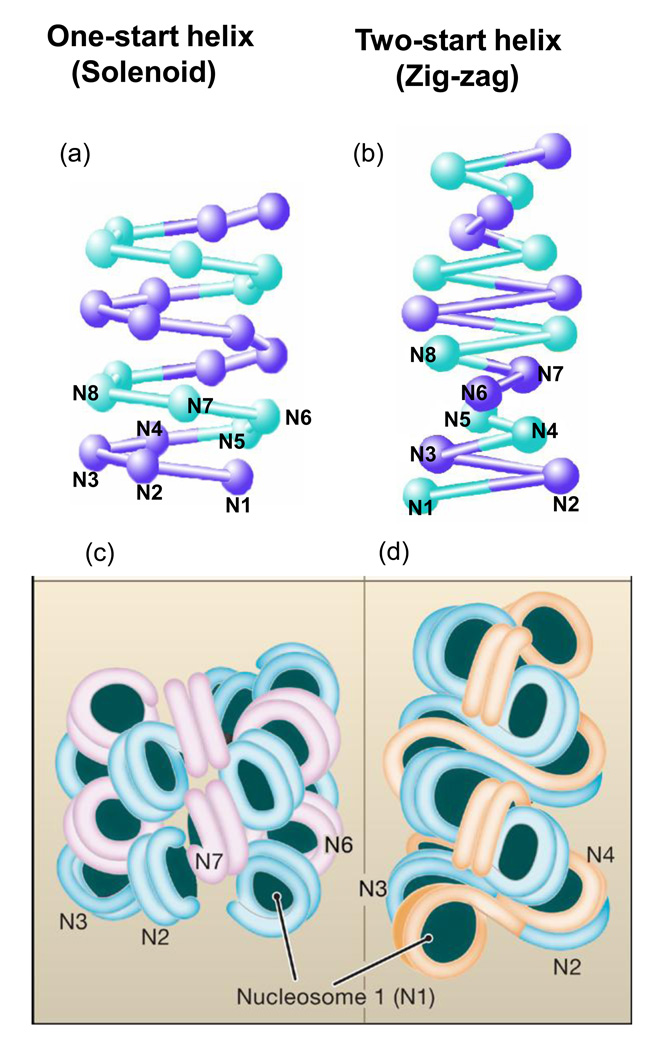
Figure 1. Models of the 30 nm Chromatin Fiber.
Two well-known structural models for 30-nm chromatin fibers are proposed: one-start helix (solenoid) (a and c) and two-start helix (zig-zag) (b and d). At the top, a schematic representation is shown for the two different topologies of 30-nm chromatin fibers (a and b). The alternative nucleosomes are numbered from N1 to N8. In the solenoid model proposed by Rhodes and colleagues, the 30-nm chromatin fiber is an interdigitated one-start helix in which a nucleosome in the fiber interacts with its fifth and sixth neighbor nucleosomes [19]. Alternative helical gyres are colored blue and magenta (c). In the zigzag model suggested by Richmond and colleagues, the chromatin fiber is a two-start helix in which nucleosomes are arranged in a zig-zag manner such that a nucleoosme in the fiber binds to the second neighbor nucleosome[18,20]. Alternative nucleosomes pairs are colored blue and orange. Image courtesy of D. Rhodes.
In another recent study, Rhodes and colleagues analyzed the structure of long and regular chromatin fibers with a range of nucleosomal repeats (from 177 to 237 bp) by EM and cryo-EM under various solution conditions. Although the detailed structure could not be resolved, the dimensions measured allowed them to propose a one-start interdigitated solenoid structure [19] (Figure 1). In this case, the chromatin fibers were analyzed under more native conditions including the presence of linker histone, a wide range of linker DNA length found in nature, and low divalent salt concentration (1.6 mM Mg++). Recent studies using reconstituted short nucleosome arrays have concluded that the linker histone is dispensable for chromatin compaction under high concentrations of Mg++ [18,20,21]. However, Rhodes and colleagues found that both the linker histone and the nucleosome repeat length (NRL) determine chromatin higher-order structure and that in the absence of linker histone, the 167-bp NRL array displays a highly ordered “ladder”-like structure consisting of the two-start helix arrangement of stacked nucleosomes [22]. This nucleosome arrangement is reminiscent of that previously observed by Richmond and colleagues in the EM analysis of the cross-linked 167-bp NRL nucleosome array [18].
Chromatin structures beyond 30-nm fiber
Inter-fiber nucleosome-nucleosome interactions
As summarized in Figure 2, Mg2+-dependent intra-fiber compaction and inter-fiber oligomerization of nucleosomal arrays is thought to recapitulate essential aspects of higher-order folding transitions that stabilize chromosome structure beyond the 30-nm chromatin fiber, and mimics fiber-fiber interaction observed in native chromatin. The compaction of nucleosome arrays into chromatin structures involves the inter-nucleosome interactions mediated by the core histone tail domains and their modifications [23–25]. Numerous studies have revealed that it is the inter-nucleosome interactions within a nucleosomal array that primarily contribute to the compaction of model nucleosome arrays into 30-nm fiber [26], while the long-range inter-array interactions are involved in the formation of tertiary chromatin structure via self-association [27].
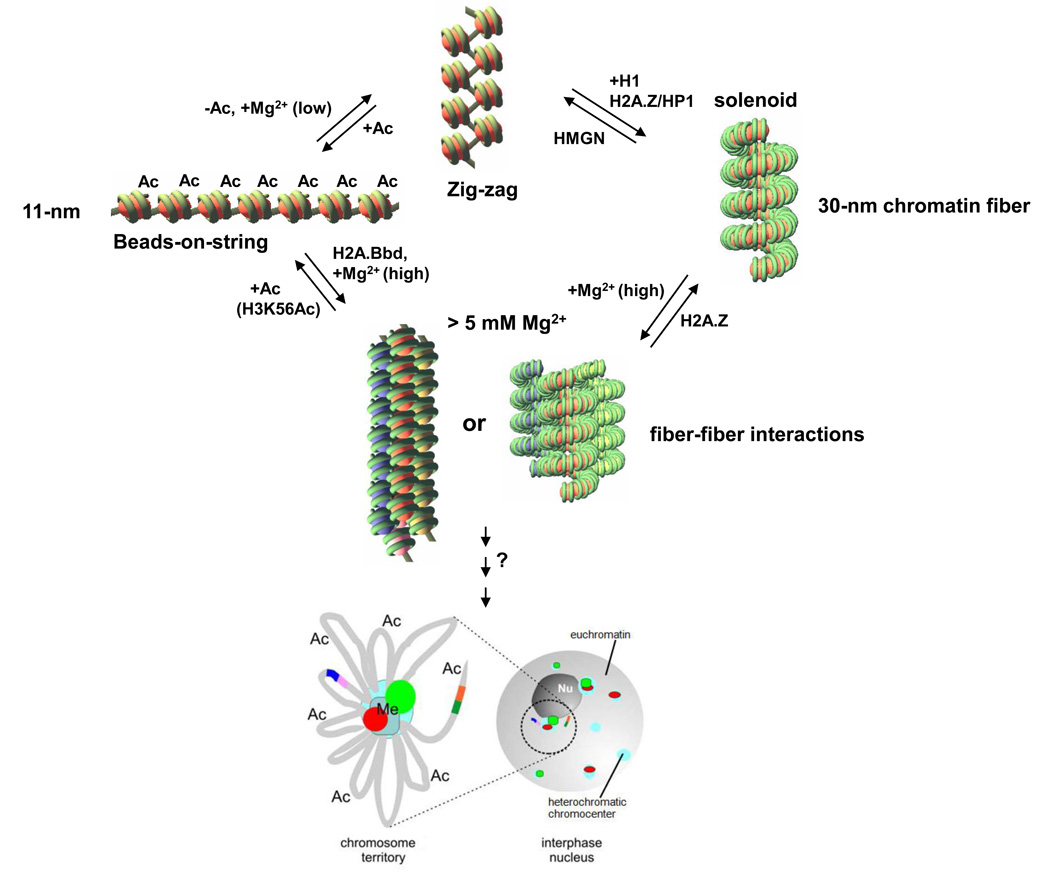
Figure 2. Hierarchical folding and Plasticity of higher-order chromatin structures.
A general scheme represent the folding of chromatin from 11-nm nucleosomal arrays (beads-on-string) to higher order chromatin structures including 30-nm chromatin fiber, fiber-fiber association, chromatin looping and positioning. Factors proposed to affect changes between different chromatin structures are shown.
According to the crystal contacts between nucleosomes observed in the first high resolution structure of the nucleosome, it was proposed that the histone H4 tail of one nucleosome can directly interact with the acidic patch interface of H2A and H2B in adjacent nucleosomes [1]. Indeed, subsequent biochemical analysis and cross-linking experiments validated that the histone H4 tail has the most pronounced effect on both inter-nucleosome and inter-array association [25,28,29], in a manner dependent on the charge [30], modification states [21,31,32], and position of the H4 tail [32]. It was proposed that oligomerization may compete with fiber folding, as promotion of fiber folding by histone variant H2A.Z inhibits inter-fiber associations [28,33] (Figure 2). If less folding does indeed favor oligomerization, then H2A.Bbd arrays would be the best substrate for the oligomerization process as these arrays essentially cannot fold. Consistent with this, H2A.Bbd arrays do oligomerize more efficiently than H2A arrays [34] (Figure 2). Of note, Dubochet and colleagues have observed a relaxed chromatin structure rather than compact 30-nm chromatin fibers in condensed mitotic chromosome. They proposed that these flexible nucleosomal fibers may compact into the 30-nm chromatin fibers through intra-array nucleosomal interactions under diluted conditions (interphase), and that an increase in chromatin concentration during chromosome condensation (mitosis) results in inter-array nucleosomal associations that interfere with the intra-array nucleosomal contacts [35].
Long-range chromatin interactions and chromatin looping
The transcription of genes in higher eukaryotes is frequently regulated by communication between cis-regulatory DNA elements such as promoters and enhancers, which can be located hundreds of kilobases away from each other along the linear chromatin fiber in vivo (Figure 2). How these enhancers communicate with the promoters of the genes being regulated is largely unknown. With the recently developed techniques, such as FISH (Fluorescence in situ Hybridization) and 3C (Chromosome Conformation Capture) and its derivatives, long-range chromatin interactions (chromatin loops) were analyzed in a wide variety of gene loci [36].
Two factors were found to promote the formation and maintenance of chromatin loops in the β-globin locus, Erythroid Kruppel-like factor (EKLF) and GATA-1. These factors function in the recruitment of the BRG1 catalytic subunit of SWI/SNF-related complexes, to both the far-upstream major-regulatory element (MRE) and the alpha2 promoter in murine fetal liver cells [37]. Interestingly, chromatin looping between nuclear receptor response elements and activated promoters was shown to occur in a dynamic, cyclical manner [38]. The dynamics of chromatin looping and transcriptional activation induced by ligand is regulated by various factors, such as mediator protein MED1, histone deacetylation (HDAC4) and histone demetylation (LSD1) [38]. Mediator and cohesin cooperatively facilitate the formation of chromatin loops between the enhancers and core promoters of active genes in murine embryonic stem (ES) cells [39]. Apart from chromatin looping between enhancer and promoter, a gene looping mechanism involving promoter and terminator sequences was also observed in the yeast genes FMP27 and SEN1 [40] or BUD3 and SEN1 genes [41], respectively. Recently, Proudfoot and colleagues further demonstrated that chromatin loops are formed on BRCA1 between the promoter and terminator region in both human cell lines and mouse mammary tissue, and are highly dynamic phenomena that occur upon transcription activatation by estrogen stimulation and during lactational development [42].
It has been proposed that PcG-mediated long-range chromosomal interactions provide a mechanism for epigenetic memory of the PcG-dependent repressed state. Verrijzer and colleagues found that Ezh2-dependent chromatin looping regulates INK4a and INK4b during human progenitor cell differentiation and cellular senescence [43]. In addition, Baylin and colleagues demonstrated that in undifferentiated embryonic carcinoma (EC) Tera-2 cells, the upstream region of the GATA-4 promoter exhibits a complex chromatin organization comprising multi-chromatin loop conformations. These species are composed of multiple internal long-range interactions that are mediated by PcG proteins including Ezh2 and the histone mark it catalyzes, H3K27me3 [44].
The affect of gene positioning
Accumulating evidence suggest that the spatial organization of chromosomes within the nucleus has an essential regulatory function on transcriptional activity. The interphase nucleus chromosomes and individual genes have been found to occupy specific locations relative to one another and to landmarks within the cell nucleus (Figure 2). The nuclear lamina, a filamentous protein network that provides a structural scaffold for the inner nuclear membrane, has been shown to dynamically interact with specific chromatin domains and regulate gene expression. Early EM and recent high-resolution light microscopy images of cell nuclei revealed that the nuclear lamina connects with highly compacted heterochromatin [45], while the nuclear pore complexes are associated with structurally open, active euchromatin. In addition, recent genome-wide DamID mapping studies have identified large genomic domains with distinct epigenetic signatures that are associated with the nuclear lamina in fly and mammalian cells [46,47, respectively]. Moreover, these lamina-associated domains (LADs) are shown to be at low gene-density and associate with some specific epigenetic signatures, marked by specific sequence elements. The majority of those genes within LADs exhibit very low expression levels [47]. More interestingly, molecular mapping indicated that lamina-genome interactions are dynamic and play essential roles in the regulation of gene expression programs during lineage commitment and terminal differentiation [48•].
Recent studies demonstrated that nuclear pore complexes might also contribute to gene regulation via interacting with specific genomic loci. For example, studies in yeast and Drosophila reveal that the nuclear pore complex might facilitate transcription by recruiting chromatin to the nuclear periphery [49]. Genome-wide mapping shows that Nup153 and Megator (Mtor) bind to hundreds of large genomic domains extending from 10 kb to 500 kb in Drosophila melanogaster [38,50–51,52•]. In contrast to LADs, these Nucleoporin-Associated Regions (NARs) correlate with markers of transcriptional activity, including high levels of RNA polymerase II and histone H4K16 acetylation. Interestingly, two other recent studies demonstrated that a subset of nucleoporins freely diffuses and binds to genes located inside of the nucleoplasm, facilitating transcriptional activation of genes involved in regulating development and the cell cycle [51,52•]. Proximity to the nuclear envelope in yeast [53] and association with the mammalian nuclear lamina [54•] promotes transcription silencing of some genes. However, recent work shows that peripheral localization is not always repressive. A number of inducible yeast genes were found to be targeted to the nuclear periphery upon transcriptional activation [55]. Intriguingly, these genes can “remember” their localization at the nuclear periphery after being silenced, and even after multiple cell generations. More importantly, the localization of GAL1 and INO1 at the nuclear periphery plays a critical role in the rapid reactivation of previously expressed genes even after multiple generations [55].
Plasticity/variation of higher-order chromatin structure
Controlling the degree of higher-order chromatin folding is a key element in partitioning the metazoan genome into functionally distinct chromosomal domains. However, the mechanisms of this fundamental process are poorly understood. Elucidating just how a nucleosomal array can be compacted into higher-order chromatin structures is central to understanding how the dynamics of chromatin structure functionally translates into the regulation of gene expression. It has long been known that core histone modifications and histone variants play important roles in modulating higher-order chromatin structures and in regulating transcription. For example, acetylation of histones prevents chromatin from folding into the 30-nm fiber and reduces the ability of chromatin fiber to self-assemble into higher-order structures (Figure 2). Peterson and colleagues reported that acetylation of histone H4 on lysine 16 (H4K16ac), a mark with a functional role in transcription activation, can inhibit higher-order chromatin folding induced by Mg2+, in the absence of histone H1 [21]. The acetylation of H3K56 has no effect on the compaction of model nucleosomal arrays in cis [56•,57], whereas it does inhibit the oligomerization of sub-saturated nucleosomal arrays that contain nucleosome-free regions and thus plays an important role in keeping chromatin with nucleosome-free regions accessible to the DNA replication and repair machinery [57].
Among the histone variants, H2A.Z promotes formation of the higher-order chromatin fiber in a manner dependent upon just two amino acid residues, which subtly extend the acidic patch of H2A.Z compared to that of H2A, and cooperate with heterochromatin protein HP1α to establish or maintain a specialized conformation at constitutive heterochromatic domains [33] (Figure 2). H3.3 was generally regarded as an active histone mark in that its presence correlated with gene activation. However, a few recent studies showed that H3.3 was also involved in heterochromatin formation in ES cells [58–61]. Allis and colleagues identified DAXX as a novel H3.3 specific histone chaperone that can cooperate with ATRX chromatin remodeling complex to assemble chromatin at telomeres [60•,61]. Interestingly, both K27 and K4 residues of H3.3 are important for formation of heterochromatin in ES cells [58,59]. In addition, both histone variants H2A.Z and H3.3 are enriched at the target genes of Polycomb Repressive Complex 2 (PRC2) in ES cells [60•,62•], but not in differentiated cells. Together, these results suggest that histone variants H2A.Z and H3.3 may function cooperatively with polycomb proteins in the formation of facultative heterochromatin in ES cells. It is interesting to note that histone variant H2A.Z antagonizes DNA methylation along the whole genome in plants and animals [63•,64, 65]. Accordingly, it has been proposed that DNA methylation results in gene silencing by inhibiting H2A.Z deposition, and vice versa, deposition of H2A.Z protects genes from DNA methylation by DNA methyltransferase. Interestingly, DNA methylation at PcG target genes is largely absent in ES cells, whereas the promoters of these PcG targeted genes bear DNA hypermethylation in adult cancer cells [66].
The linker histones have long been proposed as being involved in the formation of higher-order chromatin structure and gene regulation [67,68]. The discrepancy between the two-start Zig-Zag and one-start solenoid models may actually be a consequence of the different levels of chromatin compaction that form as a function of the presence of linker histone H1 (Figure 2). The binding properties of the H1 histones are mainly determined by their long, C-terminal tails that are responsible for substantial differences in the binding affinity of the individual subtypes [69]. FRAP and AFM experiments demonstrated that the H1 subtypes exhibit different affinities for chromatin and different abilities to promote chromatin compaction [70]. A number of novel, modified residues in H1 proteins have been identified, including phosphorylation, acetylation, methylation, and ubiquitination [71,72]. Phosphorylation of H1 histones results in weakening their binding to the nucleosome [69]. However, the affect of acetylation and methylation of histone H1 on higher-order chromatin dynamics it is still largely unknown. Most of the acetylated lysines identified in the globular domain of H1.4 have been considered as being directly involved in DNA binding and chromatin compaction [71]. Deacetylation of the highly conserved lysine-26 (K26) in H1.4 by the histone deactylase SirT1 could be linked to the formation of facultative heterochromatin [73]. SirT1 physically interacts with the PRC2 complex that methylates H1.4 at K26 [74], and this product provides the binding site for HP1 to repress transcription [75]. We, and others, recently reported that different histone H1 subtypes could be methylated by G9a/HMT1c and Glp1/KMT1D at different lysine residues [76,77].
The binding of non-histone architectural proteins with chromatin also plays a critical role in chromatin structural dynamics. The HMG proteins decrease the compactness of the chromatin fiber and enhance the accessibility of chromatin targets to regulatory factors [78] (Figure 2). HMGN5 is a recently identified member of the HMGN family of nucleosome-binding proteins, the members of which contain a functional nucleosome-binding domain (NBD) and a negatively charged C-terminus of various lengths. Bustin and colleagues demonstrated that the negatively charged C-terminal domain of HMGN5 interacts with the positively charged C-terminal domain of linker histone H5, thereby counteracting the linker histone-mediated compaction of a nucleosomal array which in turn facilitates transcriptional activation [79]; this finding is consistent with another recent report that HMGB1 interacts with linker histone H1 [80].
We recently demonstrated that the malignant-brain-tumor (MBT) protein L3MBTL1 compacts nucleosomal arrays dependent on the presence of mono- and dimethylated versions of histone H4 lysine 20 and histone H1b lysine 26, and this results in negatively regulating the expression of a subset of E2F-target genes [81•]. However, recent high-resolution genome-wide analyses revealed that the role of H4K20me1 in transcription might not be straightforward [82,83]. Nonetheless, a number of recent studies demonstrated that the removal of the mono-methyl mark at H4K20 by PHF8 can abolish L3MBTL1 binding to the promoter with resultant chromatin opening for transcription [84,85], further substantiating a functional interaction between H4K20me1 and L3MBTL1 in the silencing of certain specific genes.
Other chromatin architectural proteins, such as PcG proteins including Polycomb Repressive Complex 1 (PRC1) [86] and PRC2 [87•], myeloid and erythroid nuclear termination stage-specific protein (MENT) [88], MeCP2 [89,90], and SIR complex [91•,92], were also found to play important roles in the formation of heterochromatin and gene silencing via chromatin compaction. Interestingly, chromatin compaction mediated by PRC1 is independent of its ability to ubiquitylate K119 of histone H2A in mouse ES cells (ESC) [93•]. Figure 3 outlines the basic features of the interactions between chromatin and its architectural proteins. Collectively, these architectural protein-mediated chromatin compactions are very important for higher-order chromatin structure formation and therefore, for gene regulation. Most recently, van Steensel and colleagues employed integrative computational analysis of genome-wide location maps for 53 widely selected chromatin proteins and 4 key histone modifications. Their strategy demonstrated that the genome is assembled into at least five distinctive chromatin types that are defined and characterized by unique combinations of chromatin architectural proteins and histone modifications in Drosophila Kc167 cells [94••]. Aside from two previously described heterochromatin types that correspond to facultative heterochromatin marked by PcG proteins and H3K27me3 and classic heterochromatin marked by HP1, SU(VAR)3–9 and H3K9me2, a prevalent type of repressive chromatin was identified as being a distinctive silent type of chromatin that covers a large part of the genome and is marked by unique combinations of seven proteins: histone H1, D1, IAL, SUUR, SU(HW), LAM, and EFF. Interestingly, this newly identified prevalent type of repressive chromatin is largely gene-poor and encompasses most silent genes in the genome. However, it is still unclear which histone modifications correspond to this prevalent heterochromatin, although H4K20me3 might play a critical role in its formation. With respect to euchromatin, this particular structure can also be classified into distinct types, in this case two, based on the presence of unique combinations of binding proteins and histone modifications. In contrast to heterochromatin, both types of euchromatin show hallmarks of transcriptionally active histone modifications (H3K4me3 and H3K79me3) and share various chromatin proteins involved in histone modifications and chromatin de-condensation. Aside from these notable similarities, these two types of euchromatin display striking differences in molecular organization, protein composition, and H3K36 methylation. Moreover, they are replicated at different times and regulate distinct classes of genes. Using a similar strategy, nine prevalent combinatorial states of chromatin were identified in Drosophila S2 and BG3 cells according to integrative analysis of genome-wide location maps of eighteen histone modifications [95••].
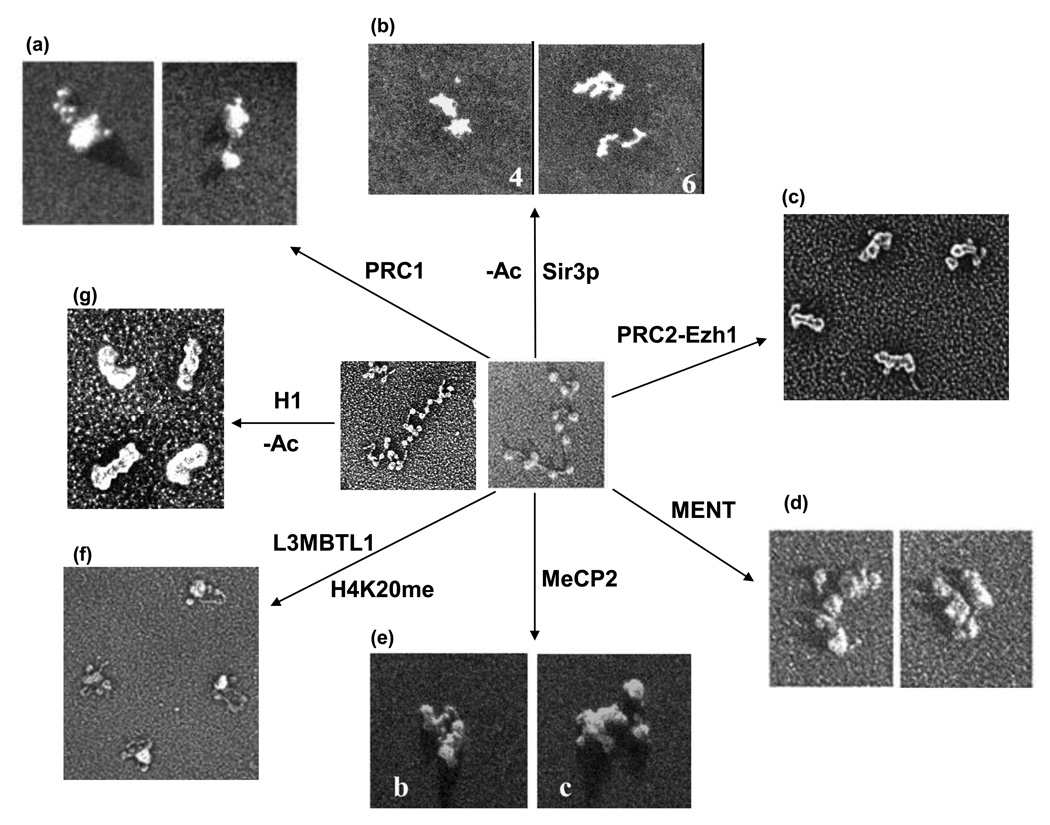
Figure 3. Chromatin compaction mediated by architectural proteins.
Electron microscopy images of the different types of chromatin compaction mediated by various chromatin architectural proteins discussed in the text. Untreated 12-mer and 25-mer reconstituted nucleosomal arrays was shown in the center (left, 25-mer arrays [G Li and D Reinberg, unpublished]; right, 12-mer arrays reproduced with permission from [88]). From a to f, 12-mer nucleosomal arrays bound with PRC1 (reproduced with permission from [86]), Sir3p (reproduced with permission from [92]), PRC2-Ezh1 (reproduced with permission from [87•]), MENT (reproduced with permission from [88]), MeCP2 (reproduced with permission from [89]), and L3MBTL1 (reproduced with permission from [81•]); g, Hypo-acetylated 25-mer nucleosomal arrays bound with H1 (G Li and D Reinberg, unpublished), all at approximately the same magnification.
Inheritance of higher-order chromatin structure
It is now recognized that various integrants, such as DNA methylation, histone modifications, histone variants and architectural proteins, are involved in the formation and regulation of higher-order chromatin structure that carries stable and inheritable epigenetic information. To date, however, it is still obscure as to how chromatin signatures or higher-order chromatin organization is transmitted from generation to generation during cell division. Based on the occurrence of histone modifications on the yeast silent mating-type region with 60 nucleosomes, Thon and colleagues described a simplified mathematical model for epigenetic inheritance of a chromatin domain and its histone modifications [96]. In this model, a number of parameters, such as positive feedback strength, cooperative effects, recruitment mechanisms, and long-distance nucleosomal interactions, are required for the bistability (i.e. stable existence in either of two states) of an epigenetically modified domain. More importantly, this bistability can be stably inherited through chromatin replication during the cell cycle given that parental histone modifications are randomly apportioned to two daughter strands and that newly deposited nucleosomes are unmodified [96]. In this regard, studies by Zhu and colleagues revealed that nearly all H3.1-H4 tetramers remained intact through DNA replication during cell division, and only a small fraction of histone variant H3.3-H4 tetramers seemed to be split [97••]. These results suggested that only very small portions of replicated genome are deposited with a mixture of newly synthesized unmodified H3.3-H4 dimer and H3.3-H4 dimer derived from a split paternal tetramer with putative epigenetic marks. According to the theoretical model above, the incorporation of these mixed nucleosomes results in about 50-fold destabilization, while a significant stability is still retained [96]. Another interesting feature of this model is that bistability was very difficult to archive when feedback only occurred within neighboring nucleosomes, thus long-range nucleosomal interaction within higher-order chromatin might facilitate the establishment of bistability and epigenetic inheritance of a chromatin state.
Using a novel metabolic labeling approach, Henikoff and colleagues recently showed that the nucleosomes modified by the PRC2 subunit EZ (Enhancer of Zeste), turn over faster than one cell cycle in Drosophila, arguing against models for epigenetic inheritance of PRC2-mediated histone H3K27me3. However, according to the theoretical model mentioned above, it is predicted that the epigenetic modifications of histone and/or chromatin states within a region composing 60 nucleosomes (i.e. a chromatin domain) can be stably maintained given the generation time of 30 (the average nucleosome conversions per nucleosome per cell cycle). Indeed, high-resolution mapping in Drosophila revealed that both PRC2 [98,99] and H3K27me3 [100] tend to form large contiguous domains. It is likely then that PRC2-mediated facultative heterochromatin composed of hundreds of nucleosomes can be stably inherited even when individual nucleosomes exhibit high dynamics in this region. The exact mechanism for the epigenetic inheritance of PcG-mediated transcriptional silencing, as in the case of PRC2, is still obscure. A mechanism proposed for epigenetic memory in a number of eukaryotic systems is based on positive feedback loops in nucleosome modifications [101•• ,102]. However, it is still unclear whether this positive-feedback loop mediates propagation of H3K27me3 in a neighbor-limited manner to stimulate conversion of an adjacent nucleosome, or a long-range interactative manner to stimulate the methylation of distant nucleosomes. The theoretical model predicts that neighbor-limited propagation of histone epigenetic marks may result in poor bistability, thus the propagation of H3K27me3 mediated by PRC2 may be neighbor-unlimited.
Higher-order chromatin dynamics and transcription
Although it is believed that the interconversion between permissive and refractory chromatin structures is important in regulating gene transcription, this process is poorly understood. Recent genome-wide chromatin immunoprecipitation (ChIP) analyses indicate that not only are promoter and enhancer associated nucleosomes highly dynamic [103,104], but also higher-order chromatin organization is also rather dynamic [15]. Most studies addressing the dynamic changes in chromatin organization during gene expression have thus far been focused on changes at the nucleosome level. It is now clear that chromatin structure exhibits a highly dynamic equilibrium between an open conformation exemplified by the 11-nm beads-on-a-string structure and a compact 30-nm fiber. As discussed above, histone modifications, histone variants, and architectural proteins play important roles in higher-order chromatin dynamics.
To address the roles of higher-order chromatin dynamics in transcriptional activation, we recently developed an in vitro system to reconstitute highly compacted 30-nm chromatin fiber using RSF/NAP-1 [105••]. Using this system, we demonstrated that linker histone H1 and deacetylation of core histones are required for formation of 30-nm chromatin fiber, which is refractory to transcription by RNAP II. While the NF1 transcription factor could not access the PEPCK promoter when embedded in such highly compacted chromatin in vitro, interestingly, transcription factors RAR/RXR could bind to this embedded promoter. Following RAR/RXR binding, the recruitment of histone-modifying enzymes (p300) and ATP-dependent chromatin-remodeling activities (SWI/SNF) resulted in the opening of the compacted 30-nm chromatin fiber, and nucleosomal rearrangement now allowed access to transcription factor NF1 and activation of the PEPCK gene after tRA induction [105••]. A very similar mechanism was reported in the case of glucocorticoid receptor (GR)-mediated activation of the mouse mammary tumor virus (MMTV) promoter [106]. Our ChIP results revealed that the pertinent transcription factors, cofactors, and the transcriptional machinery are recruited to the PEPCK promoter in a specific sequential and cyclical manner [105••]. Moreover, similar to our findings with the PEPCK promoter, Gannon and colleagues also showed that nucleosome organization over the pS2 promoter is also dynamically remodeled by synergism between SWI/SNF and p300 [107]. Recently, Gaudreau and colleagues demonstrated that incorporation of H2A.Z into the promoter regions of ERα target genes occurs cyclically when the gene is activated by estrogen induction [108]. H2A.Z is also required for FoxA1 association with the enhancers of ERα-regulated genes, suggesting that H2A.Z may contribute to gene activation through modulating the chromatin higher-order structure given that FoxA1 enables compacted higher-order chromatin to open for gene activation [109]. The function of H2A.Z in transcriptional regulation is very controversial [8], so that further studies are necessary to address the underlying mechanism by which H2A.Z, in cooperation with H3.3, modulates nucleosome/chromatin dynamics and transcription activity. An in vitro purified chromatin/transcription system may prove to be fruitful in this regard [110].
Perspectives and conclusions
To date, much of the detailed structural information for the “30-nm” chromatin fiber remains controversial. Most of our recent knowledge of “30-nm” chromatin structure comes from in vitro studies on a well-defined reconstituted nucleosomal array system that incorporates a strong “widom 601 nucleosome positioning” sequence [18,19]. However, it has been shown that the native chromatin fiber in vivo may bear irregular structure due to the variations of linker length, histone and DNA components, and architectural chromatin proteins. In order to elucidate the structure of the “30-nm” chromatin fiber carrying natural genetic (DNA sequence) and epigenetic (histone modifications or variants) information, we recently developed an in vitro system to reconstitute highly compacted 30-nm chromatin fiber using natural DNA sequence and histones carrying different modifications [105••]. This system will likely generate more natural “30-nm” chromatin fiber for subsequent structural and functional analysis, for example using cryo-electron microscopy. As well, single molecular FRET techniques should be highly informative for analyses of the structure of chromatin fiber and the mechanisms of its dynamics at the level of the single molecule.
The next key question concerns the structure of chromatin fiber in vivo. Cryo-electron microscopy of vitreous sections is a relatively new technique, enabling direct high-resolution observation of the cell structures in a close-to-native state. Using this technique, Dubochet and colleagues demonstrated that 30-nm chromatin fibers were not detectable in mitotic chromosomes in situ [35]. In keeping with this, careful analyses of the chromatin fiber structure during interphase are warranted. In addition, some super-resolution optical imaging techniques, such as dual-color photoactivated localization microscopy (PALM), and stimulated emission depletion (STED), are also suitable for analyzing higher-order chromatin structure and its dynamic changes during transcription activation and repression in living cells.
During each cell cycle, there are, inherently, two periods when chromatin organization undergoes global changes: DNA-replication in S-phase and chromosome condensation in M-phase. Yet the maintenance of defined chromatin organization through many cell generations is essential during cell differentiation and cell proliferation as well. Recently, Francis and colleagues found that PcG proteins can remain stably bound to chromatin during DNA replication, suggesting that the PcG protein-mediated chromatin structure may be inherited via the process of DNA replication [111••]. Another important question at the moment is how genomic chromatin domains find their place in the nucleus after mitosis. As major structural proteins within the nucleus, the nuclear lamins and associated proteins were proposed to play critical roles in the re-establishment of chromatin organization and nuclear architecture following mitosis [112]. However, it is still obscure how chromatin organization faithfully maintains its spatial memory after this process. The chromatin field is now in a solid position to address these open and key questions, the resolution of which will likely redirect our options for tackling medical-related goals that rest on advances in chromatin biology.
Acknowledgements
We thank Drs. P. Chen, R. Margueron and E. Campos for their critical reading of the manuscript. We are grateful to Dr. L. Vales for critical reading of this manuscript and active discussions. We apologize to authors whose studies could not be cited due to space limitations. G.L. is funded by the grants from the Ministry of Science and Technology (2011CB966300), the Natural Science Foundation of China (91019007 and 31071147), and Chinese Academy of Sciences (KSCX2-YW-R-234). D.R. is funded by the US National Institutes of Health (grants RO1GM064844 and 4R37GM037120) and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the "30-nm" chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci U S A. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos EI, Reinberg D. New chaps in the histone chaperone arena. Genes Dev. 2010;24:1334–1338. doi: 10.1101/gad.1946810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choy JS, Wei S, Lee JY, Tan S, Chu S, Lee TH. DNA methylation increases nucleosome compaction and rigidity. J Am Chem Soc. 2010;132:1782–1783. doi: 10.1021/ja910264z.. ▪In this paper, the single-molecular fluorescence resonance energy transfer (FRET) method is used to show that DNA methylation increases the compaction and rigidity of nucleosomes.
- 5.Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J Biol Chem. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 6.Hoch DA, Stratton JJ, Gloss LM. Protein-protein Forster resonance energy transfer analysis of nucleosome core particles containing H2A and H2A.Z. J Mol Biol. 2007;371:971–988. doi: 10.1016/j.jmb.2007.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thakar A, Gupta P, Ishibashi T, Finn R, Silva-Moreno B, Uchiyama S, Fukui K, Tomschik M, Ausio J, Zlatanova J. H2A.Z and H3.3 histone variants affect nucleosome structure: biochemical and biophysical studies. Biochemistry. 2009;48:10852–10857. doi: 10.1021/bi901129e. [DOI] [PubMed] [Google Scholar]
- 9.Angelov D, Molla A, Perche PY, Hans F, Cote J, Khochbin S, Bouvet P, Dimitrov S. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell. 2003;11:1033–1041. doi: 10.1016/s1097-2765(03)00100-x. [DOI] [PubMed] [Google Scholar]
- 10.Angelov D, Verdel A, An W, Bondarenko V, Hans F, Doyen CM, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, et al. SWI/SNF remodeling and p300-dependent transcription of histone variant H2ABbd nucleosomal arrays. EMBO J. 2004;23:3815–3824. doi: 10.1038/sj.emboj.7600400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao Y, Konesky K, Park YJ, Rosu S, Dyer PN, Rangasamy D, Tremethick DJ, Laybourn PJ, Luger K. Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. EMBO J. 2004;23:3314–3324. doi: 10.1038/sj.emboj.7600316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyen CM, Montel F, Gautier T, Menoni H, Claudet C, Delacour-Larose M, Angelov D, Hamiche A, Bednar J, Faivre-Moskalenko C, et al. Dissection of the unusual structural and functional properties of the variant H2A.Bbd nucleosome. EMBO J. 2006;25:4234–4244. doi: 10.1038/sj.emboj.7601310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyen CM, An W, Angelov D, Bondarenko V, Mietton F, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, Dimitrov S. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol Cell Biol. 2006;26:1156–1164. doi: 10.1128/MCB.26.3.1156-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Widom J, Klug A. Structure of the 300A chromatin filament: X-ray diffraction from oriented samples. Cell. 1985;43:207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- 15.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams SP, Athey BD, Muglia LJ, Schappe RS, Gough AH, Langmore JP. Chromatin fibers are left-handed double helices with diameter and mass per unit length that depend on linker length. Biophys J. 1986;49:233–248. doi: 10.1016/S0006-3495(86)83637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Holde K, Zlatanova J. Chromatin fiber structure: Where is the problem now? Semin Cell Dev Biol. 2007;18:651–658. doi: 10.1016/j.semcdb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 19.Robinson PJ, Rhodes D. Structure of the '30 nm' chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 21.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 22.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci U S A. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fletcher TM, Hansen JC. Core histone tail domains mediate oligonucleosome folding and nucleosomal DNA organization through distinct molecular mechanisms. J Biol Chem. 1995;270:25359–25362. doi: 10.1074/jbc.270.43.25359. [DOI] [PubMed] [Google Scholar]
- 24.Schwarz PM, Felthauser A, Fletcher TM, Hansen JC. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- 25.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 26.Zheng C, Lu X, Hansen JC, Hayes JJ. Salt-dependent intra- and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J Biol Chem. 2005;280:33552–33557. doi: 10.1074/jbc.M507241200. [DOI] [PubMed] [Google Scholar]
- 27.Kan PY, Lu X, Hansen JC, Hayes JJ. The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol Cell Biol. 2007;27:2084–2091. doi: 10.1128/MCB.02181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 29.Gordon F, Luger K, Hansen JC. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J Biol Chem. 2005;280:33701–33706. doi: 10.1074/jbc.M507048200. [DOI] [PubMed] [Google Scholar]
- 30.McBryant SJ, Klonoski J, Sorensen TC, Norskog SS, Williams S, Resch MG, Toombs JA, 3rd, Hobdey SE, Hansen JC. Determinants of histone H4 N-terminal domain function during nucleosomal array oligomerization: roles of amino acid sequence, domain length, and charge density. J Biol Chem. 2009;284:16716–16722. doi: 10.1074/jbc.M109.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol Cell Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kan PY, Caterino TL, Hayes JJ. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol Cell Biol. 2009;29:538–546. doi: 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol. 2002;9:172–176. doi: 10.1038/nsb767. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat Struct Mol Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 35.Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci U S A. 2008;105:19732–19737. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadauke S, Blobel GA. Chromatin loops in gene regulation. Biochim Biophys Acta. 2009;1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SI, Bultman SJ, Kiefer CM, Dean A, Bresnick EH. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc Natl Acad Sci U S A. 2009;106:2259–2264. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saramaki A, Diermeier S, Kellner R, Laitinen H, Vaisanen S, Carlberg C. Cyclical chromatin looping and transcription factor association on the regulatory regions of the p21 (CDKN1A) gene in response to 1alpha,25-dihydroxyvitamin D3. J Biol Chem. 2009;284:8073–8082. doi: 10.1074/jbc.M808090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010 doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 41.Ansari A, Hampsey M. A role for the CPF 3'-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci U S A. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kheradmand Kia S, Solaimani Kartalaei P, Farahbakhshian E, Pourfarzad F, von Lindern M, Verrijzer CP. EZH2-dependent chromatin looping controls INK4a and INK4b, but not ARF, during human progenitor cell differentiation and cellular senescence. Epigenetics Chromatin. 2009;2:16. doi: 10.1186/1756-8935-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, Baylin SB. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 47.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–948. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 48. Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016.. ▪ In this paper, a high-resolution molecular maps of the reorganization of genome-nuclear lamina interactions reveals that lamina-genome interactions are widely involved in the regulation of gene expression programs during differentiation and cell lineage commitment.
- 49.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 50.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011.. ▪This study and ref [51] demonstrate that a number of nucleoporins can directly interact with chromatin and stimulate expression of developmental and cell-cycle regulated genes in drosophila.
- 53.Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- 54. Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727.. ▪This paper demonstrates the repositioning of genes to the nuclear lamina can result in the transcriptional repression using three-dimensional DNA-immunoFISH.
- 55.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027.. ▪ In this paper, using a general method to produce site-specifically and homogeneously acetylated recombinant histone H3 at lysine 56 residue by genetically encoding acetyl-lysine, the authors demonstrate that acetylation of H3 K56 has little effect on the compaction of chromatin, but increases the DNA breathing in mononucleosomes.
- 57.Watanabe S, Resch M, Lilyestrom W, Clark N, Hansen JC, Peterson C, Luger K. Structural characterization of H3K56Q nucleosomes and nucleosomal arrays. Biochim Biophys Acta. 2010;1799:480–486. doi: 10.1016/j.bbagrm.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol. 2010;12:853–862. doi: 10.1038/ncb2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003.. ▪ In this paper, ATRX and DAXX are found to associate with H3.3 in a HIRA-independent manner. The authors demonstrate that ATRX is required for the HIRA-independent incorporation of H3.3 at telomeres and for the repression of telomeric RNA.
- 61.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056.. ▪ In this paper, the genome-wide analysis reveals that H2A.Z is enriched at the promoters of polycomb proteins target genes in ES cells. The authors demonstrate that H2A.Z, together with polycomb proteins, is necessary for ES differentiation and cell fate transition.
- 63. Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324.. ▪ This paper shows that DNA methylation and histone H2A.Z are mutually antagonistic distribution in the whole genome of the plant Arabidopsis thaliana.
- 64.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 65.Conerly ML, Teves SS, Diolaiti D, Ulrich M, Eisenman RN, Henikoff S. Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome Res. 2010 doi: 10.1101/gr.106542.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zlatanova J, Caiafa P, Van Holde K. Linker histone binding and displacement: versatile mechanism for transcriptional regulation. FASEB J. 2000;14:1697–1704. doi: 10.1096/fj.99-0869rev. [DOI] [PubMed] [Google Scholar]
- 68.Catez F, Ueda T, Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol. 2006;13:305–310. doi: 10.1038/nsmb1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hendzel MJ, Lever MA, Crawford E, Th'ng JP. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- 70.Clausell J, Happel N, Hale TK, Doenecke D, Beato M. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS One. 2009;4:e0007243. doi: 10.1371/journal.pone.0007243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wisniewski JR, Zougman A, Kruger S, Mann M. Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol Cell Proteomics. 2007;6:72–87. doi: 10.1074/mcp.M600255-MCP200. [DOI] [PubMed] [Google Scholar]
- 72.Deterding LJ, Bunger MK, Banks GC, Tomer KB, Archer TK. Global changes in and characterization of specific sites of phosphorylation in mouse and human histone H1 Isoforms upon CDK inhibitor treatment using mass spectrometry. J Proteome Res. 2008;7:2368–2379. doi: 10.1021/pr700790a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 74.Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J Biol Chem. 2005;280:38090–38095. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- 76.Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T, Reinberg D. Dynamic Histone H1 Isotype 4 Methylation and Demethylation by Histone Lysine Methyltransferase G9a/KMT1C and the Jumonji Domain-containing JMJD2/KDM4 Proteins. J Biol Chem. 2009;284:8395–8405. doi: 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weiss T, Hergeth S, Zeissler U, Izzo A, Tropberger P, Zee BM, Dundr M, Garcia BA, Daujat S, Schneider R. Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenetics Chromatin. 2010;3:7. doi: 10.1186/1756-8935-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rochman M, Malicet C, Bustin M. HMGN5/NSBP1: a new member of the HMGN protein family that affects chromatin structure and function. Biochim Biophys Acta. 2010;1799:86–92. doi: 10.1016/j.bbagrm.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, McNally JG, Wu X, Bubunenko NA, Grigoryev S, et al. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol Cell. 2009;35:642–656. doi: 10.1016/j.molcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cato L, Stott K, Watson M, Thomas JO. The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. J Mol Biol. 2008;384:1262–1272. doi: 10.1016/j.jmb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 81. Trojer P, Li G, Sims RJ, 3rd, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048.. ▪ In this paper, the malignant-brain-tumor (MBT) protein L3MBTL1 has been found to form a protein complex with a number of chromatin proteins including core histones, histone H1, HP1γ and Rb. The authors demonstrate that the L3MBTL1 MBT domains exhibit chromatin compaction activity in a histone-methylation-dependent manner.
- 82.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 83.Congdon LM, Houston SI, Veerappan CS, Spektor TM, Rice JC. PR-Set7-mediated monomethylation of histone H4 lysine 20 at specific genomic regions induces transcriptional repression. J Cell Biochem. 2010;110:609–619. doi: 10.1002/jcb.22570. [DOI] [PubMed] [Google Scholar]
- 84.Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 87. Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004.. ▪ In this paper, the mammalian homologs Ezh1 and Ezh2 are found to form similar PRC2 complexes but exhibit quite different repressive roles through different mechanisms in which PRC2-Ezh2 catalyzes di- and tri-methylation of histone H3 lysine 27 (H3K27me2/3) and PRC2-Ezh1 directly and robustly represses transcription through compacting chromatin templates.
- 88.Springhetti EM, Istomina NE, Whisstock JC, Nikitina T, Woodcock CL, Grigoryev SA. Role of the M-loop and reactive center loop domains in the folding and bridging of nucleosome arrays by MENT. J Biol Chem. 2003;278:43384–43393. doi: 10.1074/jbc.M307635200. [DOI] [PubMed] [Google Scholar]
- 89.Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 90.Ghosh RP, Horowitz-Scherer RA, Nikitina T, Shlyakhtenko LS, Woodcock CL. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol Cell Biol. 2010 doi: 10.1128/MCB.00379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Johnson A, Li G, Sikorski TW, Buratowski S, Woodcock CL, Moazed D. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol Cell. 2009;35:769–781. doi: 10.1016/j.molcel.2009.07.030.. ▪ In this paper, the authors successfully reconstitute SIR-dependent heterochromatin in vitro using purified components and demonstrate that the SIR complex can efficiently mediate transcriptional gene silencing through deacetylating chromatin templates and promoting the alteration of chromatin structures.
- 92.McBryant SJ, Krause C, Woodcock CL, Hansen JC. The silent information regulator 3 protein, SIR3p, binds to chromatin fibers and assembles a hypercondensed chromatin architecture in the presence of salt. Mol Cell Biol. 2008;28:3563–3572. doi: 10.1128/MCB.01389-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032.. ▪ In this paper, the authors demonstrate that the effect of Ring1B on the compaction of chromatin structure and repression of Hox gene is not dependent on its histone ubiquitination activity.
- 94. Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009.. ▪In this paper, the authors employ integrative computational analysis of genome-wide location maps for chromatin proteins and histone modifications. They demonstrate that the genome is assembled into five distinctive chromatin types that are defined and characterized by unique combinations of chromatin architectural proteins and histone modifications in Drosophila Kc167 cells.
- 95. Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2010 doi: 10.1038/nature09725.. ▪In this paper, nine prevalent combinatorial states of chromatin were identified in drosophila S2 and BG3 cells using a similar strategy in Ref [94].
- 96.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 97. Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994.. ▪In this paper, the authors demonstrate that comparing to the canonical H3.1–H4 tetramers, the H3.3–H4 tetramers exhibit different partitioning pattern during DNA replication-dependent chromatin assembly. They show that certain fractions of H3.3–H4 tetramers split during DNA replication, whereas most H3.1–H4 tetramers remain intact.
- 98.Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 100.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 101. Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398.. ▪ In this paper, the authors find that the EED subunit of Polycomb Repressive Complex 2 (PRC2) can specifically bind to the histone tails harboring trimethyl-lysine residues associated with repressive chromatin marks and these interactions can lead to the stimulation of histone methyltransferase activity of PRC2. Based on these findings, they propose a model for the propagation of the H3K27me3 mark.
- 102.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 104.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Li G, Margueron R, Hu G, Stokes D, Wang YH, Reinberg D. Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol Cell. 2010;38:41–53. doi: 10.1016/j.molcel.2010.01.042.. ▪In this paper, the authors develope an in vitro system to reconstitute 30 nm chromatin fiber using purified components. Using this novel in vitro-assembled highly compacted chromatin, the authors demonstrate the dynamic process of folding and unfolding of higher-order chromatin during the transcription activation in vitro and in vivo.
- 106.Di Croce L, Koop R, Venditti P, Westphal HM, Nightingale KP, Corona DF, Becker PB, Beato M. Two-step synergism between the progesterone receptor and the DNA-binding domain of nuclear factor 1 on MMTV minichromosomes. Mol Cell. 1999;4:45–54. doi: 10.1016/s1097-2765(00)80186-0. [DOI] [PubMed] [Google Scholar]
- 107.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 108.Gevry N, Hardy S, Jacques PE, Laflamme L, Svotelis A, Robert F, Gaudreau L. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 2009;23:1522–1533. doi: 10.1101/gad.1787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 110.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 111. Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017.. ▪In this paper, using an in vitro chromatin replication system, the authors demonstrate that the Polycomb Repressive Complex 1 (PRC1) remains bound to chromatin and DNA during DNA replication in vitro.
- 112.Martin C, Chen S, Jackson DA. Inheriting nuclear organization: can nuclear lamins impart spatial memory during post-mitotic nuclear assembly? Chromosome Res. 2010;18:525–541. doi: 10.1007/s10577-010-9137-8. [DOI] [PubMed] [Google Scholar]