Abstract
Hepatic cytochrome P450 (P450) gene and protein expression are modulated during inflammation and infection. Oral infection of C57BL/6 mice with Citrobacter rodentium produces mild clinical symptoms while selectively regulating hepatic P450 expression and elevating levels of proinflammatory cytokines. Here, we explored the role of cytokines in the regulation of hepatic P450 expression by orally infecting tumor necrosis factor-α (TNFα) receptor 1 null mice (TNFR1−/−), interleukin-1 (IL1) receptor null mice (IL1R−/−), and Kupffer cell depleted mice with C. rodentium. CYP4A mRNA and protein levels and flavin monooxygenase (FMO)3 mRNA expression levels were down-regulated, while CYP2D9 and CYP4F18 mRNAs remained elevated during infection in wild-type, receptor knockout, and Kupffer cell depleted mice. CYPs 3A11 and 3A25 mRNA levels were down-regulated during infection in wild-type mice but not in TNFR1−/− mice. Consistent with this observation, CYPs 3A11 and 3A25 were potently down-regulated in mouse hepatocytes treated with TNFα. Oral infection of IL1R−/− mice and studies with mouse hepatocytes indicated that IL1 does not directly regulate CYP3A11 or CYP3A25 expression. Uninfected mice injected with clodronate liposomes had a significantly reduced number of Kupffer cells in their livers. Infection increased the Kupffer cell count, which was attenuated by clodronate treatment. The P450 mRNA and cytokine levels in infected Kupffer cell depleted mice were comparable to those in infected mice receiving no clodronate. The results indicate that TNFα is involved in the regulation of CYPs 3A11 and 3A25, but IL1β and Kupffer cells may not be relevant to hepatic P450 regulation in oral C. rodentium infection.
Keywords: Cytochrome P450, Inflammation, Kupffer cells, Interleukin-1, TNFα
1. Introduction
Cytochromes P450 are a superfamily of drug-metabolizing enzymes that catalyze the oxidation of a variety of exogenous and endogenous compounds [1]. Infection and inflammation can cause down-regulation or induction of hepatic P450 expression and activity [2, 3], which in turn can alter drug clearance. Lipopolysaccharide (LPS), a Gram-negative bacterial endotoxin widely used as a sterile model of inflammation to study P450 expression during infection or inflammation, has been shown to alter P450 expression and activity and increase inflammatory cytokine levels in rodents [4–10]. Models of live infection and inflammation have also demonstrated similar trends [11–14]. Citrobacter rodentium is a natural murine pathogen that is equivalent to human enteropathogenic Escherichia coli (EPEC), with similar colonic pathology displaying attaching and effacing lesions on intestinal cells of the host.
C. rodentium can serve as a live model for human EPEC infection and colitis, and inflammatory bowel disease (IBD) for humans and mice [15, 16]. Oral C. rodentium infection in C57BL/6 mice demonstrated mild symptoms, selective regulation of hepatic P450 expression, and elevated levels of some serum cytokines, including interleukin (IL)-6, tumor necrosis factor alpha (TNFα), and IL2 [14]. IL1 was not elevated in the serum but its mRNA was induced in the liver [13, 14]. In vivo and in vitro administrations of cytokines (IL6, TNFα, IL1β and interferon (IFN)γ) have demonstrated selective regulation of P450 expression [17–20]. However, the in vivo roles of cytokines in hepatic P450 regulation in the C. rodentium model are not known. In this work, we tested the role of TNFα and IL1β on the known regulation of hepatic P450 expression during oral C. rodentium infection in mice, by comparing the effects of infection in WT and TNFR−/− and IL1−/− mice.
Two receptors, TNFR1 and TNFR2, mediate the cellular effects of TNFα [21]. TNFR1 is ubiquitously expressed and is the main receptor that mediates the effects of soluble TNFα on cells; whereas, TNFR2 is expressed mainly in hematopoietic cells and has the membrane-bound form of TNFα as its primary ligand [22]. For these reasons, we chose to focus on TNFR1 for the current study on the effects of TNF in the liver.
Because Kupffer cells are a source of cytokines such as TNFα, IL6, and IL1β in the liver [23, 24], we hypothesized that Kupffer cells may be activated and release cytokines during C. rodentium infection, leading to increased cytokine levels and selective regulation of P450 levels. To address this question, we studied the effect of depleting Kupffer cells with clodronate liposomes, on the responses of hepatic P450 expression to oral C. rodentium infection. In this work, we demonstrate that IL1β and Kupffer cells do not play a significant role in the regulation of P450 expression during oral C. rodentium infection, but TNFα is an important factor in regulating the expression of CYPs 3A11 and 3A25 in this inflammation model.
2. Materials and Methods
2.1. Bacteria
A wild-type strain of C. rodentium (#51116) was obtained from the American Type Culture Collection (Manassas, VA). Before infection, C. rodentium was grown in Luria broth without shaking overnight. Bacteria were resuspended in sterile phosphate-buffered saline, and the nominal concentration was calculated spectrophotometrically. Actual concentrations were determined by retrospective plating on MacConkey agar, on which C. rodentium forms small pink colonies with white rims.
2.2. Chemicals, Animals, and Treatments
Unless otherwise specified, reagents and chemicals were obtained from Sigma-Aldrich (St. Louis, MO). Dichloromethylene bisphosphonate (clodronate) was a gift of Roche Diagnostics GmbH (Mannheim, Germany) and was encapsulated in liposomes as described previously [25, 26]. Female wild-type C57BL/6 and TNFR1−/− mice were obtained from Jackson Laboratory (Bar Harbor, ME). The mice were acclimatized to the animal facility for at least 1 week, and mice were 8–9 weeks of age at the time of infection. Two groups of mice (6 C57BL/6 mice and 6 TNFR1−/− mice) were housed, infected with C. rodentium, and sacrificed in a Biosafety Level-2 facility to prevent transmission of infection to other mouse colonies. Oral administration of C. rodentium was achieved by infection of the drinking water (containing 20% sucrose) for 24 h at a nominal concentration calculated to result in a bacterial dose of 1.05 × 109 cells/mouse. Two sets of control groups (6 C57BL/6 mice and 6 TNFR1−/− mice) received 20% sucrose in their drinking water for 24 h as well. Mice were sacrificed 7 days after administration of sucrose or bacteria. Livers and colons were collected. Livers were rinsed in cold 1.15% potassium chloride, and stored at −80 °C for later RNA or microsome preparation. Distal colon samples were washed for fecal content in cold 1.15% potassium chloride and fixed in 10% neutral buffered formalin solution until they were processed for histology by the Emory Healthcare Department of Histology. They were stained with hematoxylin and eosin for histopathological evaluation. The remainder of the colon was sectioned to measure myeloperoxidase (MPO) activity and to determine C. rodentium CFUs. Blood was also collected from all the animals, and allowed to clot for ~30 minutes. Serum was separated by centrifugation and stored at −80 °C until analyzed.
IL1R−/− mice were a gift from Dr. Daniel Kalman (Emory University), who obtained them from Jackson Laboratory (Bar Harbor, ME). C576BL/6 wild-type mice (Charles River Laboratories (Wilmington, MA)) and ILR−/− mice were infected in parallel with C. rodentium at a bacterial dose of 5.00 × 108 cells/mouse Treatment, sacrifice, dissection and collection of organs for ILR−/− and wild-type mice form were as described above for TNFR p55−/− mice and their wild-type controls.
For studies requiring depletion of Kupffer cells, 9-week-old C57BL/6 mice were initially injected intraperitoneally with 100 μL of clodronate liposomes 2 days before treatment with sucrose or bacteria. Control mice were injected i.p. with 100 μL of sterile saline. Subsequent injections occurred every 4 days since Kupffer cells were found to repopulate the liver 5 days after administration of the clodronate liposomes [27]. Mice were administrated sucrose or bacteria (9.15 × 108 cells/mouse) as described above and were sacrificed 9 days later. Livers, colons, and blood were collected and stored as described above. Liver sections were also stained for detection of Kupffer cells. All the protocols were reviewed and approved by the Institutional Animal Care and Use Committee of Emory University.
2.3. Preparation of Total RNA
Total liver RNA was prepared using RNA-Bee isolation reagent according to the manufacturer's instructions (Tel-Test, Friendswood TX). Total RNA concentration was estimated spectrophotometrically at 260 nm, and RNA purity and integrity were confirmed by formaldehyde-agarose gel electrophoresis followed by visualization with ethidium bromide.
2.4. Microsome Preparation
Liver microsomes were prepared by differential centrifugation as described previously [14] and stored at −80 °C. Microsomal protein concentrations were determined using bicinchoninic acid (BCA) protein assay kit from Thermo Fisher Scientific (Rockford, IL), with bovine serum albumin as the protein standard.
2.5. cDNA Synthesis
Purified total RNA was reverse-transcribed using the Applied Biosystems High Capacity cDNA Reverse Transcription Kit (Foster City, CA), according to the manufacturer's protocol.
2.6. PCR Primer Sequences
All primers for mouse P450s, cytokines, acute phase proteins, and glyceraldehyde dehydrogenase (GAPDH) have been described and used by our laboratory previously [8, 13, 28]. All the primers were custom-synthesized on a 50-nmol scale by MWG Biotech, Inc. (High Point. NC), and obtained desalted and lyophilized. Primers were diluted to 100 μM in deionized water and stored at −80°C.
2.7. Quantitative Real-Time RT-PCR
Real-time RT-PCR was performed using the ABI PRISM 7000 Sequence Detection System and Power SyBr Green to determine the expression of mRNAs of interest in mouse liver, as described previously [13, 14]. Duplicate reactions were performed in a total volume of 25 μl using Power SyBr Green Master Mix reagent (Applied Biosystems, Warrington, UK); 5 μl of cDNA was used as template for the reaction, with 10 μM forward and reverse primers. Thermal cycling conditions included 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 95°C for 15 s, and 1 min at the appropriate annealing temperature. Results are expressed as relative levels of target mRNA, normalized to levels of the housekeeping gene GAPDH, as calculated by the ΔΔCt method [29]. The expression level in control samples was arbitrarily set at 1.
2.8. Immunoblotting
P450 protein levels in mouse hepatic microsomes were measured by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting, as described previously [14]. Equal amounts of protein (10 μg) were loaded on the gels for each assay. Antibodies to rat CYP4A and 2E, and mouse CYP2D were generously provided by Dr. Gordon Gibson (University of Surrey, Guildford, UK), Dr. Magnus Ingelman-Sundberg (Karolinska Institute, Stockholm, Sweden), and Dr. Masahiko Negishi (NIEHS, Research Triangle Park, NC), respectively. Antibodies to rat CYP2B and 3A were provided by Dr. James Halpert (University of Texas Medical Branch, Galveston, TX). Polyclonal antibodies to rat CYPs 2E, 3A and 4A proteins and to mouse CYP2D were diluted 1:5,000, while 2B and 2C antibodies were diluted 1:20,000. Secondary antibodies were as follows: goat anti-rabbit for 2B and 2E were diluted 1:10,000; 1:5000 for 2C, and 1:3000 for 2D; rabbit anti-goat for 3A was diluted 1:10000; rabbit anti-sheep for 4A was diluted to 1:5000. All assays were performed within a linear range, and the intensity of stained bands was measured by laser densitometry.
2.9. Determination of Tissue Bacterial Loads
To measure the CFU of C. rodentium in colon and liver, tissue samples weighing approximately 0.1 to 0.3g were homogenized at low speed with a Tissuemizer (Fisher Scientific, Pittsburgh, PA) in 1ml of PBS. The lysate was plated on MacConkey agar plates at various dilutions. C. rodentium colonies were recognized as pink with a white rim as previously described [30] and were counted after 20 hours of incubation at 37°C to determine the CFU per gram of tissue.
2.10. Colon Pathology
Paraffin-embedded colonic tissue sections were stained with hematoxylin and eosin. For crypt height measurements, three measurements of well oriented crypts were taken in the distal colon for each mouse, using micrometry by a Zeiss (Thornwood, NY) 200M microscope with a 20× NA1.4 lens, and Slidebook (Intelligent Imaging Innovations, Denver CO).
2.11. Myeloperoxidase (MPO) Assay)
Colonic tissue samples weighing between 0.2–0.4 g were homogenized on ice in 50mM potassium phosphate buffer, pH 6.0, containing 150 mg of hexadecyltrimethlammonium bromide (HTAB). The homogenates were subjected to 3 freeze-thaw cycles, sonicated (Microson™ Ultrasonic cell disruptor, Misonix Inc, Farmingdale, NY) on ice for 10s (power level 15), and centrifuged at 14,000 rpm for 15 min at 4°C. The supernatant was collected and the protein concentration was measured using the BCA protein assay kit from Thermo Fisher Scientific (Rockford, IL). Aliquots of each supernatant or MPO standard (10 μL) were added to 200 μL of reactive buffer (0.280 mg/mL o-dianisidine, 0.0005% H2O2, in potassium phosphate buffer), and absorbance was measured at 450 nm with a plate reader (ThermoMax Microplate Reader, Molecular Devices, Sunnyvale, CA). One unit of enzyme activity was defined as the amount that consumes 1mmol of H2O2 /min.
2.12. Kupffer Cell Detection
Paraffin-embedded liver tissue sections were mounted on ion positive slides and incubated with rat anti-mouse F4/80-antibody diluted 1:100 (Abcam, Cambridge, MA) by the Emory Healthcare Department of Histology. Samples were developed using horseradish peroxidase (HRP) conjugated secondary antibody and 3,3-diaminobenzidine (DAB) and were counterstained with hematoxylin. Kupffer cells were visualized using Zeiss Axioplan 2 Upright Microscope with Zeiss Axiovision Software. Images were taken at 10× magnification and cells were counted using Image J software (NIH, Bethesda, MD).
2.13. Cytokine Analysis
Serum samples were analyzed for cytokines in 25 μL of serum from each sample using the MILLIPLEX™ MAP kit (Millipore Corp, St. Charles, MO), based on Luminex® xMAP technology (Luminex, Austin, TX). The cytokines IL1β, IL2, IL6, TNFα, IFNγ, granulocyte colony stimulating factor (G-CSF), macrophage inflammatory protein 1 alpha (MIP-1α), monocyte chemotactic protein -1 (MCP-1), and chemokine CXCL1 (KC) were analyzed by following the protocol supplied with the kit. Briefly, 96-well plate filter plates were washed with wash buffer, and samples (1:1 dilution, 50 μl final volume) were applied to each well. Antibody-coated beads were added to the wells, and the samples were incubated for 18 h at 4°C. After incubation, the plate was washed twice with buffer supplied with the kit. Biotinylated antibodies against the different cytokines and chemokines from the kit were added, and the mixture was incubated for 1 h at room temperature. Then the cytokine antibody complexes were detected by addition of streptavidin coupled to phycoerythrin. For determination of the number of positive complexes, each sample was read in a Luminex XYP platform. Data were analyzed using MasterPlex 1.2 (Mirai-Bio, Hitachi Solutions Americ Ltd, San Francisco, CA) and the concentrations are expressed in picograms per milliliter.
2.14. Mouse Hepatocyte Isolation and Treatment
Mouse hepatocytes were isolated by a two-step in situ collagenase perfusion procedure previously described [19]. After female C57BL/6 mice (20 – 25 g) were anesthetized with ketamine/xylazine solution, the liver was perfused in situ via the portal vein with 120 – 150 mls of Krebs-Ringer bicarbonate buffer at a flow rate of 14 – 15 mls/min and then with 50 – 70 mls of 0.3 mg/ml collagenase type IV (Sigma, MO) for 5 min. Hepatocyte preparations that were greater than 80% viable were plated on 12-well collagen-coated plates (BD Bioscience, San Jose, CA) in plating medium (Williams E medium containing 10mM Hepes pH 7.4, 150 nM insulin, 50 nM dexamethasone, 10mg/mL Pen/Strep, and 10% fetal bovine serum (Atlanta Biologicals, Atlanta, GA) at a density of 0.5 × 106 cells/well. 4 h later, cells were overlaid with new CWE medium (Williams E containing 10mM Hepes pH 7.4, 150 nM insulin, 50 nM dexamethasone and 10mg/mL Pen/Strep) containing Matrigel (0.23 mg/mL, BD Biosciences). Next day, the medium was changed to CWE medium. Cultures were maintained for 6 days at 37°C in 5% CO2 with regular media changes. The primary cultures were treated on the 6th day with TNFα or IL1β.
2.15. Statistical Analysis
Differences between groups were determined by Students T-test or by one-way analysis of variance followed by Tukey's test as appropriate. The level of significance was set at P < 0.05.
3. Results
3.1 Oral Infection with C. rodentium in TNFR1 −/− Mice
The effects of C. rodentium expression on hepatic P450 and FMO expression in WT mice have been described in previous publications from this laboratory [13, 14, 28]. The earliest time point at which we previously demonstrated significant alterations in P450 expression in response to C. rodentium was 7 days after the initiation of infection [14]. We therefore chose this time point to study the roles of TNFα and IL1 in P450 regulation in this model.
3.1.1. Bacterial loads and myeloperoxidase activities in the liver and colon
TNFR1−/− mice infected with C. rodentium were reported to have a higher bacterial load and increased crypt length in the colons compared to wild-type mice infected with the bacteria [16]. In this work, the colonic bacterial load was similar between treated C57BL/6 and TNRFR1−/− mice (Supplemental Fig. 1), while the crypt lengths for each infected group were significantly higher than the crypt lengths of the corresponding control groups (Supplemental Table 1). Myeloperoxidase (MPO) activity was measured as an assessment of neutrophil recruitment to the colon [31]. MPO activity (U/g) in the colons of both infected groups tended to increase ~2.5 fold, but only the levels in the TNFR1−/− infected mice were significantly different from untreated (Supplemental Table 1). The liver masses in infected mice of either genotype were statistically higher than in control mice (Supplemental Table 1). C. rodentium cells were found in the livers of infected groups but the bacterial loads were 10,000 and 430,000 times lower than in the colons of wild-type infected mice and TNFR1−/− infected mice, respectively (Supplemental Fig. 1).
3.1.2. Serum Cytokines
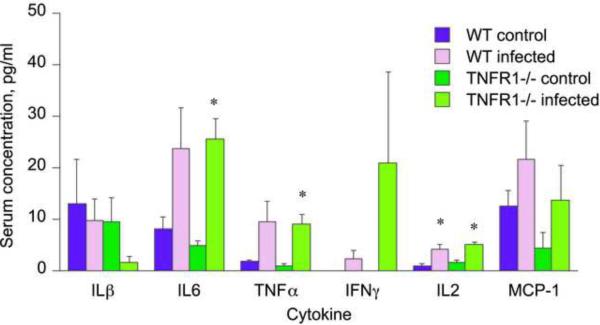
Overall, the pattern of serum cytokines was very similar in infected mice of either genotype. Serum levels of MCP-1, KC, MIP-1α, IFNγ, and IL1β were not significantly affected by infection in mice of either strain (Fig. 1). Serum levels of IL2 (Fig. 1) and granulocyte colony stimulating factor (not shown) were elevated to similar extents in both infected groups when compared to their own controls. Significant elevations in serum concentrations of IL6 and TNFα were observed in the TNFR1−/− infected mice compared to their uninfected counterparts (Fig. 1). A similar tendency in WT mice failed to achieve significance (Fig. 1).
Fig. 1.
Serum cytokine profile during oral Citrobacter rodentium infection in wild-type and TNFR1−/− mice. Blood was collected at sacrifice for serum cytokine analysis as described under Materials and Methods. Values represent means ± S.E.M. of 4 mice per group. *, P<0.05 compared with control group of same genotype. Differences between groups were determined by Student's t test
3.1.3. Hepatic P450 and Flavin Monooxygenase Expression
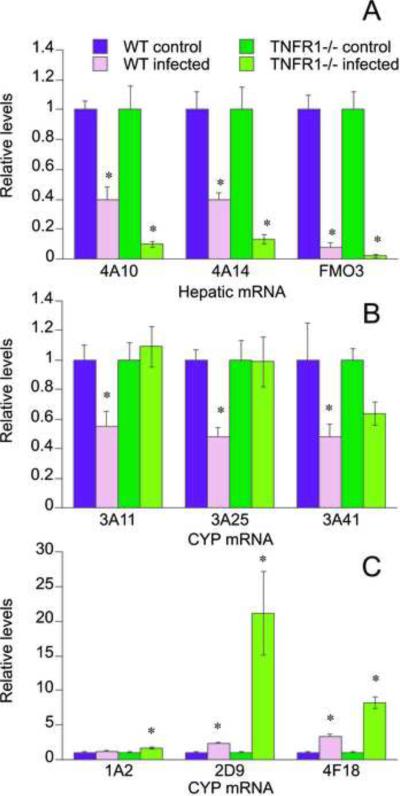
The regulation of hepatic mRNA expression during C. rodentium infection in the two genotypes exhibited four distinct patterns. The first pattern featured mRNAs (CYP4A10, CYP4A14, FMO3) whose down-regulation was greater in the knockout mice than in the wild-type animals (Fig. 2A).
Fig. 2.
Effect of TNFR1 gene deletion on responses of P450 mRNAs in mouse liver to oral C. rodentium infection. Mice were orally infected with C. rodentium, and livers were harvested 7 days later for measurement of P450 mRNA levels as described under Materials and Methods. Values represent means ± S.E.M. of 6 mice per group. A, CYPs 4A10 and 4A14 and flavin monooxygenase 3 (FMO3); B, CYPs 3A11, 3A25, and 3A41; C, CYPs 1A2, 2D9, and 4F18. *, P<0.05 compared with control group of same genotype. Differences between groups were determined by Student's t test.
The second pattern was exhibited by P450s whose down-regulation was blocked or attenuated in TNFR1−/− mice (Fig 2B). CYPs 3A11 and 3A25 were both down-regulated in wild-type infected mice to approximately 50% of uninfected levels, but the down-regulation of both CYPs was blocked in TNFR1−/− infected mice. CYP3A41 was significantly down-regulated in the livers of WT mice. Although there was also a tendency for CYP3A41 to be suppressed to a lesser degree in TNFR1−/− infected mice, this was not statistically significant (Fig. 2B).
The third distinct pattern of regulation observed was for P450s that were induced by infection (Fig.2C). CYP2D9 and CYP4F18 were induced 2-fold and 3.3-fold in wild-type infected mice, and these inductions were even greater in mice lacking the TNFR1 receptor (18-fold and 8-fold, respectively, Fig 2C). CYP1A2 was induced 1.7-fold, but only in the knockout mice (Fig 2C).
Finally, a group of enzymes comprising CYPs 2A5, 2B9, 2C29, 2D22, 2E1, and 3A13 showed no significant differences between untreated and infected mice of either genotype (Suppl. Fig. 2).
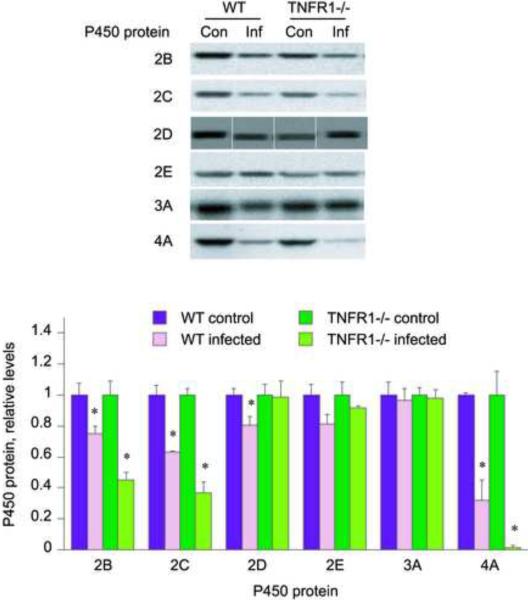
Hepatic P450 protein levels were measured by Western blotting to determine if the observed mRNA regulation was manifested by similar changes in the cognate proteins. CYP2E and 3A protein levels (Fig. 3) showed no significant changes in either wild-type or TNFR1−/− infected groups. CYPs 2B, 2C, and 4A protein levels were all down-regulated in the wild-type infected mice, and the down-regulation was potentiated in the TNFR1−/− treated mice, especially for CYP4A. Thus, CYP2B, 2C and 4A protein levels in wild-type infected mice were 75%, 63% and 43%, respectively, of the levels of the uninfected mice, but CYP2B, 2C and 4A expression fell to 45%, 37% and 4%, respectively in TNFR1−/− infected mice when compared to their control groups. CYP2D protein levels were significantly down-regulated in the wild-type infected group (80% of untreated) but remained unchanged in TNFR1−/− infected mice.
Fig. 3.
Effect of TNFR1 gene deletion on responses of P450 proteins in mouse liver to oral C. rodentium infection. Mice were orally infected with C. rodentium, and livers were harvested 7 days later for measurement of P450 protein levels by Western blotting as described under Materials and Methods. Values represent means ± S.E.M. of 6 mice per group. A, Western blots of samples; B, quantitative analysis of data in A. *, P<0.05 compared with control group of same genotype. Differences between groups were determined by Student's t test.
3.1.4. Hepatic cytokine and acute-phase mRNAs
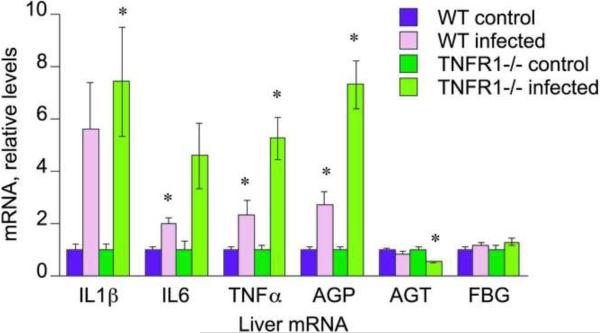
IL1β, IL6 and TNFα mRNAs, as well as the acute phase protein α1-acid glycoprotein (AGP), all showed similar patterns of regulation, with their induction by infection being potentiated in the TNFR1−/− mice (Fig. 4). Angiotensinogen (AGT) was significantly down-regulated in the TNFR1−/− treated mice (50% of untreated levels), with no significant change observed in the wild-type infected group. Fibrinogen (FGB) levels were unaffected by treatment (Fig. 4).
Fig. 4.
Effect of oral C. rodentium infection on P450 cytokine and acute-phase protein mRNAs in WT and TNFR1−/− mouse liver. Mice were orally infected with C. rodentium, and livers were harvested 7 days later for measurement of P450 mRNA levels as described under Materials and Methods. Values represent means ± S.E.M. of 6 mice per group. *, P<0.05 compared with control group of same genotype. Differences between groups were determined by Student's t test.
3.2. Oral Infection with C. rodentium in IL1R−/− Mice
To test the role of IL1β in hepatic P450 regulation, a separate analogous study was conducted in IL1R−/− mice. The results are presented in Supplemental Fig. 3. In this study, the effects of C. rodentium in WT mice were not as robust as we usually observe. For example, down-regulation of the CYP4A mRNAs was not statistically significant. However, dramatic down-regulation of CYP4A10 and 4A14 was observed in the livers of the IL1−/− mice. CYPs 1A2, 2B9, 2D22, 2E1, 3A11, 3A25 and 3A41 mRNAs were also significantly down-regulated only in the IL1−/− mice. CYP2A5 was induced in the WT mice, but was unaffected in the IL1−/− mice
3.3. Effects of Cytokines on P450 Expression in Cultured Mouse Hepatocytes
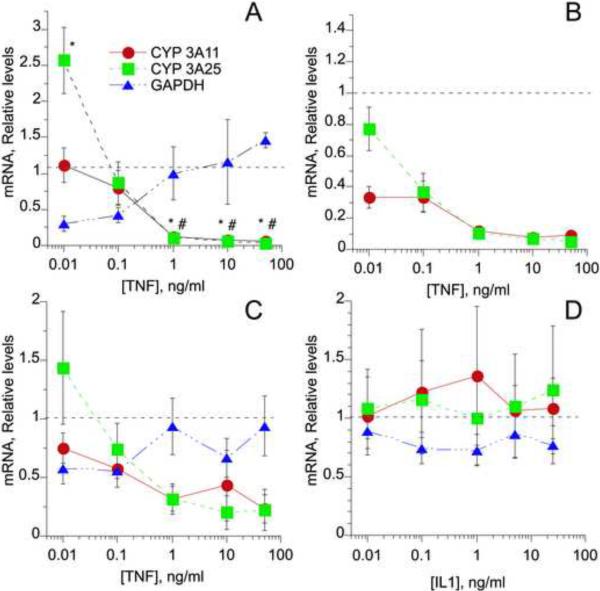
Since the preceding results provided evidence for participation of TNFα in the regulation of CYP3A11 and 3A25 during C. rodentium infection, we tested the abilities of TNFα and IL1β to regulate these P450s in cultured mouse hepatocytes. Several experiments were conducted to optimize the experimental conditions. Time course experiments revealed that hepatocyte exposure to TNFα for 24 hours caused reliable and significant regulation of several P450 mRNA levels (data not shown). Concentration dependence studies monitored the effects of hepatocyte exposure to various concentrations of TNFα over a 24 hour period (Fig. 5). At low concentrations of TNFα (0.01 ng/mL), mRNA levels of CYP3A25 were significantly induced, 2.5-fold over control levels (Fig5A), whereas increasing concentrations of TNFα caused significant and almost complete down-regulation of CYPs 3A11 and 3A25, with an approximate EC50 of between 0.1 and 1 ng/ml in each case. GAPDH mRNA levels are commonly used as a reference for RT-QPCR measurements. In our hands, 10 and 100 pg/ml TNFα down-regulated GAPDH mRNA, and this could be responsible for the apparent increase in CYP3A25 as these concentrations. When relative levels of CYP3A11 and 3A25 mRNA were calculated without GAPDH normalization, both mRNAs were seen to be potently down-regulated in the hepatocytes (Fig 5B). The effects seen in the experiment in Fig. 5A were also observed when the results of 4 independent experiments were pooled (Fig. 5C).
FIG. 5.
Cytokine regulation of CYP3A mRNAs in mouse hepatocyte cultures. Hepatocytes cultured as described in the text were incubated with different concentrations of TNFα or IL1β for 24 h, and hepatic mRNA expression was measured by quantitative RT-PCR. A, Data from a single TNFα experiment, in which each data point is the mean ± S.E.M. of measurements from 6 separate culture plates.; B, Data from the same experiment as in A, in which CYP3A11 and 3A25 expression was calculated without normalization to GAPDH mRNA; C, Averaged results from 4 independent experiments with hepatocytes from different mice. D. Averaged results from 4 independent experiments with hepatocytes treated with IL1. *, P<0.05 significantly different from untreated for 3A25; #, P<0.05 significantly different from untreated for 3A11. Differences between groups were determined by Student's t test.
In contrast to the potent effects of TNFα on CYP3A11 and 3A25 expression, IL1β treatment had no consistent effect on the expression of these mRNAs (Fig. 5D).
3.4. Effects of Kupffer Cell Depletion on Responses of P450 Genes to C. rodentium Infection in Mice
In the LPS model of sterile inflammation, depletion of Kupffer cells with gadolinium chloride blocked hepatic mRNA induction of IL6, IL1β, and TNFα, as well as the down-regulation of protein, catalytic activity, and mRNA levels of CYP3A11 [32]. These findings suggest that Kupffer cell-derived cytokines may play a role in the down-regulation of CYP3As in the LPS model of inflammation [32]. Therefore, we set out to determine whether or not activation of Kupffer cells might contribute to the regulation of hepatic P450 infection during C. rodentium infection. Because gadolinium chloride has been reported to transiently activate Kupffer cells [33], we used liposome-encapsulated dichloromethylene diphosphonate (clodronate) liposomes as they do not cause this complication [26]. Clodronate does not cross cell membranes, has a short half-life in the circulation and body fluids, and does not affect non-phagocytic cells. Also, gadolinium chloride has to be given intravenously, whereas clodronate liposomes are effective when injected i.p. [26]. This was important for our study because multiple treatments were necessary to ensure continued Kupffer cell ablation over the time course of infection.
A preliminary experiment involving 2 groups of mice (3 mice injected with 200 μL sterile saline and 3 mice injected with 200 μL liposomes), was conducted to verify depletion of Kupffer cells. Since Kupffer cells begin to repopulate 5 days after depletion [27], the injections were repeated every 4th day. Kupffer cells were present in mice injected with sterile saline but were severely depleted in liposome-injected mice (data not shown). Therefore, mice were injected i.p. with the clodronate liposomes 2 days before receiving a drinking solution containing either sucrose or C. rodentium, and injections were repeated every 4th day after the initial injection until the end of the C. rodentium infection period. The C. rodentium infection period was extended to 9 days because our previous work had shown that maximal colon inflammation and hepatic P450 down-regulation occurs between 7 and 10 days.
3.4.1 Depletion of Kupffer cells and Bacterial Loads in the Liver and Colon
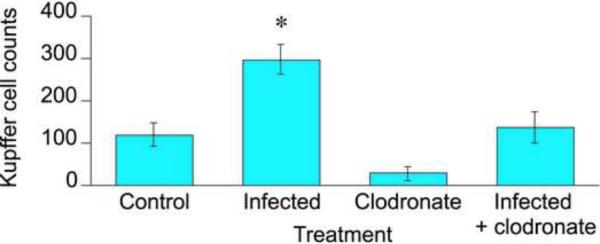
To assess the efficacy of the clodronate treatment in depletion of hepatic Kupffer cells, we subjected livers from the various groups to immunohistochemical analysis using an antibody that detects the glycoprotein F4/80 that is expressed on the surface of Kupffer cells [34]. Micrographs are shown in Supplemental Fig. 4. Clodronate treatment effectively depleted the Kupffer cells in the livers of uninfected mice (Fig. 6). C. rodentium infection significantly increased the number of Kupffer cells 2.5-fold in the livers of saline-injected mice. In clodronate-treated mice, there was a similar tendency towards an increase in the number of Kupffer cells during infection (4.5-fold), but the effect was not statistically significant (Fig. 6). Clodronate treatment significantly reduced the number of Kupffer cells in livers of infected mice, but not to the level of uninfected, clodronate-treated mice.
Fig. 6.
Depletion of Kupffer cells by clodronate liposomes in mice orally infected with C. rodentium. Mice were injected i.p. with 100 μL of sterile saline or clodronate liposomes two days before receiving sucrose (Uninfected) or C. rodentium (Infected) in the drinking water. Mice were injected with sterile saline or clodronate liposomes every 4 days during C. rodentium infection, and livers were harvested 9 days after initial treatment with sucrose or C. rodentium for detection of Kupffer cells as described under Materials and Methods. Values represent means ± S.E.M. of 4 mice per group. *, P<0.05 compared with untreated. Differences between groups were determined by Student's t test.
Clodronate-treated infected mice had half the number of bacterial colonies in their colons compared to saline-injected, infected mice (Supplemental Fig. 5). Conversely, the number of bacterial colonies in livers of in clodronate injected, infected mice was one quarter that of the saline-injected mice (Supplemental Fig. 5). The number of colonies in colons of infected mice of either genotype was 8,000–10,000 times as high as in the number of colonies in their own livers (Supplemental Fig. 5).
3.4.2. Serum Cytokines
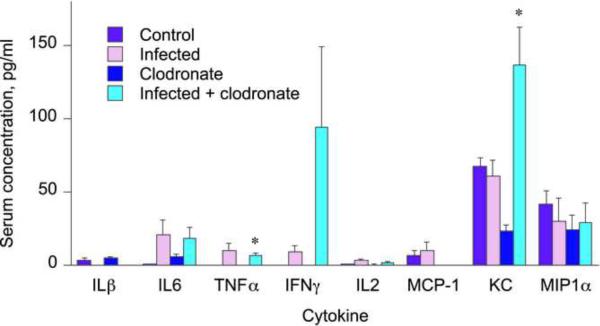
In this experiment C. rodentium infection was not associated with significant increases in MCP-1, KC, MIP-1α, INFγ, IL1β, IL2, IL6, or TNFα serum levels (Fig. 7), although there were trends towards increases for IL6, TNFα, IFNγ and IL2. The clodronate injected, infected mice showed significant increases in the levels of KC and TNFα compared to clodronate injected, uninfected mice (Fig. 7). Granulocyte-colony stimulating factor serum levels were increased by infection in both clodronate-treated and–untreated mice, with the levels being slightly lower in the clodronate-treated mice (not shown).
Fig. 7.
Serum cytokine profile during oral Citrobacter rodentium infection in Kupffer cell depleted mice. Blood was collected at sacrifice for serum cytokine analysis as described under Materials and Methods. Values represent means ± S.E.M. of 4 mice per group. *, P<0.05 compared with untreated control group of same genotype Differences between groups were determined by Student's t test
3.4.3. Hepatic mRNA Expression Levels
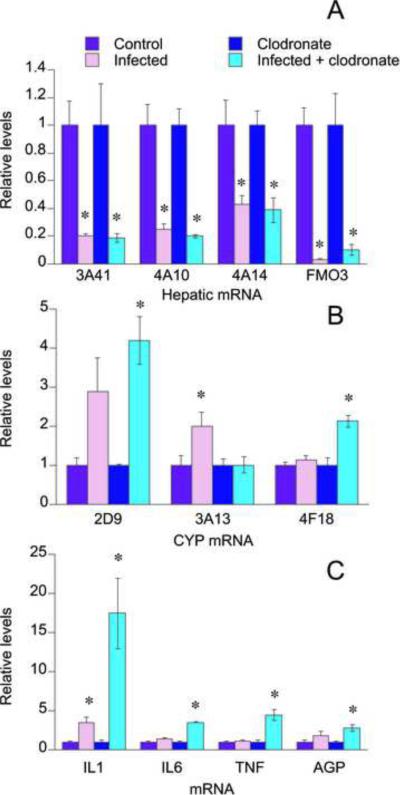
The mRNA levels for FMO3 as well as CYPs 3A41, 4A10, and 4A14 were all significantly down-regulated to a similar extent in the saline injected and clodronate injected infected mice (Fig. 8A). CYP3A11 and 3A25 mRNAs also tended to show a down-regulation in both infected groups, but in this study it did not achieve significance (not shown). CYP3A13 mRNA levels were significantly induced (2-fold increase over control) in the saline injected, infected groups, while the induction was absent in the clodronate injected, infected group (Fig. 8B). CYP4F18 and CYP2D9 mRNA levels were only significantly elevated in the clodronate injected, infected mice. CYP1A2 as well as acute phase proteins FBG and AGT, did not show a significant change in mRNA levels (not shown). Hepatic IL6, TNFα, and the acute phase protein AGP mRNA levels were only significantly elevated in the clodronate injected, infected mice at levels that were approximately 3.5, 4.5, and 3 fold higher, respectively, over control levels (Fig. 8C). IL1β mRNA was significantly elevated in both groups, but to a higher extent in the clodronate-treated mice (Fig 8C).
Fig. 8.
Liver mRNA levels in Kupffer cell depleted mice orally infected with C. rodentium. Mice were orally infected with C. rodentium 2 days after i.p. injection of clodronate liposomes or sterile saline as described in Fig. 6, and livers were harvested 9 days later for measurement of mRNA levels as described under Materials and Methods. Values represent means ± S.E.M. of 6 mice per group. A, CYPs 3A41, 4A10, 4A14 and flavin monooxygenase 3 (FMO3); B, CYPs 2D9, 3A13, and 4F18; C, Cytokines interleukin 1 (IL1), interleukin 6 (IL6), and tumor necrosis factor alpha (TNFα), and acute phase protein α1-acid glycoprotein. *, P<0.05 compared with uninfected control (with or without clodronate as appropriate). Differences between groups were determined by Student's t test.
4. Discussion
The current studies investigated the role of TNFα, IL1β and Kupffer cells in the regulation of murine hepatic cytochrome P450 during oral infection with C. rodentium. Studies in the TNFR1−/− mice provided clear evidence for an in vivo role of TNFα and its p55 receptor TNFR1 in the down-regulation of CYP3A11, 3A25 and 3A41 mRNAs in this mouse model of human food poisoning. The down-regulation of CYP3A11 and 3A25 was ablated, and that of CYP3A41 was attenuated, in the mice lacking TNFR1. This was supported by demonstration of potent and complete down-regulation of P450s 3A11 and 3A25 by TNFα in cultured mouse hepatocytes. In general, C. rodentium infection exhibited greater effects in mice lacking the IL1R or mice treated with clodronate liposomes, than in WT mice, suggesting that neither IL1 nor Kupffer cells are necessary for hepatic P450 regulation in C. rodentium infection.
The lack of effect of C. rodentium infection on CYP3A11, 3A25 and 3A41 mRNAs in TNFR1−/− mice was clearly specific for these mRNAs, and this observation was not due to reduced colonic inflammation or bacterial colonization in the TNFR1 −/− animals. In fact, it was reported that TNFR1−/− mice infected with C. rodentium developed more severe pathology compared to WT mice, with increased bacterial loads and longer crypt lengths indicating mucosal hyperplasia in their colons [16]. Here, we found similar numbers of C. rodentium colonies in the colons of WT and TNFR1−/− infected mice, and significantly higher crypt lengths in both groups when compared to their own controls (Supplemental Fig. 1). MPO activity was only elevated significantly in the TNFR1−/− infected group (Supplemental Table 1). Thus, our indices of colon pathology suggest similar levels of colonic inflammation and colonization in wild-type and TNFR1−/− infected mice, The lower level of hepatic bacterial colony counts in TNFR1−/− infected mice than in wild-type infected mice (Supplemental Fig. 1) could contribute to the differences between the strains in their CYP3A responses. However, hepatic cytokine mRNAs and the acute-phase mRNA AGP were elevated to a greater extent in the TNFR1−/− mice, suggesting a higher degree of liver inflammation in this strain. This point is emphasized by the fact that the down-regulation of FMO3 and CYPs 4A10, 4A14, as well as the induction of CYPs 2D9 and 4F18, were potentiated in the knockout mice indicating a lack of a direct role for TNFα in their regulation. In this respect, it is also important to note that the absence of TNFR1 did not significantly affect the profile of cytokines in the plasma during infection.
The notion that TNFα is the primary factor down-regulating CYPs 3A11 and 3A25 is further supported by our observation of potent down-regulation of these P450s in cultured mouse hepatocytes. Nevertheless, the concentrations of TNFα required to down-regulate these P450s in culture (Fig. 5) were 10-100-fold higher than concentrations observed in plasma during infection (Figs 1, 7). Plasma cytokine levels are a measure of TNF production, but are not necessarily the same as those the hepatocytes experience during an infection. Because this is a GI infection, we would expect portal blood cytokine levels to be higher than those in the general circulation. It is also possible that cultured hepatocytes are less sensitive to TNF than are hepatocytes in vivo.
In the experiment using TNFR1−/− mice, CYPs 2A5, 2B9, 2C29, 2D22, 2E1, and 3A13 mRNA levels (Fig. 2) were not affected at statistically significant levels by infection with C. rodentium in either wild-type or TNFR1−/− mice, even though clear signs of infection and inflammation were documented. In our previous publication, CYPs 2B9, 2C29, and 2D22 were down-regulated in infected C57BL/6 mice [14], while CYP3A13 was slightly induced. It is germane to note that the magnitudes of down-regulation of CYP4A10, CYP4A14 and FMO3, and of induction of CYP2D9 and CYP4F18, were all smaller in the current study than in our previous one [14]. Therefore, we believe that the generally reduced effects on WT mice in the current study (also in the ensuing clodronate and IL1R experiments) is due to a nonspecific decrease in the severity of infection. For this reason, the potential role of TNFα in the regulation of CYPs 2B9, 2C29, 2D22 and CYP3A13 could not be determined. CYP1A2 mRNA levels were induced in TNFR1−/− infected mice while no changes were observed in infected wild-type mice. This could suggest that in WT mice, TNFα exerts a suppressive effect that masks the inductive effects of other inflammatory or infectious mediators.
Studies in TNFα-null mice previously suggested a role for TNFα in the down-regulation of CYP3A11 and CYP2C29 that occurs during aseptic inflammation caused by injection of the tuberculosis vaccine Bacillus Calmette-Guerin (BCG) [6]. Down-regulation of CYP3A11 mRNA expression was attenuated in TNFα knock-out mice treated with BCG compared to WT (46% of control versus 21% of control) [6]. CYP2C29 mRNA expression levels were significantly down-regulated in wild-type BCG treated mice (52% of controls), but not in TNFα knock-out mice treated with BCG [6]. Our work demonstrates the role of TNFα in the regulation of CYP3A11 as well as CYP3A25 and 3A41 in a different model of infectious inflammation, and further shows that this occurs via TNFR1. On the other hand, studies in double receptor knock-out mice (TNFR1−/−/TNFR2−/−) [4] concluded that TNFα is not required for the effects of LPS injection on CYPs 1A, 2B, 3A, and 4A expression in mouse liver, and studies involving TNFα null mice suggested that TNFα may play a protective role in the regulation of hepatic CYP3A and CYP2C proteins in LPS-induced inflammation [35]. The involvement of TNFα in regulating expression of murine CYPs 3A11 and 3A25 during C. rodentium infection and CYPs 3A11 and 2C29 after BCG administration, as well as conflicting results in LPS models, clearly demonstrates the selective nature of cytokine-dependent regulation on P450 expression in different models of inflammation. Our study does not exclude the participation of TNFα, acting through TNFR2, in the regulation of other P450s during C. rodentium infection, although this seems unlikely given the restricted expression and function of TNFR2
Western blots were performed to observe whether the changes in mRNA expression of specific mouse P450s were also reflected in the expression of P450 proteins. Unfortunately, the lack of specific antibody tools makes solid conclusions difficult. CYP2E1 (unaffected) and CYP4A (down-regulated) protein expression levels followed a similar trend as the mRNA expression levels for CYP2E1 and CYPs 4A10 and 4A14. However, C. rodentium infection had no effect on protein levels detected by the CYP3A antibody, despite 50% down-regulation of mRNA levels for CYP 3A11, 3A25 and 3A41. This could indicate that either the antibody preferentially recognizes other CYP3As in mouse liver, or that up-regulation or no effect on other mouse CYP3As (e.g. CYP3A13 in this study) masks the down-regulation. On the other hand, protein levels for CYPs 2B, and 2D were down-regulated in infected wild-type mice, and the down-regulation is potentiated in infected TNFR1−/− mice. This could suggest that C. rodentium infection may lead to post-translational regulation of the above mentioned proteins, or that the P450 mRNAs measured from these particular subfamilies are not the predominant forms expressed at the protein level. Finally, the large relative increase in CYP2D9 mRNA expression, especially in pTNFR1−/− mice, is not reflected in an increase in CYP2D protein. The reason for this is not known, but may reflect that CYP2D9 is a male-specific gene, and so even a 20-fold induction of its mRNA in females may not achieve a high absolute level of expression.
In a previous publication, we reported that the down-regulations of CYP3A11 and CYP3A25 were attenuated in livers of IFNγ-null mice [36]. Based on the facts that the infected IFNγ-null mice lacked TNFα in their sera, and that IFNγ was ineffective in regulation of these P450 mRNAs in cultured hepatocytes, we postulated that the lack of down-regulation of the CYP3As in IFNγ-null mice might reflect an in vivo role of TNFα [36]. The current work strongly supports this idea.
The studies in IL1R-null mice were hampered by very weak observed effects in WT animals. Nevertheless, it was clear that for those P450s that are commonly down-regulated in C. rodentium infection, the absence of IL1R potentiated the down-regulation. Previous studies with IL1R −/− mice infected with C. rodentium revealed increased colonic damage and increased mortality after 7 days of infection [37]. While the infected IL1R −/− mice in the current study demonstrated colonic bleeding but no premature death. These observations, together with the inconsistent effects of IL1 on CYP3A11 and 3A25 expression in mouse hepatocytes and the lack of an increase in circulating levels of IL1β in infected mice (this study and [14]), all argue against a significant role of IL1β in hepatic P450 regulation in this model. The possible exception was CYP2A5, which was induced in the WT mice but not in the IL1R null mice. Our previous study in C57BL/6 mice did not observe induction of this mRNA [14] but we did observe this previously in C3H/HeOuJ mice [13].
TNFα, as well as IL1β and IL6, are released from activated Kupffer cells [23, 24]. We hypothesized that activation of Kupffer cells derived P450 expression during C. rodentium infection. Therefore, we tested the effect of ablating Kupffer cells in this model of infection. While injection of clodronate liposomes in wild-type mice dramatically reduced the number of Kupffer cells in the livers of uninfected mice, infection with C. rodentium caused an increase in the number of Kupffer cells in the liver (Fig. 6). Although clodronate treatment reduced the number of Kupffer cells in the livers of infected mice (Fig. 7), these mice still had numbers of Kupffer cells that were similar to untreated mice, and this complicated the interpretation of our results. The down-regulations of CYP3A41, CYP4A10, CYP4A14 and FMO3 and the induction of CYP2D9 by infection were similar with or without clodronate treatment. These data suggest that Kupffer cells are not involved in the regulation of at least these mRNAs in the C. rodentium model. Also arguing against a significant activation of Kupffer cells in the C. rodentium model is the inconsistent and low-level induction of hepatic cytokine and acute-phase mRNAs that we observed previously [14] and in this series of studies. Indeed, down-regulation of CYP3A41, CYP4A10, CYP4A14 and FMO3 in wild-type mice was observed in the absence of significant induction of hepatic cytokine mRNAs (Fig. 8). Although C. rodentium infection increases the number of Kupffer cells, this increase may not correlate with more activated cells or with induction of cytokine levels. Clodronate treatment potentiated the induction of the cytokine mRNAs even though it lowered the Kupffer cell numbers in infected mice. These observations suggest that cytokines produced in the liver are not a major factor in this model of infection, and that these cytokines may be produced by a cell type other than Kupffer cells. A solid conclusion awaits a more efficient method to ablate Kupffer cells in infected mice.
In conclusion, TNFα plays a role in the regulation of hepatic mRNA levels for CYPs 3A11, 3A25 during oral infection with Citrobacter rodentium, while IL1 does not appear to be relevant to hepatic P450 levels in this model of inflammation. Though Kupffer cells are induced during oral C. rodentium infection, they may not be the source of TNFα involved in the regulation of hepatic P450s. Additional studies are needed to explore the source and mechanism of TNFα in hepatic P450 regulation during C. rodentium infection.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants DK072372 and DK072372-S1 to ETM. We are grateful to our colleagues Beatrice Nyagode for advice on cultured mouse hepatocytes; Daniel Kalman and Sara Lebeis for the IL1R−/− mice; and Malik Raynor for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lewis DFV. Guide to cytochrome P450 structure and function. Taylor & Francis; London; New York: 2001. [Google Scholar]
- [2].Renton KW. Regulation of drug metabolism and disposition during inflammation and infection. Expert Opin Drug Metab Toxicol. 2005;1:629–40. doi: 10.1517/17425255.1.4.629. [DOI] [PubMed] [Google Scholar]
- [3].Aitken AE, Richardson TA, Morgan ET. Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu Rev Pharmacol Toxicol. 2006;46:123–49. doi: 10.1146/annurev.pharmtox.46.120604.141059. [DOI] [PubMed] [Google Scholar]
- [4].Warren GW, Poloyac SM, Gary DS, Mattson MP, Blouin RA. Hepatic cytochrome P-450 expression in tumor necrosis factor-alpha receptor (p55/p75) knockout mice after endotoxin administration. J Pharmacol Exp Ther. 1999;288:945–50. [PubMed] [Google Scholar]
- [5].Siewert E, Bort R, Kluge R, Heinrich PC, Castell J, Jover R. Hepatic cytochrome P450 down-regulation during aseptic inflammation in the mouse is interleukin 6 dependent. Hepatology. 2000;32:49–55. doi: 10.1053/jhep.2000.8532. [DOI] [PubMed] [Google Scholar]
- [6].Ashino T, Oguro T, Shioda S, Horai R, Asano M, Sekikawa K, et al. Involvement of interleukin-6 and tumor necrosis factor alpha in CYP3A11 and 2C29 down-regulation by Bacillus Calmette-Guerin and lipopolysaccharide in mouse liver. Drug Metab Dispos. 2004;32:707–14. doi: 10.1124/dmd.32.7.707. [DOI] [PubMed] [Google Scholar]
- [7].Richardson TA, Morgan ET. Hepatic cytochrome P450 gene regulation during endotoxin-induced inflammation in nuclear receptor knockout mice. J Pharmacol Exp Ther. 2005;314:703–9. doi: 10.1124/jpet.105.085456. [DOI] [PubMed] [Google Scholar]
- [8].Chaluvadi MR, Nyagode BA, Kinloch RD, Morgan ET. TLR4-dependent and - independent regulation of hepatic cytochrome P450 in mice with chemically induced inflammatory bowel disease. Biochem Pharmacol. 2009;77:464–71. doi: 10.1016/j.bcp.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Goralski KB, Abdulla D, Sinal CJ, Arsenault A, Renton KW. Toll-like receptor-4 regulation of hepatic Cyp3a11 metabolism in a mouse model of LPS-induced CNS inflammation. Am J Physiol Gastrointest Liver Physiol. 2005;289:G434–43. doi: 10.1152/ajpgi.00562.2004. [DOI] [PubMed] [Google Scholar]
- [10].Ghose R, White D, Guo T, Vallejo J, Karpen SJ. Regulation of hepatic drug-metabolizing enzyme genes by Toll-like receptor 4 signaling is independent of Toll-interleukin 1 receptor domain-containing adaptor protein. Drug Metab Dispos. 2008;36:95–101. doi: 10.1124/dmd.107.018051. [DOI] [PubMed] [Google Scholar]
- [11].Armstrong S, Renton KW. Hepatic cytochrome P450 and related drug biotransformation during an outbreak of mouse hepatitis virus in a colony of Swiss BALB/c mice. Can J Physiol Pharmacol. 1993;71:188–90. doi: 10.1139/y93-027. [DOI] [PubMed] [Google Scholar]
- [12].Armstrong SG, Renton KW. Mechanism of hepatic cytochrome P450 modulation during Listeria monocytogenes infection in mice. Mol Pharmacol. 1993;43:542–7. [PubMed] [Google Scholar]
- [13].Richardson TA, Sherman M, Antonovic L, Kardar SS, Strobel HW, Kalman D, et al. Hepatic and renal cytochrome p450 gene regulation during citrobacter rodentium infection in wild-type and toll-like receptor 4 mutant mice. Drug Metab Dispos. 2006;34:354–60. doi: 10.1124/dmd.105.007393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chaluvadi MR, Kinloch RD, Nyagode BA, Richardson TA, Raynor MJ, Sherman M, et al. Regulation of hepatic cytochrome P450 expression in mice with intestinal or systemic infections of citrobacter rodentium. Drug Metab Dispos. 2009;37:366–74. doi: 10.1124/dmd.108.024240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Higgins LM, Frankel G, Douce G, Dougan G, MacDonald TT. Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect Immun. 1999;67:3031–9. doi: 10.1128/iai.67.6.3031-3039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Goncalves NS, Ghaem-Maghami M, Monteleone G, Frankel G, Dougan G, Lewis DJ, et al. Critical role for tumor necrosis factor alpha in controlling the number of lumenal pathogenic bacteria and immunopathology in infectious colitis. Infect Immun. 2001;69:6651–9. doi: 10.1128/IAI.69.11.6651-6659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Craig PI, Mehta I, Murray M, McDonald D, Astrom A, van der Meide PH, et al. Interferon down regulates the male-specific cytochrome P450IIIA2 in rat liver. Mol Pharmacol. 1990;38:313–8. [PubMed] [Google Scholar]
- [18].Muntane-Relat J, Ourlin JC, Domergue J, Maurel P. Differential effects of cytokines on the inducible expression of CYP1A1, CYP1A2, and CYP3A4 in human hepatocytes in primary culture. Hepatology. 1995;22:1143–53. [PubMed] [Google Scholar]
- [19].Sewer MB, Morgan ET. Nitric oxide-independent suppression of P450 2C11 expression by interleukin-1beta and endotoxin in primary rat hepatocytes. Biochem Pharmacol. 1997;54:729–37. doi: 10.1016/s0006-2952(97)00226-8. [DOI] [PubMed] [Google Scholar]
- [20].Aitken AE, Morgan ET. Gene-specific effects of inflammatory cytokines on cytochrome P450 2C, 2B6 and 3A4 mRNA levels in human hepatocytes. Drug Metab Dispos. 2007;35:1687–93. doi: 10.1124/dmd.107.015511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].MacEwan DJ. TNF receptor subtype signalling: differences and cellular consequences. Cell Signal. 2002;14:477–92. doi: 10.1016/s0898-6568(01)00262-5. [DOI] [PubMed] [Google Scholar]
- [22].Ernandez T, Mayadas TN. Immunoregulatory role of TNFalpha in inflammatory kidney diseases. Kidney Int. 2009;76:262–76. doi: 10.1038/ki.2009.142. [DOI] [PubMed] [Google Scholar]
- [23].Decker K. Biologically active products of stimulated liver macrophages (Kupffer cells) Eur J Biochem. 1990;192:245–61. doi: 10.1111/j.1432-1033.1990.tb19222.x. [DOI] [PubMed] [Google Scholar]
- [24].Ramadori G, Armbrust T. Cytokines in the liver. Eur J Gastroenterol Hepatol. 2001;13:777–84. doi: 10.1097/00042737-200107000-00004. [DOI] [PubMed] [Google Scholar]
- [25].Van Rooijen N, Van Nieuwmegen R. Elimination of phagocytic cells in the spleen after intravenous injection of liposome encapsulated dichloro-methylenediphosphonate. An enzyme-histochemical study. Cell and Tissue Research. 1984;238:355–8. doi: 10.1007/BF00217308. [DOI] [PubMed] [Google Scholar]
- [26].Van Rooijen N, Sanders A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. J Immunol Methods. 1994;174:83–93. doi: 10.1016/0022-1759(94)90012-4. [DOI] [PubMed] [Google Scholar]
- [27].Fraser CC, Chen BP, Webb S, van Rooijen N, Kraal G. Circulation of human hematopoietic cells in severe combined immunodeficient mice after Cl2MDP-liposome-mediated macrophage depletion. Blood. 1995;86:183–92. [PubMed] [Google Scholar]
- [28].Zhang J, Chaluvadi MR, Reddy R, Motika MS, Richardson TA, Cashman JR, et al. Hepatic flavin-containing monooxygenase gene regulation in different mouse inflammation models. Drug Metab Dispos. 2009;37:462–8. doi: 10.1124/dmd.108.025338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [30].Vallance BA, Deng W, Jacobson K, Finlay BB. Host susceptibility to the attaching and effacing bacterial pathogen Citrobacter rodentium. Infect Immun. 2003;71:3443–53. doi: 10.1128/IAI.71.6.3443-3453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Malaviya R, Abraham SN. Role of mast cell leukotrienes in neutrophil recruitment and bacterial clearance in infectious peritonitis. J Leukoc Biol. 2000;67:841–6. doi: 10.1002/jlb.67.6.841. [DOI] [PubMed] [Google Scholar]
- [32].Xu DX, Wei W, Sun MF, Wu CY, Wang JP, Wei LZ, et al. Kupffer cells and reactive oxygen species partially mediate lipopolysaccharide-induced downregulation of nuclear receptor pregnane x receptor and its target gene CYP3a in mouse liver. Free Radic Biol Med. 2004;37:10–22. doi: 10.1016/j.freeradbiomed.2004.03.021. [DOI] [PubMed] [Google Scholar]
- [33].Ruttinger D, Vollmar B, Wanner GA, Messmer K. In vivo assessment of hepatic alterations following gadolinium chloride-induced Kupffer cell blockade. J Hepatol. 1996;25:960–7. doi: 10.1016/s0168-8278(96)80302-3. [DOI] [PubMed] [Google Scholar]
- [34].Austyn JM, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol. 1981;11:805–15. doi: 10.1002/eji.1830111013. [DOI] [PubMed] [Google Scholar]
- [35].Miyoshi M, Nadai M, Nitta A, Ueyama J, Shimizu A, Takagi K, et al. Role of tumor necrosis factor-alpha in down-regulation of hepatic cytochrome P450 and P-glycoprotein by endotoxin. Eur J Pharmacol. 2005;507:229–37. doi: 10.1016/j.ejphar.2004.11.035. [DOI] [PubMed] [Google Scholar]
- [36].Nyagode BA, Lee C-M, Morgan ET. Modulation of Hepatic Cytochrome P450s by Citrobacter rodentium Infection in Interleukin-6- and Interferon-γ-null. Mice J Pharmacol Exp Ther. 2010 doi: 10.1124/jpet.110.171488. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lebeis SL, Powell KR, Merlin D, Sherman MA, Kalman D. Interleukin-1 receptor signaling protects mice from lethal intestinal damage caused by the attaching and effacing pathogen Citrobacter rodentium. Infect Immun. 2009;77:604–14. doi: 10.1128/IAI.00907-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.