Abstract
A central problem in de novo drug design is determining the binding affinity of a ligand with a receptor. A new scoring algorithm is presented that estimates the binding affinity of a protein-ligand complex given a three-dimensional structure. The method, LISA (Ligand Identification Scoring Algorithm), uses an empirical scoring function to describe the binding free energy. Interaction terms have been designed to account for van der Waals (VDW) contacts, hydrogen bonding, desolvation effects and metal chelation to model the dissociation equilibrium constants using a linear model. Atom types have been introduced to differentiate the parameters for VDW, H-bonding interactions and metal chelation between different atom pairs. A training set of 492 protein-ligand complexes was selected for the fitting process. Different test sets have been examined to evaluate its ability to predict experimentally measured binding affinities. By comparing with other well known scoring functions, the results show that LISA has advantages over many existing scoring functions in simulating protein-ligand binding affinity, especially metalloprotein-ligand binding affinity. Artificial Neural Network (ANN) was also used in order to demonstrate that the energy terms in LISA are well designed and do not require extra cross terms.
Keywords: Empirical scoring function, Artificial Neural Network
Introduction
Structure-based drug design, especially de novo drug design, has made significant strides in the past several decades. New lead compounds are designed based on the structure of proteins and by applying the principles of molecular recognition such as optimization of van der Waals interactions and hydrogen bonding. A good candidate for a drug molecule should have an appropriate binding affinity for its target receptor, which is typically in the low nanomolar (nM) range. While reported pKi values range between 3 and 10,1 binding affinities weaker than the nM range may result in modest efficacy while affinity for multiple targets could lead to undesired side effects. Additionally, with the development of fragment based methods,2,3 small molecules (< 200–300 MW) are being studied that generally have low affinity. Thus the chemical space of interest to medicinal chemists covers a wide range of binding affinites. Being able to accurately predict the binding affinity for these molecules is a central problem of structure-based drug design and remains a very significant scientific challenge.5–7
There are three types of scoring functions, physics based,8–11 knowledge-based,12–20 and empirical.21–25 Physics based scoring functions are parameterized using quantum mechanical calculations including available experimental information. These scoring functions are also known as “force-field” scoring functions where each term has a physical significance. However, these functions are computationally intensive relative to simpler scoring functions because the associated energy landscapes associated are generally rugged and therefore expensive to evaluate.26,27 Knowledge-based scoring functions use statistical atom pair potentials derived from structural databases to yield a score. However, these atom-pair potentials, known as the potentials of mean force (PMF) are typically pair-wise (the probability of finding atoms A and B at a distance r), which means it unfortunately ignores the influence from surrounding atoms. Another limitation arises from the inaccessibility of the reference state where the interatomic interactions are zero. Moreover, knowledge-based scoring functions assume that all of the distance space is accounted for, which given the limited dataset available is unlikely to be the case for all chemotypes encountered in the drug discovery process. Empirical scoring functions are computationally efficient, because of their simple energy functions, but this also highlights their major limitation, the training-set-dependent parameterization. Energy functions with simple forms can mask the relationship between binding affinity and the crystal structures used to build the model. Hence, the training process only derives parameters that represent a compromise between the simplicity of the energy expression and the nuances of the interactions seen in protein-ligand complexes.
In this work, we have developed a new empirical scoring function that is readily applicable to de novo drug design. This function, Ligand Identification Scoring Algorithm (LISA), aims to compensate for the common disadvantages of empirical scoring functions, with a focus towards Zn metalloproteins. Because van der Waals (VDW) interactions and hydrogen bonding are very important in protein-ligand complexes, different atom types have been introduced in order to simulate these interactions between different atoms. A desolvation term has also been included in order to capture solvation changes resulting from protein-ligand complexation. Among protein-ligand complexes with high binding affinity (pKd>7), metal chelation between active-site zinc ions and metal-binding “warheads” (e.g., carboxylate, sulphonamides, etc.) in ligands is widely observed; hence, we have also built a Zn chelation term into LISA to capture this class of interactions.
Methods
Training set
It is well appreciated that both the size and the quality of a protein-ligand training set will affect the final form and effectiveness of a scoring function. Since our scoring function focuses on the evaluation of binding affinity for de novo drug design, the ligands in our training set are required to have binding affinity data and structures that reflect what drug candidates experience upon complexation. Hence in this work, we have chosen our training set from the PDBbind v2010 refined data set28,29 and have further curated this set to further ensure quality. (1) Every complex in the training set is a crystal structure with an overall X-ray resolution ≤ 2.5 Å. Complexes solved by NMR techniques are currently not included in our selection; (2) Crystal contacts are interchain or intermolecular contacts that occur as the result of the protein crystallization process, and they are found in all X-ray structures. Crystal contacts may influence the conformation of a protein and where a ligand binds to the protein, by providing further crystal contacts or even un-natural binding pockets. In order to avoid protein-ligand complexes affected by crystal contacts in our training set, we eliminated all complexes in our training set that Søndergaard et al.30 found to have crystal contacts affecting the ligand. (3) Only the complexes with Kd and Ki values have been introduced into our training set. Complexes with only IC50 values were not included. (4) We focused on complexes with pKi or pKd's distributed from 3 to 11, because binding affinities in this range are pharmaceutically relevant. Potential inconsistencies in Kd values due to experimental conditions such as pH, temperature, etc. were not taken into account. (5) Based on pharmacokinetic considerations, as embodied by Lipinski's rule-of-five, an orally administered drug should have no more than 5 hydrogen bond donors, no more than 10 hydrogen bond acceptors, an octanol-water partition coefficient log P of less than 5, and a molecular weight under 500 daltons.4 In the present research, we did not focus on the first three rules in order to not limit our training set. For example, in prodrug design, the hydrogen bond donor or acceptor atoms can be masked, and the logP value can change once in vivo. On the other hand, we did retain the MW rule because complexes with high MW ligands could have “fooled” our scoring function to become artificially dependent on MW through the van der Waals terms. Hence, we decided that complexes containing ligands with MW>600 would be excluded. 2061 complexes from PDBbind v2010 refined data set were filtered based on the above 5 criteria and 492 complexes were left in our training set. Both ligand and protein atoms were classified (atom-typed) as defined in Table 1.
Table 1.
List of 20 atom types in LISA
| Atom Type | Description | Hydrogen Bonding Donor/Acceptor? | VDW radii |
|---|---|---|---|
| C3 | sp3 hybridized carbon | N/A | 1.94 |
| C2 | sp2 hybridized carbon | N/A | 1.90 |
| C1 | sp hybridized carbon | N/A | 1.80 |
| Car | aromatic carbon | N/A | 1.85 |
| N4 | positively charged nitrogen | N/A (if no H bonded) / Donor (if H bonded) | 1.83 |
| N3 | sp3 hybridized nitrogen | N/A (if no H bonded) / Donor (if H bonded) | 1.87 |
| N2 | sp2 hybridized nitrogen | N/A (if no H bonded) / Donor (if H bonded) | 1.86 |
| N1 | sp hybridized nitrogen | N/A | 1.85 |
| Nar | aromatic nitrogen | N/A | 1.86 |
| Nam | amide nitrogen | N/A (if no H bonded) / Donor (if H bonded) | 1.83 |
| Npl3 | trigonal planar nitrogen | N/A (if no H bonded) / Donor (if H bonded) | 1.86 |
| O3 | sp3 hybridized oxygen | Acceptor (if no H bonded) / Donor-Acceptor (if H bonded) | 1.74 |
| O2 | sp2 hybridized oxygen | Acceptor | 1.66 |
| S | sulfur | N/A | 2.09 |
| P | phosphor | N/A | 2.03 |
| F | fluorine | N/A | 1.55 |
| Cl | chlorine | N/A | 2.00 |
| Br | bromine | N/A | 2.20 |
| I | Iodine | N/A | 2.40 |
| Zn | zinc cation | N/A | 1.20 |
Scoring Function
The empirical scoring functions in current use are based on the “Master Equation” model31, where overall binding free energy (ΔGbind - see eqn. 1) can be decomposed into independent free energy contributions (see eqn. 2). Each component is the sum of a certain type of structure-related empirical energy terms (the fi(x,y,z) term) multiplied by a weighting coefficient ci (see eqn. 3). The Master Equation represents the linear combination of these components.
| (1) |
| (2) |
| (3) |
In the current research, our scoring function was decomposed into the following interaction categories: van der Waals interactions, hydrogen bonding, desolvation (hydrophobic effect) and metal chelation.
van der Waals interactions
van der Waals interactions are one of the most important interactions present in protein-ligand complexes. The computed potential energy depends on the distances between pairs of atoms. The Lennard-Jones 6–12 term is employed in this work to reflect van der Waals interactions when two atoms approach during the binding process between a protein and a ligand.
| (4) |
| (5) |
In eqn. 4 and 5, rij is the distance between atom i in the protein and atom j in the ligand. σij is the interatomic separation at which repulsive and attractive forces balance (the sum of the van der Waals radii of atom i and atom j). ε is the potential well depth, subscripts A and B refer to atom type A and B. εAB can be expressed using εA and εB according to eqn. 6. Thus, based on the εA parameters taken, from the work of Cornell et al.32, we can compare our well depth values obtained via regression with force field based values.
| (6) |
We scanned for all of the atom contacts between the ligands and their protein receptors from the 2061 complexes found in the PDBbind v2010 data set and categorized those contacts based on the atom types listed in Table 1. Some interaction types were not included in this scoring function because (1) they rarely appeared as protein/ligand contacts; (2) they added little substantive improvement to our model based on linear regression results, whose 95% confidence intervals included 0. In the present work, VDW interactions between all pairs of atoms are calculated according to the atom types listed in Table 2.
Table 2.
List of atom types for the van der Waals interaction
| Atom Type | Description |
|---|---|
| C3 | sp3 hybridized carbon |
| C2 | sp2 hybridized carbon |
| Car | aromatic carbon |
| N4 | positively charged nitrogen |
| N3 | sp3 hybridized nitrogen |
| N2 | sp2 hybridized nitrogen |
| Npl3 | trigonal planar nitrogen |
| O3 | sp3 hybridized oxygen |
| O2 | sp2 hybridized oxygen |
| S | sulfur |
We set a distance cutoff of 3 to 5.5 Å to avoid non-physical attractive or repulsive forces. Moreover, to avoid the large repulsions introduced by overlapped atom pairs, we set an upper limit for fij(x,y,z). Cutoffs of 0.5, 1, and 5 pKd units have all been tested, and a value of 0.5 was found to yield the best model. Hence, for any pair of atoms, if fij(x,y,z). exceeds the 0.5 pKd cutoff, it is set to 0.5. Finally, the εAB parameter was obtained by linear fitting using our training set.
Hydrogen bonding
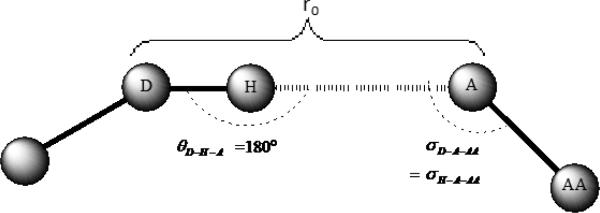
Hydrogen bonding is another important interaction found in most protein-ligand complexes. Such an interaction occurs when a lone pair on a (typically) polar group approaches a hydrogen atom bound (typically) to a polar atom like N or O. The principle variable associated with hydrogen bonding is the distance between the hydrogen bond donor and hydrogen bond acceptor, dHA, the bond angle between the hydrogen bond donor and acceptor, θD-H-A and the H---A-AA angle defined by the hydrogen bond acceptor σH-A-AA. In the present work, we modeled hydrogen bonding as defined by eqn. 7, which is an adaptation from earlier work of Vedani and co-workers (see eqn. 7).33,34 In this description of hydrogen bonding, dHA, θD-H-A and σH-A-AA have defined optimal values. Departure of dHA, θD-H-A and σH-A-AA from these optimal values destabilizes the hydrogen bond interaction.
| (7) |
The f1(dHA) distance function is modeled as a Lennard-Jones 6–12 potential with the well depths to be obtained from linear regression. In the two angle functions, the optimal angle for θD-H-A is 180°, while for σH-A-AA, the optimal angle depends on the type of acceptor atom and on the nature of the molecule in which it is embedded. Based on previous research,33,34 σo is 135° for carbonyl, carboxyl, and sulfonamide oxygen atoms, 109.5° (sp3) or 120° (sp2) for hydroxyl oxygen atoms.
The spatial orientations of hydrogen atoms are normally not revealed by X-ray crystallography at the resolutions typically seen for protein-ligand complexes (>1.5 Å). Although hydrogen atoms can be added later, energy minimization is usually required to optimally position them. Adding hydrogen atoms could become problematic especially when hydrogen atoms could be placed into multiple positions, as in cases where a drug molecule has multiple tautomeric states.35 Therefore, we modified eqn. 7 in order to resolve the hydrogen atom positioning problem (see Figure 1). For the bond distance, dHA, we use the distance between the hydrogen bond donor and the hydrogen bond acceptor dDA in place of dDA. For the two angle variables, we use the following approximations: Based on an analysis of the geometry of hydrogen bonds in which imidazole, serine, threonine, tyrosine, adenine, cytosine, water, and sulfonamide fragments act as hydrogen-bond acceptors,34,36 up to 90% of the angles θD-H-A range within 180° ± 30°, which means that cos2(θD-H-A - θ0) ranges from 0.75 to 1. Hence the assumption was made that θD-H-A can be set to 180°, which results in f2(θD-H-A) = 1. Moreover, as a result of this approximation, the value of σH-A-AA is equal to that of σD-A-AA. With these simplifying approximations with regards to the hydrogen atom positioning, the hydrogen bond interaction between a ligand and its target protein can be quantified with explicit variables.
Figure 1.
A simple description of the hydrogen bonding model
In summary, hydrogen bonding in our model is described as follows:
| (8) |
For carbonyl, carbxyl and sulfonic oxygen atoms, σ0=135°, and for hydroxyl oxygen atoms σ0=109.5°.
The εAB terms are decided upon based on the atom types found in the hydrogen bond donor and acceptor pair. Two atom types for hydrogen bonds are used in this work, O and N. So the εO-O and εO-N terms need to be derived in the fitting process. Hydrogen bond lengths are always shorter than the sum of VDW radii and longer than covalent bonds. Based on O—H⋯O and N—H⋯O bond lengths, r0 for the O⋯⋯O distance was set to 2.8 Å and the N⋯⋯O distance was set to 2.9 Å in our model, with an upper limit for rij of 5 Å. Furthermore, hydrogen bond “saturation” was also considered in our model. One lone electron pair can form one hydrogen bond with only one polar hydrogen atom and vice versa. In order to avoid over saturation in our computations, the program scans the atoms in the ligand and protein. With the labels we have assigned to each atom, the program determines the number of polar hydrogen atoms within 3.5 Å of a “potential” H-bond acceptor. Next, the program determines the formation of a hydrogen bond using the following principles: When an H-bond donor has n hydrogen atoms bonded (n ≥ 1), it will form hydrogen bonds with the nearest n H-bond acceptors; When an H-bond acceptor has n lone pairs (n ≥ 1), it will form hydrogen bonds with the nearest n H-bond donors.
Desolvation
Desolvation causes changes in the entropy as well as in enthalpy of the ligand and its target protein. This effect is very difficult to accurately characterize since it involves complicated ligand-water, protein-water, and water-water interactions before and after binding. Different algorithms have been used in other empirical scoring functions. In this work, we associated the free energy change caused by the desolvation effect with the binding surface area. Significant advancements have been made over the last several decades in the computation of molecular surfaces,37,38 but most are computationally too expensive for this work because we will evaluate thousands of protein-ligand complexes. Thus, a novel method was created to reflect the binding surface area with a grid-based algorithm.
First, the effective distance between the ligand and its target protein, within which the desolvation effect occurs, is set to 5 Å. An atom from the ligand (protein) would be judged to be “within the binding surface” if any atom from the protein (ligand) is less than 5 Å from it. In the second step of the computation, the program defines a box to cover the atoms from both the ligand and protein marked as “within the binding surface”, and create regularly-spaced grids within the box. The grid spacing used is 0.5 Å. Distances between the grids and every single atom in the box are computed. If a distance between a grid and an atom is less than the van der Waals (VDW) radius of the atom, the grid is marked as “within the atom”, otherwise, the grid is marked as “outside the atom”. Third, grid points marked as “within the atom” are translated by 0.5 Å along the Cartesian axes and if a grid point is re-identified as “outside the atom” after one of these translations, the grid point is labeled as a “boundary atom” of either the ligand or protein. Because the grid points are closely spaced, we identify the sum of the grid points marked as “boundary atoms” as qualitatively reflecting the binding surface area of the either ligand or protein. Hence, the mean value of the sum of boundary atom grid points, of both the ligand and protein, represents the binding surface area used in this work.
| (9) |
Metal Chelation
Metal chelates are observed in numerous metalloprotein-ligand complexes as metal binding “warheads“.39 Scanning the pdbbind v2010 database, numerous chelates between ligands and Cu, Fe, Mg can be found for protein-ligand complexes where the pKd is higher than 6, but interestingly these metal binding warheads do not show as a significant effect on the binding affinity as was observed in the case of Zn. Ligands use different “warheads“ to chelate zinc ions such as O, N, S. We organized the ligands into different categories and assigned different parameters for their corresponding models. It is very important to design an appropriate model for the Zn chelation term. We started with ligands with N as “warhead” because they show the strongest Zn chelation effect.
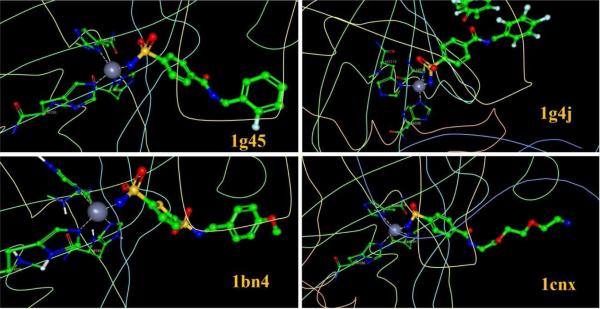
When the observed pKd is higher than 7 for a metalloenzyme-ligand complex, N-Zn chelation is generally observed. For pKd values ranging from 7 to 11, we found 38 acceptable (no crystal contacts, etc.) complexes containing Zn-N chelation interactions in the PDBbind v2010 database. Ultimately, all these complexes consisted of Carbonic anhydrase II as receptor and a sulfonamide as ligand. The structure of the tetrahedral active site is formed by three nitrogen atoms from imidazole groups (from His residues) and a nitrogen atom from sulfonamide. Some examples are shown in Figure 2.
Figure 2.
four examples of protein-ligand complexes with Zn-N ligand chelation. Fragments are shown as ball-and-stick models.
Although VDW interactions, hydrogen bonding and desolvation effects also exist in these complexes, Zn chelation is still a significant effect in binding. From the calculation of the interactions listed above, we found that for complexes without Zn chelation, who have high binding affinity (pKd>7), the sum of VDW interactions between C3 and C2 () was always high (see atom types in Tables 1 and 2). However, for those complexes with Zn chelation whose pKd ranged above 7, VDW interactions between C3 and C2 was not as high (see Table 3). This phenomena shows that Zn-N chelation is balanced with VDW interactions in high binding affinity complexes, which to some degree demonstrates that Zn chelation may be the dominant interaction in metalloenzyme complexes with high pKds. A more detailed comparison is shown in Table 3.
Table 3.
Some example complexes showing the relations between sp3 hybridized Carbon & sp2 hybridized Carbon VDW interaction, Zn chelation and pKd.
| PDB Code | VDW C3_C2 | Zn Chelation | pKd |
|---|---|---|---|
| 1vkj | −16.885 | N/A | 4.85 |
| 1a99 | −12.084 | N/A | 5.70 |
| 1apb | −15.76 | N/A | 5.82 |
| 1b58 | −37.926 | N/A | 6.59 |
| 1lrh | −28.539 | N/A | 6.82 |
| 1h2t | −23.873 | N/A | 7.89 |
| 1fcy | −60.245 | N/A | 8.52 |
| 1kdk | −27.797 | N/A | 9.05 |
| 2fgu | −45.694 | N/A | 9.18 |
| 1mrw | −39.35 | N/A | 9.7 |
| 1df8 | −42.823 | N/A | 9.92 |
| 1xpz | −12.559 | Y | 7.08 |
| 1cny | −15.87 | Y | 7.85 |
| 1ydb | −3.7781 | Y | 8.24 |
| 1cim | −12.885 | Y | 8.82 |
| 1cil | −14.889 | Y | 9.43 |
| 1bnt | −14.195 | Y | 9.89 |
| 1bnn | −3.683 | Y | 10.00 |
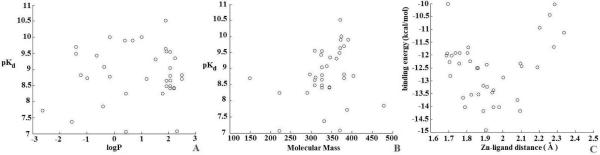
Given the importance of Zn chelation in metalloprotein-ligand complexes, a mathematical model for Zn-ligand chelation needs to be built. We need to figure out the relationship between all factors and their contributions to binding affinity. First, a direct comparison between the chelate structure and binding affinity were made. From Figure 3C we see that when the distance between the ligand nitrogen and Zn is around 2 Å, binding energy is likely to reach its maximum, and decreases when N---Zn distance moves away from 2 Å. This has also been shown by Vedani and Huhta in their research.39 Then, we also examined the relation between binding affinity and ligands' logP and molecular mass. From Figure 3A and 3B, we cannot see clear trends of pKd changing with logP nor molecular mass of ligands. This indicated that ligands' hydrophilicity and molecular weight factors on binding affinity were excluded. Given this insight a function was constructed to model Zn chelation:
| (10) |
Figure 3.
Plots of pKd (y axis) vs. logP (logP values are derived using xlogP program40), pKd (y axis) vs. molecular mass, and binding energy vs. the distance Å (x axis) between the ligand nitrogen and Zn for the 38 complexes containing Zn chelation.
where r is the distance between the nitrogen in a ligand and Zn, δ is the distance at which chelation affinity reaches its maximum.
We applied this model to all other Zn-ligand chelation cases. Besides Zn-N chelation, Zn-O chelations are also widely seen among metalloprotein-ligand complexes. They include monodentate ligands as carbonyl groups and phosphate groups, and bidentate ligands like hydroxamic acid groups. There are also other ligands using employing Zn-S chelation, but the available examples were few. From pdbbind v2010, we found 38 complexes with Zn-N chelation (sulfonamide as ligands), 20 complexes with Zn-O chelation, 23 complexes with Zn-O (hydroxamic acid as ligands) bidentate chelation and only 6 complexes with Zn-S chelation. The equilibrium distances δ for different chelates were obtained from Vedani and Huhta;39 δZn-O for a monodentate ligand was set to 1.961 Å, δZn-O for abidentate ligand was set to 2.068 Å, δZn-N was set to 2.041 Å and interaction between Zn and S were ignored because of too small training set.
The Final Expression
In summary our score has the following functional form:
| (11) |
and for a detailed explanation of the terms see the discussion below.
Result and Discussion
Model Training Results
LISA's mathematical model included 17 descriptors to describe corresponding interactions between certain types of atoms and 1 descriptor for the solvent effect. For the 2nd (MVDW C3−C2/Car), 3rd (MVDWC3−N3/Npl3) and 5th (MVDW C3/C2/Car−S) terms, we combined multiple interaction types and allowed them share one common weight, in order to decrease the number of parameters to be fitted. Merging these interactions in this way is sensible because they represent similar interacting atom types (In 2nd term, sp3 carbon - aromatic carbon interaction was combined with sp3 carbon - sp2 carbon interaction, etc.), so our observation is sensible.
Based on the mathematical model described in detail above, we performed linear fitting to our training set of 492 complexes. Our scoring function is able to reproduce the binding affinities of the entire training set with an R2 of 0.536 and an RMSD of 1.32 pKd, corresponding to 1.86 kcal/mol in binding affinity at physiological temperature (310 K). Leave-one-out cross-validation was also done for the training set with a Q2 of 0.503 and RMSD of 1.38 pKd (1.94 kcal/mol).
Goodness of fit and the resultant parameters are listed in Table 4. Each parameter derived from fitting reflects the weight factor for each term. We also provide the parameters for the normalized data in order to eliminate the difference in scaling for all of the terms, because (1) in most cases, carbons are more prevalent than other atoms in ligands, causing the scale of for the VDW potential for carbon-carbon interactions to be larger than that of other interactions; (2) SASA was described as surface area in LISA, while other terms in energy units, so they have different variable scalings. Each term was scaled according to the following formula:
| (12) |
where Xi and are the raw and normalized ith term values for interaction type i; Xi,min and Xi,max are the minimum and maximum values for ith descriptor, respectively.
Table 4.
Parameters and fitting results derived from linear fitting after training data normalization (without chelation terms)
| Interaction Type | Weight | 95% confidence interval | Normalized Weight | no. of contacts in training set | |
|---|---|---|---|---|---|
| VDW C3_C3 | 0.1184 | 0.0848 | 0.1520 | 0.5878 | 22556 |
| VDW C3_C2 | 0.0910 | 0.0791 | 0.1029 | 0.5938 | 53562 |
| VDW C3_N3 | 0.2457 | 0.1553 | 0.3361 | 0.2569 | 4980 |
| VDW C3_N4 | 0.4111 | 0.1907 | 0.6316 | 0.2363 | 1099 |
| VDW C_S | 0.2708 | 0.2071 | 0.3345 | 0.2576 | 2926 |
| VDW C2_C2 | 0.1197 | 0.0491 | 0.1903 | 0.2123 | 4140 |
| VDW C2_O3 | 0.1658 | 0.0830 | 0.2487 | 0.2775 | 5161 |
| VDW C2_O2 | 0.0991 | 0.0226 | 0.1757 | 0.1800 | 8779 |
| VDW C2_Npl3 | 0.2402 | 0.0936 | 0.3868 | 0.0987 | 1781 |
| VDW Car_Car | 0.0660 | 0.0498 | 0.0821 | 0.4020 | 6091 |
| VDW Car_O2 | 0.0765 | 0.0243 | 0.1286 | 0.1714 | 10340 |
| VDW Car_N3 | 0.2807 | 0.0249 | 0.5364 | 0.2790 | 1790 |
| VDW Car_N2 | 0.1398 | 0.0391 | 0.2405 | 0.1363 | 1572 |
| VDW O_N | 0.1658 | 0.0611 | 0.2705 | 0.2075 | 7861 |
| H bond between O & O | 0.2716 | 0.1179 | 0.4252 | 0.2074 | 9421 |
| H bond between O & N | 0.2383 | 0.0578 | 0.4188 | 0.1693 | 5370 |
| SASA | 0.0120 | 0.0063 | 0.0176 | 0.0453 | - |
From the result shown in Table 4, we can compare the contribution of each interaction to the predicted binding affinity. We find that the VDW interaction between sp3 hybridized carbon atoms and unsaturated carbon atoms is the most important term in predicting binding affinity. However, they outnumber other interactions by a factor of two or more. This reflects the crucial effect of hydrophobic interactions in protein-ligand binding. However, contributions of other interactions are not proportional to their contact number. Some types of interactions, which are not so obvious, like VDW C3_N4 and VDW Car_Car, etc. have considerable contributions to the binding affinity. This shows that for the design of new ligands, simply increasing contact numbers in some cases may not increase binding affinity effectively.
VDW interactions between all other atom pairs are neglected in this work for three reasons: (1) εABS for some atom pairs show an unphysical negative contribution to the total binding affinity based on preliminary fitting results, and as a result adding them into the scoring function significantly lowered the correlation coefficient R2. Hence, they were neglected for both a lack of physical interpretation and a lowering the goodness of fit. (2) 95% confidence intervals derived for each descriptor should not include 0, otherwise the descriptor would be regarded as statistically insignificant in the mathematical model. (3) VDW interactions between some atom pairs like O and S, N and S etc. are rare in the training set. Adding them into the scoring function would lead to the derivation of misleading parameters.
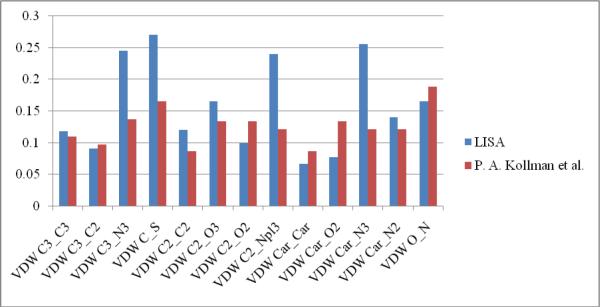
Presented in kcal/mol, εAB can be compared between our parameters and those derived by Cornell et al for use in a force field designed for the simulation of proteins and nucleic acids.32 From Table 5 we see that except for the well depths for VDW C3_N3, VDW C_S, VDW C2_Npl3 and VDW Car_N3, most of our εAB parameters are generally very similar to those derived by Cornell and co-workers. Graphical comparisons are shown in Figure 4. This demonstrates that the VDW parameters derived herein are physically meaningful and reliable to be used to estimate binding energies of protein-ligand complexes.
Table 5.
Comparison between εAB derived in this work and ε’AB derived by Cornell, et al.
| Interaction Type | εAB (kcal/mol)a | ε’AB (kcal/mol)b | Interaction Type | εAB (kcal/mol) | ε’AB (kcal/mol) |
|---|---|---|---|---|---|
| VDW C3_C3 | 0.11842 | 0.1094 | VDW C2_O2 | 0.09914 | 0.134 |
| VDW C3_C2 | 0.091029 | 0.097 | VDW C2_Npl3 | 0.24021 | 0.121 |
| VDW C3_N3 | 0.24569 | 0.1364 | VDW Car_Car | 0.06596 | 0.086 |
| VDW C3_N4 | 0.41112 | - | VDW Car_O2 | 0.076465 | 0.134 |
| VDW C_S | 0.27081 | 0.1654 | VDW Car_N3 | 0.25577 | 0.121 |
| VDW C2_C2 | 0.1197 | 0.086 | VDW Car_N2 | 0.13977 | 0.121 |
| VDW C2_O3 | 0.16581 | 0.134 | VDW O_N | 0.16579 | 0.1889 |
εAB is derived in this work
ε’AB is derived by W. D. Cornell et al.32
Figure 4.
VDW potential well depth (εAB) by LISA vs. VDW potential well depth (ε'AB) by QM
The SASA term is also important to binding affinity prediction. To further test the SASA term, we tried grid densities of 0.1 Å and 1 Å. When using 1 Å as our grid density, the correlation coefficient (R2) fell to 0.502 and RMSD fell to 1.38 for our training set (R2=0.536 and the RMSD was 1.32 with our default 0.5 Å grid density). Using 0.1 Å as our grid density, the correlation coefficient (R2) becomes 0.542 with a standard deviation of 1.31. Fitting results for the two grid densities (0.5 Å and 0.1 Å) were similar, but a 0.1 Å grid density was much more computationally intensive than a 0.5 Å grid density. Hence, in LISA we used 0.5 Å grid density throughout to simulate SASA.
Parameters for our chelation model are listed in Table 6. We can see both zinc-oxygen and zinc-nitrogen chelation show significant contributions to the observed binding affinity. However, compared to Zn-O chelation, Zn-N chelation plays a bigger role in the binding affinity when the latter interaction is present. The reason for this is unclear, but geometrically most of complexes containing zinc-nitrogen coordination retain structures closer to tetrahedral than zinc-oxygen complexes. For Zinc-sulfur chelation, we couldn't obtain reliable parameters due to the limited training set.
Table 6.
Parameters and fitting results for Zinc chelation terms
| interaction type | weight | 95% confidence interval | normalized weight for chelation term | normalized weight for sum of other terms | |
|---|---|---|---|---|---|
| Zn-O monodentate | 0.60154 | 0.26073 | 0.94235 | 0.88654 | 0.2189 |
| Zn-O bidentate | 0.45576 | 0.29547 | 0.61605 | 0.77997 | 0.31438 |
| Zn-N monodentate | 1.3063 | 1.1673 | 1.4454 | 1.1666 | 0.18926 |
Validation of LISA
Lisa was validated for its ability to predict experimentally measured binding affinities using three benchmarks. Then Artificial Neural Network (ANN) analysis was employed to determine whether cross terms were needed or can improve LISA. ANN helps to analyze correlations between LISA terms, and whether each term in LISA was well defined.
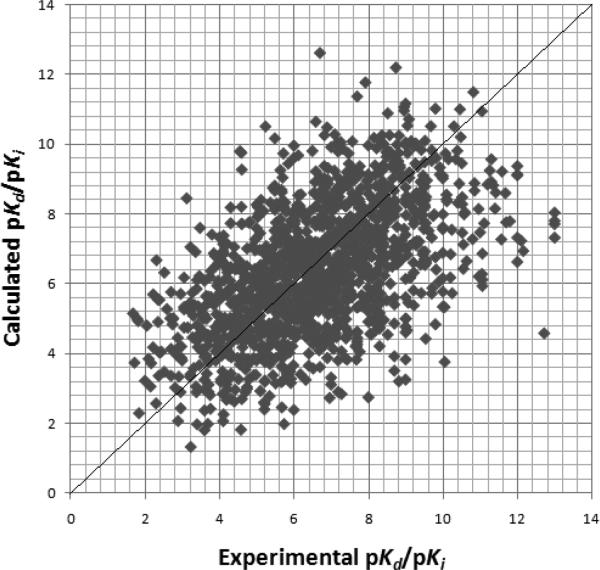
First, we introduced the entire PDBbind v2010 database with a total of 6772 protein-ligand complexes. Using the same 5 criteria we used to choose our training set (see above), 2047 complexes were selected. Eliminating the complexes we used in our training set and other test sets, 1399 complexes were left in our test set. Using this test set, LISA gave a Pearson correlation coefficient r of 0.534 with a RMSD of 2.65 kcal/mol (see Figure 5)
Figure 5.
LISA calculated pKi/pKd vs the experimental pKi/pKd for the PDBbind v2010 test set of 1399 protein-ligand complexes.
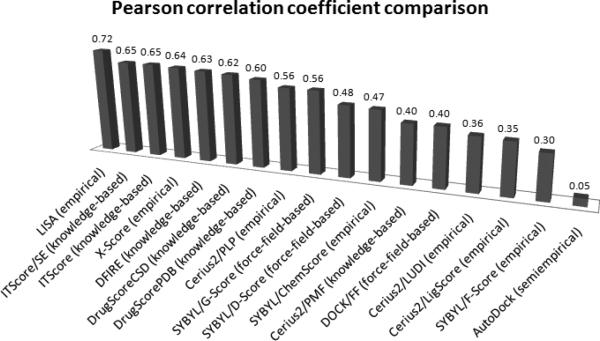
We utilized two already constructed and widely used test sets built by Wang et al.41 and by Muegge and Martin12. Wang's test set contains 100 diverse protein-ligand complexes, and we obtained r=0.72, RMSD=2.32 kcal/mol using LISA. Comparison of LISA to other score functions is presented in Figure 6, showing that LISA performs quite well on this test set.
Figure 6.
With the test set built by Wang,41 binding affinity comparison was done for LISA and some well-known scoring functions, ITScore/SE,20 ITScore,19 X-Score,5 DFIRE,42 DrugScoreCSD,18 DrugScorePDB,15 Cerius2/PLP,43,44 SYBYL/G-Score,45 SYBYL/D-Score,46 SYBYL/ChemScore,47 Cerius2/PMF,12 DOCK/FF,46 Cerius2/LUDI,31,48 Cerius2/LigScore,49 SYBYL/F-Score,50 AutoDock.51
Muegge and Martin's test set contains 77 diverse protein-ligand complexes from five protein classes. Using our model we obtained an R2 of 0.68 and a RMSD of 1.42 or 2.01 kcal/mol. For a detailed breakdown of each protein class, see Table 7. Complexes from class 1 were chosen from serine proteases, where carbon-carbon interactions dominate. LISA yielded a good correlation coefficient of R2=0.91, and a good RMSD value of 0.97 expressed in pKd units or 1.38 kcal/mol. This shows that LISA models protein-ligand complexes dominated by carbon-carbon contacts. Class 2 consisted of 15 Zn-O monodentate chelation complexes. The correlation coefficient R2 derived from LISA was 0.93 and the RMSD was 0.89 expressed in pKd unit or 1.26 kcal/mol. Compared with other scoring functions, we believe that LISA is more reliable in predicting metalloprotein-ligand binding affinities. Muegge and Martin's class 3 test set consisted of complexes with very small ligands (L-arabinoses), while class 4 contains larger ligands. LISA had comparatively poor results for these two classes (R2=0.43 and RMSD=1.87 or 2.65 kcal/mol for class 3, R2=0.33 and RMSD=1.81 or 2.57 kcal/mol for class 4). This result to some degree proved that ligand size affect the prediction of protein-ligand binding affinity and suggests future modifications of LISA should consider very small and very large fragments in the training set. Several other score functions struggled with these two classes, so this seems to be a general problem. For class 5, we obtained R2=0.83 and RMSD=1.89 or 2.35 kcal/mol.
Table 7.
Comparison of R2 and RMSD between LISA and other scoring functions for the Muegge and Martin's test set
| correlation (R2) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | set | no. of complexes | LISA | ITScore/SE20 | ITScore19 | PMF9912 | DrugScore15 | BLEEP52 | SMoG0116 | SCORE131 | SMOG53 |
| 1 | serine protease | 16 | 0.91 | 0.89 | 0.87 | 0.87 | 0.86 | 0.79 | 0.81 | 0.76 | 0.76 |
| 2 | metalloprotease | 15 | 0.93 | 0.71 | 0.71 | 0.58 | 0.70 | 0.59 | 0.64 | 0.41 | 0.58 |
| 3 | L-arabinose binding protein | 18 | 0.43 | 0.48 | 0.49 | 0.48 | 0.22 | 0.14 | 0.06 | 0.00 | 0.04 |
| 4 | endothiapepsin | 11 | 0.33 | 0.36 | 0.35 | 0.22 | 0.30 | 0.04 | 0.03 | 0.39 | 0.05 |
| 5 | others | 17 | 0.83 | 0.8 | 0.7 | 0.69 | 0.43 | 0.49 | 0.50 | 0.53 | 0.25 |
| 6 | sets 1–5 | 77 | 0.68 | 0.76 | 0.65 | 0.61 | n/a | 0.28 | 0.46 | 0.30 | 0.21 |
| RMSD | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | set | no. of complexes | LISA | ITScore/SE20 | ITScore19 | PMF9912 | DrugScore15 | BLEEP52 | SMoG0116 | SCORE131 | SMOG53 |
| 1 | serine protease | 16 | 0.97 | n/a | n/a | 0.96 | 0.95 | n/a | 1.09 | 1.39 | 1.34 |
| 2 | metalloprotease | 15 | 0.89 | n/a | n/a | 2.31 | 1.53 | n/a | 1.62 | 3.27 | 2.29 |
| 3 | L-arabinose binding protein | 18 | 1.87 | n/a | n/a | 0.86 | 0.75 | n/a | 0.8 | 69.7 | 4.06 |
| 4 | endothiapepsin | 11 | 1.81 | n/a | n/a | 1.89 | 0.94 | n/a | 1 | 1.26 | 4.18 |
| 5 | others | 17 | 1.66 | n/a | n/a | 1.56 | 1.85 | n/a | 1.63 | 2.21 | 4.05 |
| 6 | sets 1–5 | 77 | 1.42 | n/a | n/a | 1.84 | n/a | n/a | 1.69 | 3.47 | 4.43 |
We note that there is an overlap between the Wang and Muegge and Martin test sets and our training set. For Wang's test set 12 complexes overlap, and for the Muegge and Martin test set 10 complexes overlap. Both test sets have less than 10% overlap with the training set, ensuring the reliability of our test set.
Following their earlierstudy, Wang and co-workers published another test of several widely used scoring functions with a larger test set, the PDBbind v2002 refined set of 800 protein-ligand complexes54 in 2004. We examined this test set to make a more comprehensive comparison with other scoring functions using a much larger data set. Binding affinity data was calculated by LISA and the performance was evaluated using the Pearson correlation coefficient r, standard deviation (SD) and unsigned mean error (ME). Comparison between the performance of LISA and other scoring functions can be seen in Table 8. In addition, we found 123 complexes including metal-ligand contacts in this test set. For these metalloprotein-ligand complexes, LISA reproduced an r of 0.77, SD of 1.09 and ME of 0.69. From the overall three test result comparisons, we believe that LISA perfoms well in predicting binding affinity, especially for metalloprotein-ligand complexes.
Table 8.
Comparison of r, SD and ME from LISA and other scoring functions to the work of Wang et al.
| Scoring Functions | r | SD | ME |
|---|---|---|---|
| LISA | 0.610 | 1.92 | 1.47 |
| X-Score::HPScore | 0.514 | 1.89 | 1.47 |
| X-Score::HMScore | 0.566 | 1.82 | 1.42 |
| X-Score::HSScore | 0.506 | 1.90 | 1.48 |
| DrugScore::Pair | 0.473 | 1.94 | 1.51 |
| DrugScore::Surf | 0.463 | 1.95 | 1.53 |
| DrugScore::Pair/Surf | 0.476 | 1.94 | 1.50 |
| Sybyl::D-Score | 0.322 | 2.09 | 1.67 |
| Sybyl::PMF-Score | 0.147 | 2.16 | 1.74 |
| Sybyl::G-Score | 0.443 | 1.98 | 1.56 |
| Sybyl::ChemScore | 0.499 | 1.91 | 1.50 |
| Sybyl::F-Score | 0.141 | 2.19 | 1.77 |
| Cerius2::LigScore | 0.406 | 2.00 | 1.57 |
| Cerius2::PLP1 | 0.458 | 1.96 | 1.52 |
| Cerius2::PLP2 | 0.455 | 1.96 | 1.53 |
| Cerius2::PMF | 0.253 | 2.13 | 1.71 |
| Cerius2::LUDI1 | 0.334 | 2.08 | 1.66 |
| Cerius2::LUDI2 | 0.379 | 2.04 | 1.62 |
| Cerius2::LUDI3 | 0.331 | 2.08 | 1.67 |
| GOLD::GoldScore | 0.285 | 2.16 | 1.72 |
| GOLD::GoldScore_opt | 0.365 | 2.06 | 1.63 |
| GOLD::ChemScore | 0.423 | 2.00 | 1.56 |
| GOLD::ChemScore_opt | 0.449 | 1.96 | 1.52 |
| HINT | 0.330 | 2.08 | 1.65 |
We also employed an Artificial Neural Network (ANN) to test the energy terms in our scoring function. ANN training result can clearly reflect how much the interaction terms in a scoring function correlate with binding affinity when judiciously controlling the number of nodes in the hidden layers. While the ANN lacks a direct physical interpretation of the interaction terms, and the training results do not provide access to the parameters, it can be very useful in validating a scoring function model. Because ANN tries all possible combinations between the input values to build up a relationship with the output value, if ANN training shows a far better result than a linear fitting method, we can determine that some cross terms need to be introduced into the scoring function. Thus, by observing the convergence speed and comparing the goodness of fit, we can judge whether the energy terms are well designed in our scoring function.
We used the LISA training set of 492 complexes to train our network, and designed a new test set to test both LISA and the trained network. For the new test set, (1) We used the same 5 criteria in choosing complexes for the training set. (2) pKd values for the complexes in the test set were distributed evenly from 3 to 11. Because for any fitting process, the goodness of fit for the function varies in different regions of the output range, the test set should be distributed evenly across its range of applicability in order to avoid false positive test results. We searched for protein-ligand complexes related to the complexes in our training set, which had similar ligands or pocket structures but different binding affinities compared to the complexes in our training set. This was done with Binding MOAD.55 Finally we selected 41 complexes to be our test set.
ANN with a Broyden–Fletcher–Goldfarb–Shanno (BFGS) algorithm, a model of the nonlinear fitting process, was employed in this work. Briefly, the ANN model is composed of connections of the processing elements (nodes). The processing elements transfer data from one bias to the next through activation functions until the output bias. A three-bias model was used in the present work, for its higher tolerance to error and a comparatively simple structure for training. For the number of hidden nodes, we employ the so-called “self-generation of hidden nodes” method to include the fewest hidden nodes while maintaining training precision. This analysis determined that five hidden nodes was the optimal choice. The network passes through activation functions defined as sigmoid functions; the tan-sigmoid function was selected as the activation function that defines the transference from the input bias to the hidden bias. The log-sigmoid function was selected as the activation function that defines the transference from the hidden bias to the output bias.
Using our new test set, LISA can reproduce pKd's with an R2 of 0.52 and RMSD of 1.20 or 1.71 kcal/mol. Trained ANN test result got R2 of 0.48 and RMSD of 1.92 kcal/mol. From Table 9, we see that the ANN test result is no better than that of LISA, from which we can conclude that the energy terms in LISA were well chosen and that no cross terms were needed.
Table 9.
ANN vs. LISA training and test results for the test set with 41 samples we have designed
| training R2 | test R2 | training RMSD (kcal/mol) | test RMSD (kcal/mol) | |
|---|---|---|---|---|
| ANN | 0.60 | 0.48 | 1.44 | 1.92 |
| LISA | 0.54 | 0.52 | 1.86 | 1.71 |
Conclusions
We have developed a new empirical based scoring function to evaluate binding affinity between different protein and ligand pairs. The scoring function uses different models to simulate van der Waals interactions, hydrogen bonds between different atom types and desolvation. We have also included an explicit term to model metal ion chelation between zinc and coordinating N and O ligand atoms. Our analysis suggests that we obtained acceptable parameters for van der Waals interactions (using a 6–12 Lennard-Jones model) through a comparison with standard force field parameters. We also collected data to represent the chelation between Zn and different type of ligands. Interaction between Zn and ligands containing N chelators was shown to be quite important is defining the observed binding affinity. Zn-O chelation also contributed to the observed binding affinity, especially in the case of bidentate chelators like hydroxamic acids. This is an important feature in our model that is largely ignored in other score functions. We also included desolvation into the current model using a fast and novel surface area algorithm. When compared with other scoring functions, LISA generally shows improved performance. An Artificial Neural Network analysis was also used to confirm the goodness of fit and to demonstrate that cross terms were not needed to improve our scoring function. The result suggests that differentiating van der Waals interactions and hydrogen bonding by atom types may be a good choice for empirical scoring functions, and that adding a metal chelation term significantly improved the prediction of binding affinity to metalloprotein.
Supplementary Material
Acknowledgments
We would like to thank the NIH (GM044974 and GM066859) for supporting the present research. Mark Benson is also acknowledged for numerous helpful discussions.
Footnotes
Supporting Information Supporting Information is available including the training set and test set PDB id, experimental pKi/pKd and calculated pKi/pKd data. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- (1).Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: A Web-Accessible Database of Experimentally Determined Protein-Ligand Binding Affinities. Nucl. Acids Res. 2007;35:198–201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Congreve M, Chessari G, Tisi D, Woodhead AJ. Recent Developments in Fragment-Based Drug Discovery. J. Med. Chem. 2008;51:3661–3680. doi: 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- (3).Mobley DL, Dill KA. Binding of Small-Molecule Ligands to Proteins: “What You See” Is Not Always “What You Get”. Structure. 2009;17:489–498. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- (5).Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J. Comput.-Aided Mol. Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- (6).Merz KM. Limits of Free Energy Computation for Protein–Ligand Interactions. J. Chem. Theory Comput. 2010;6:1769–1776. doi: 10.1021/ct100102q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kolb P, Irwin JJ. Docking Screens: Right for the Right Reasons? Current Topics in Medicinal Chemistry. 2009;9:755–770. doi: 10.2174/156802609789207091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ewing TJA, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: Search Strategies for Automated Molecular Docking of Flexible Molecule Databases. J. Comput.-Aided Mol. Des. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- (9).Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- (10).Weiner SJ, Kollman PA, Case DA. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984;106:765–784. [Google Scholar]
- (11).Weiner SJ, Kollman PA, Nguyen DT, Case DA. An all atom force field for simulations of proteins and nucleic acids. J. Comput. Chem. 1986;7:230–252. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- (12).Muegge I, Martin YC. A General and Fast Scoring Function for Protein–Ligand Interactions: A Simplified Potential Approach. J. Med. Chem. 1999;42:791–804. doi: 10.1021/jm980536j. [DOI] [PubMed] [Google Scholar]
- (13).Muegge I. A knowledge-based scoring function for protein-ligand interactions: Probing the reference state. Perspect. Drug Discovery Des. 2000;20:99–114. [Google Scholar]
- (14).Muegge I. Effect of ligand volume correction on PMF scoring. J. Comput. Chem. 2001;22:418–425. [Google Scholar]
- (15).Gohlke H, Hendlich M, Klebe G. Knowledge-based scoring function to predict protein-ligand interactions. J. Mol. Biol. 2000;295:337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- (16).Ishchenko AV, Shakhnovich EI. Small molecule growth 2001 (SMoG2001): An improved knowledge-based scoring function for protein-ligand interactions. J. Med. Chem. 2002;45:2770–2780. doi: 10.1021/jm0105833. [DOI] [PubMed] [Google Scholar]
- (17).Mitchell JBO, Laskowski RA, Alex A, Thornton JM. BLEEP—potential of mean force describing protein-ligand interactions: I. Generating potential. J. Comput. Chem. 1999;20:1165–1176. [Google Scholar]
- (18).Velec HFG, Gohlke H, Klebe G. DrugScoreCSD-knowledgebased scoring function derived from small molecule crystal data with superior recognition rate of near-native ligand poses and better affinity prediction. J. Med. Chem. 2005;48:6296–6303. doi: 10.1021/jm050436v. [DOI] [PubMed] [Google Scholar]
- (19).Huang S-Y, Zou X. An iterative knowledge-based scoring function to predict protein-ligand interactions: II. Validation of the scoring function. J. Comput. Chem. 2006;27:1876–1882. doi: 10.1002/jcc.20505. [DOI] [PubMed] [Google Scholar]
- (20).Huang S-Y, Zou X. Inclusion of Solvation and Entropy in the Knowledge-Based Scoring Function for Protein–Ligand Interactions. J. Chem. Inf. Model. 2010;50:262–273. doi: 10.1021/ci9002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Jones G, Wilett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- (22).Rarey M, Kramer B, Lengauer T, Klebe GA. A Fast Flexible Docking Method using an Incremental Construction Algorithm. J. Mol. Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- (23).Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- (24).Wang R, Liu L, Lai L, Tang Y. SCORE: A New Empirical Method for Estimating the Binding Affinity of a Protein-Ligand Complex. J. Mol. Model. 1998;4:379–394. [Google Scholar]
- (25).Korb O, Stützle T, Exner TE. Empirical Scoring Functions for Advanced Protein–Ligand Docking with PLANTS. J. Chem. Inf. Model. 2009;49:84–96. doi: 10.1021/ci800298z. [DOI] [PubMed] [Google Scholar]
- (26).Teramoto R, Fukunishi H. Supervised Consensus Scoring for Docking and Virtual Screening. J. Chem. Inf. Model. 2007;47:526–534. doi: 10.1021/ci6004993. [DOI] [PubMed] [Google Scholar]
- (27).Deng Y, Roux B. Computations of Standard Binding Free Energies with Molecular Dynamics Simulations. J. Phys. Chem. B. 2009;113:2234–2246. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wang R, Fang X, Lu Y, Yang C-Y, Wang S. The PDBbind Database: Methodologies and Updates. J. Med. Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- (29).Wang R, Fang X, Lu Y, Wang S. The PDBbind Database: Collection of Binding Affinities for Protein–Ligand Complexes with Known Three-Dimensional Structures. J. Med. Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- (30).Søndergaard CR, Garrett AE, Carstensen T, Pollastri G, Nielsen JE. Structural Artifacts in Protein–Ligand X-ray Structures: Implications for the Development of Docking Scoring Functions. J. Med. Chem. 2009;52:5673–5684. doi: 10.1021/jm8016464. [DOI] [PubMed] [Google Scholar]
- (31).B□hm HJ. The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J. Comput.-Aided Mol. Des. 1994;8:243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- (32).Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- (33).Vedani A. YETI: An interactive molecular mechanics program for small-molecule protein complexes. J. Comput. Chem. 2004;9:269–280. [Google Scholar]
- (34).Vedani A, Dunitz JD. Lone-pair directionality in hydrogen-bond potential functions for molecular mechanics calculations: the inhibition of human carbonic anhydrase II by sulfonamides. J. Am. Chem. Soc. 1985;107:7653–7658. [Google Scholar]
- (35).Martin YC. Let's not forget tautomers. J. Comput.-Aided Mol. Des. 2009;23:693–704. doi: 10.1007/s10822-009-9303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Sarkhel S, Desiraju GR. N–H…O, O–H…O, and C–H…O hydrogen bonds in protein–ligand complexes: Strong and weak interactions in molecular recognition. Proteins: Struct. Funct. Bioinf. 2004;54:247–259. doi: 10.1002/prot.10567. [DOI] [PubMed] [Google Scholar]
- (37).Connolly ML. The molecular surface package. J. Mol. Graphics. 1993;11:139–141. doi: 10.1016/0263-7855(93)87010-3. [DOI] [PubMed] [Google Scholar]
- (38).Huang S-Y, Kuntz ID, Zou X. Pairwise GB/SA Scoring Function for Structure-based Drug Design. J. Phys. Chem. B. 2004;108:5453–5462. [Google Scholar]
- (39).Vedani A, Huhta DW. A new force field for modeling metalloproteins. J. Am. Chem. Soc. 1990;112:4759–4767. [Google Scholar]
- (40).Wang R, Fu Y, Lai L. A New Atom-Additive Method for Calculating Partition Coefficients. J. Chem. Inf. Comput. Sci. 1997;37:615–621. [Google Scholar]
- (41).Wang R, Lu Y, Wang S. Comparative Evaluation of 11 Scoring Functions for Molecular Docking. J. Med. Chem. 2003;46:2287–2303. doi: 10.1021/jm0203783. [DOI] [PubMed] [Google Scholar]
- (42).Zhang C, Liu S, Zhu Q, Zhou Y. A Knowledge-Based Energy Function for Protein–Ligand, Protein–Protein, and Protein–DNA Complexes. J. Med. Chem. 2005;48:2325–2335. doi: 10.1021/jm049314d. [DOI] [PubMed] [Google Scholar]
- (43).Gehlhaar DK, Verkhivker GM, Rejto PA, Sherman CJ, Fogel DB, Freer ST. Molecular recognition of the inhibitor AG-1343 by HIV-1 Protease: Conformationally flexible docking by evolutionary programming. Chem. Biol. 1995;2:317–324. doi: 10.1016/1074-5521(95)90050-0. [DOI] [PubMed] [Google Scholar]
- (44).Gehlhaar DK, Bouzida D, Rejto PA. In: Rational Drug Design: Novel Methodology and Practical Applications. Parrill L, Reddy MR, editors. Vol.719. American Chemical Society; Washington, DC: 1999. pp. 292–311. [Google Scholar]
- (45).Jones G, Willett P, Glen RC, Leach AR, Talor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- (46).Meng EC, Shoichet BK, Kuntz ID. Automated docking with grid-based energy approach to macromolecule-ligand interactions. J. Comput. Chem. 1992;13:505–524. [Google Scholar]
- (47).Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J. Comput.-Aided Mol. Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- (48).B□hm HJ. Prediction of binding constants of ptotein ligands: A fast method for the polarization of hits obtained from de novo design or 3D database search programs. J. Comput.-Aided Mol. Des. 1998;12:309–323. doi: 10.1023/a:1007999920146. [DOI] [PubMed] [Google Scholar]
- (49).CERIUS2 LigandFit User Manual. Accelrys Inc.; San Diego, CA: 2000. pp. 3–48. [Google Scholar]
- (50).Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- (51).Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- (52).Nobeli I, Mitchell JBO, Alex A, Thornton JM. Evaluation of a knowledge-based potential of mean force for scoring docked proteinligand complexes. J. Comput. Chem. 2001;22:673–688. [Google Scholar]
- (53).DeWitte RS, Shakhnovich EI. SMoG: de novo design method based on simple, fast, and accurate free energy estimates. 1. Methodology and supporting evidence. J. Am. Chem. Soc. 1996;118:11733–11744. [Google Scholar]
- (54).Wang R, Lu Y, Fang X, Wang S. An Extensive Test of 14 Scoring Functions Using the PDBbind Refined Set of 800 Protein-Ligand Complexes. J. Chem. Inf. Comput. Sci. 2004;44:2114–2125. doi: 10.1021/ci049733j. [DOI] [PubMed] [Google Scholar]
- (55).Hu L, Benson ML, Smith RD, Lerner MG, Carlson HA. Binding MOAD (Mother Of All Databases) Proteins. 2005;60:333–400. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.