Abstract
The survival motor neuron (SMN) protein plays an essential role in the assembly of uridine-rich small nuclear ribonuclear protein complexes. Phosphorylation of SMN can regulate its function, stability, and sub-cellular localization. This study shows that protein kinase A (PKA) phosphorylates SMN both in vitro and in vivo. Bioinformatic analysis predicts 12 potential PKA phosphorylation sites in human SMN. Mass spectrometric analysis of a tryptic digest of SMN after PKA phosphorylation identified five distinct phosphorylation sites in SMN (serines 4, 5, 8, 187 and threonine 85). Mutagenesis of this subset of PKA-phosphorylated sites in SMN affects association of SMN with Gemin2 and Gemin8. This result indicates that phosphorylation of SMN by PKA may play a role in regulation of the in vivo function of SMN.
Keywords: Spinal muscular atrophy, survival motor neuron protein, protein kinase A, phosphorylation site
1. Introduction
Deficiency of the survival motor neuron (SMN) protein causes spinal muscular atrophy (SMA), a devastating neuromuscular disorder characterized by spinal motor neuron degeneration and muscle atrophy [1]. Accumulating evidence indicates that SMN plays a role in the biogenesis of uridine-rich small nuclear ribonuclear proteins (snRNPs), pre-mRNA splicing, axonal transport, and cell survival [2-4]. Ubiquitously expressed, SMN localizes in the nucleus, cytoplasm, and neuronal processes. In the nucleus, SMN is concentrated in sub-cellular structures called gems, which often co-localize with Cajal bodies [2, 4, 5]. The function for SMN in snRNP assembly is performed by the SMN complex composed of SMN, Gemins2-8, and unrip [2-5]. The SMN protein displays a compartment-specific phosphorylation pattern [6]. Cytoplamsic SMN appears to be hyper-phosphorylated while nuclear SMN seems to be hypo-phosphorylated [6]. The cytoplasmic form of SMN is the key assembler of snRNPs [2, 5], and recent studies suggest that phosphorylation of SMN regulates its function in snRNP assembly, affects its stability, and controls its sub-cellular distribution [6-8]. In this study, we describe phosphorylation of the SMN protein by protein kinase A (PKA) and identification of a subset of the PKA phosphorylation sites in this protein by mass spectrometric analysis.
2. Materials and Methods
2.1. Cell culture, transfection, and drug treatment
HEK293 cells were cultured in DMEM supplemented with 10% fetal bovine serum. Approximately 1 × 106 cells were seeded in 100-mm dishes, and 24 hours later they were transfected with 10 μg of PKA catalytic subunit or dominant negative constructs using Lipofectamine2000 following the manufacturer's instruction (Invitrogen, Carlsbad, CA). Seventy-two hours later, transfected cells were harvested and analyzed by two-dimensional western blotting (see below). For glutathione S-transferase (GST) pull-down assay, GST or GST-SMN constructs driven by SRα promoter were transfected into COS-7 cells to maximize the yield of protein expression. Note that COS-7 cells were SV40 large T antigen-transformed cells. For transfection, COS-7 cells were seeded in 100-mm dishes, and 24 hours later they were transfected with 10 μg of GST or GST-SMN constructs using Lipofectamine2000. Transfection efficiency for each experiment was approximately 40% as determined by expressing the pEGFP plasmid (data not shown).
For drug treatments, cells were plated and 24 hours later were either left untreated or treated with 1 mM sodium vanadate (Sigma, St. Louis, MO) for 5 hours, 50 μM calyculin A (Cell Signaling, Danvers, MA) for 1 hour, 50 μM forskolin (Cell Signaling) for 1 hour, or 0.1 μM PKA inhibitor KT5720 (Sigma) for 1 hour. Cells were harvested and analyzed by two-dimensional western blotting (see below).
2.2. Expression and purification of recombinant proteins
Control GST protein was expressed from the GST expression vector pGEX-6P (GE Healthcare, Piscataway, NJ) in Escherichia coli strain BL21(DE3) and purified using glutathione-agarose according to the manufacturer's instruction (Sigma). GST-SMN was expressed in Spodoptera frugiperda sf9 cells using the Bac-to-Bac baculovirus expression system (Invitrogen). The donor pFastBac-GST plasmid was constructed by cutting the GST fragment from pGEX-6P with StuI and XhoI restriction enzymes and ligating to the same restriction enzymes-digested pFastBac™1 vector. Human SMN cDNA was amplified by polymerase chain reaction with a primer set of 5’-CGGAATTCCATGGCGATGAGCAGCGGC -3’ and 5’-TCCCAAGCTTGTACAATGAACAGCCATGTCCACC -3’and sub-cloned into pFastBac-GST using EcoRI and HindIII sites. The sequence of pFastBac-GST-SMN was confirmed by sequencing, and the recombinant plasmid was transformed into E. coli DH10Bac™ (Invitrogen). Recombinant bacmids were selected, sequence confirmed, and transfected into sf9 cells using Cellfectin (Invitrogen) to generate recombinant baculovirus GST-SMN stock (P1). The baculovirus GST-SMN stock was further amplified in sf9 cells to obtain P3 viral stock, and the titers of the viral stocks were determined by immunofluorescence staining of virus-infected sf9 cells using antibodies against GST and SMN (data not shown).
For large-scale preparation of GST-SMN protein, 30 × 106 cells were seeded in 150-mm dishes, and 1 hour later they were infected with GST-SMN baculovirus stock (P3, 1:50-100). Seventy-two hours after infection, the cells were harvested and processed for purification. Ten dishes of baculovirus-infected sf9 cells were re-suspended in 2.5 volumes of lysis buffer (20 mM Tris-HCl, pH 7.5 containing 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 100 mM NaF, 1 mM Na3VO4, 1 mM sodium β-glycerol phosphate, 2.5 mM sodium pyrophosphate, and protease inhibitor cocktail [Roche, Indianapolis, IN]). Lysates were cleared by centrifuging at 10,000 rpm at 4°C for 20 min. Cell lysates were incubated with 0.5 ml of pre-washed glutathioneagarose at 4°C for 1 hour with constant agitation. The agarose beads were then washed three times with 15 ml of lysis buffer followed by twice with 15 ml of phosphate-buffered saline (PBS) containing protease inhibitor cocktail. The bound GST-SMN protein was eluted in 50 mM Tris, pH 8.0 containing 50 mM reduced glutathione (Sigma), and 20 μM E64. Eluted protein was dialyzed against PBS containing 0.1 mM phenylmethylsulphonyl fluoride (PMSF) overnight with changing dialysis buffer three times. The purified GST-SMN protein was concentrated using Ultracel-10 filter (Millipore, Billerica, MA), and glycerol and DTT were added to a final concentration of 10% and 0.1 mM, respectively. An aliquot of the purified GST-SMN was analyzed for purity by 10% SDS-PAGE, and its concentration was determined by Coomassie™ blue staining (R-250, Sigma) using bovine serum albumin as a standard. The identity of the purified GST-SMN was confirmed by western blotting using anti-SMN (1:1000, BD Bioscience, San Jose, CA) and anti-GST (Santa Cruz, Santa Cruz, CA) antibodies.
2.3. Two-dimensional western blotting
Drug-treated or PKA construct-transfected cells were harvested and lysed in 30 mM Tris, pH 8.5 containing 7 M urea, 2 M thiourea, and 4% CHAPS. Lysates were cleared by centrifuging at 13,200 rpm at 4°C for 15 min. The protein concentration of lysates was measured by Bio-Rad protein assay (Bio-Rad, Hercules, CA). One hundred micrograms of proteins were mixed and loaded onto IPG strips (Bio-Rad, Hercules, CA, USA) via rehydration in the presence of the same lysis buffer (see above) containing 0.5% Pharmalyte 3-10 (Bio-Rad). Samples were run on pH 3-10 linear IPG strips. After isoelectric focusing, IPG strips were equilibrated for 15 min with 75 mM Tris, pH 8.8, 6 M Urea, 30% Glycerol, 2% SDS, and 65 mM DTT followed by 15 min with 75 mM Tris, pH 8.8, 6 M Urea, 30% Glycerol, 2% SDS, and 135 mM iodoacetamide (Sigma). The IPG strips were loaded onto 12% SDS-PAGE gels, and proteins were separated by electrophoresis. The gels were subsequently subjected to western blot analysis using anti-SMN antibody (BD Bioscience).
2.4. In vitro PKA phosphorylation
Purified GST-SMN or GST (0.1 μg) was incubated with the catalytic subunit of PKA (0.1 μg, Calbiochem, Gibbstown, NJ) for 1 hour at 30°C in a buffer containing 25 mM Tris-HCl, pH 7.0, 3 mM MgCl2, 1mM EGTA, 0.5 mM EDTA, and 0.2 mM DTT in the presence of 100 μM ATP and [γ-32P]ATP. To confirm the specificity of in vitro phosphorylation of SMN, PKA was pre-incubated with 0.5, 5, and 10 μM of PKA inhibitor KT5720 at 30°C for 1 hour before the addition of GST-SMN protein. For phosphatase treatment, 1 μl lambda phosphatases (400 U/μl, New England Biolab, Ipswich, MA) was added to each sample and incubated at 30°C for another hour. All reactions were terminated by the addition of SDS-PAGE sample buffer and resolved by 10% SDS-PAGE. Gels were dried and exposed to a film at -80°C. Signals were quantified as described previously [9, 10]. For mass spectrometry analysis, ~200 μg of purified GST-SMN was phosphorylated in the absence or presence of 24 μg of PKA catalytic subunit for 24 hours at 30°C. Samples were resolved on SDS-PAGE, and GST-SMN proteins were cut from the gels and digested with trypsin.
2.5. Mass spectrometry (MS) analysis
Trypsin digested samples were dried in a vacuum concentrator and dissolved with 0.1% formic acid. Each sample was then separated on a Dionex 3000 nLC system (Sunnyvale, CA) equipped with an Acclaim PepMap 100 C18 trap column (300 μm × 5 mm, 5 μm, Dionex) and an Acclaim PepMap 100 C18 analytical column (75 μm × 15 cm, 3 μm, Dionex). Peptides were resolved using a gradient between A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile) at 250 μL/min over 100 min. The column eluent was mixed with 80% isopropanol and introduced into an AB SCIEX 4000 QTRAP mass spectrometer (Foster City, CA) through a nano-electrospray ionization source. Data acquisition consisted of a precursor scan targeting -79 m/z in negative ion mode and MS/MS of the corresponding five most intense ions from each survey scan in positive ion mode. Data were searched against a custom protein database using Mascot search engine (v. 2.2, Matrix Science, London, UK) for peptide/protein identification (at a confidence interval ≥ 95%) and were also verified manually.
2.6. Plasmids and GST pull-down assay
Mammalian GST expression vector was constructed by first cutting the GST fragment from pGEX-6P with StuI and XhoI restriction enzymes and sub-cloning into pBluescript KS(-) vector (Stratagene, Santa Clara, CA) using SmaI and XhoI sites. The GST fragments were then released from pBluescript vector with SacI and HindIII restriction enzymes and ligated into the same restriction enzymes-digested mammalian expression vector pcDEBdelta [11]. Human SMN cDNA was amplified by PCR as described above and subcloned into pcDEBdelta-GST using EcoRI and HinIII sites. All point mutations for SMN PKA sites were created in pcDEBdelta–GST-SMN by QuikChange Lightning Site-Directed Mutagenesis following the manufacturer's instruction (Stratagene). Subcloned GST and GST-SMN cDNA constructs were confirmed by sequencing.
For GST pull-down analyses, COS-7 cells transfected with GST or GST-SMN constructs were harvested 48 hours after transfection. Lysates from transfected cells were prepared in lysis buffer containing 20 mM Tris-HCl, pH 7.5 containing 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 100 mM NaF, 1 mM Na3VO4, 1 mM sodium β-glycerol phosphate, 2.5 mM sodium pyrophosphate, and protease inhibitor cocktail (Roche, Madison, WI). Approximately 1.5 mg of protein lysate was mixed with GST-binding resin (Sigma) and incubated for 3 hours at 4°C. The resin was washed five times with the same buffer, and the bound proteins were eluted by boiling the samples in 1x SDS sample buffer. Presence of Gemin proteins in GST pull-down samples was analyzed by western blotting [9, 10, 12] using anti-Gemin2 (1:1000, Abcam, Cambridge, MA) and anti-Gemin8 (1:500, Santa Cruz) antibodies.
3. Results and Discussion
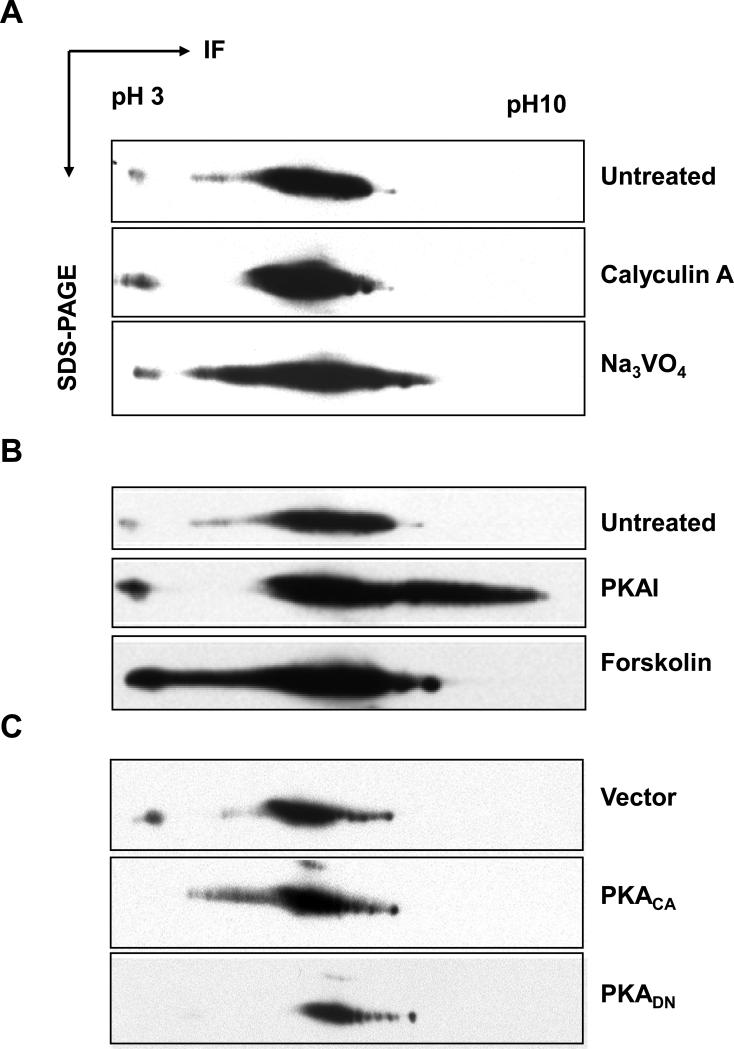
Previous studies indicate that phosphorylation of SMN regulates its function in snRNP assembly, affects its stability, and controls its sub-cellular distribution [6-8]. To investigate phosphorylation of SMN in more detail, we used bioinformatic programs (PhosphoMotif Finder and NetPhosK) to locate potential phosphorylation sites and corresponding kinases for these sites in SMN. Although sequence-based prediction is hypothetical, this mathematical approach could provide us with leads to possible phosphorylation sites in SMN. PhosphoMotif Finder predicts 12 possible PKA sites in human SMN (Fig. 1) whereas NetPhosK predicts only 2 PKA sites (threonine 25 with a NetPhosK score of 0.72 and threonine 122 with a NetPhosK score of 0.61). Note that prediction scores for NetPhosK range from 0 (no probability) to 1 (high probability). The kinase motif used to predict phosphorylation site FRRGT25GQSD is [R/K][R/K]X[pS/pT], and the motif used for the site FKRET122CVVV is KXX[pS/pT]. In addition to PKA, more than 12 other kinases were predicted to phosphorylate SMN on serine, threonine, and tyrosine sites. Table 1 displays those non-PKA sites that are predicted by both bioinformatic tools, and these sites are conserved in human, mouse, and rat SMN (see Fig. 1 for sequence alignment). These sites in SMN are putative substrates for protein kinase C (PKC), casein kinase I/II (CKI/II), cell division cycle 2 (cdc2), mitogen-activated protein kinase p38 (p38 MAPK), epidermal growth factor receptor (EGFR), insulin receptor (INSR), and ataxia telangiectasia mutated (ATM) (Table 1). To determine if endogenous SMN is indeed phosphorylated, we treated HEK293 cells with phosphatase inhibitors (calyculin A for serine/threonine phosphatases and sodium vanadate for tyrosine phosphatases). We then examined changes in SMN phosphorylation using two-dimensional western blotting. Lysate proteins were first separated by isoelectric focusing, followed by SDS-PAGE and western blotting using anti-SMN antibody (Fig. 2). Upon treatment with phosphatase inhibitors, more acidic SMN signals were detected (Fig. 2A), indicating that endogenous SMN is phosphorylated on serine/threonine and tyrosine residues. Since phosphorylation of SMN by PKA has been implicated in stabilization of SMN protein [7], we next analyzed the effects of PKA agonist forskolin and antagonist KT5720 on phosphorylation of endogenous SMN. Forskolin activates adenylyl cyclase to stimulate PKA activity, while KT5720 (EMD Chemicals) acts as a potent ATP-competitive inhibitor of PKA (Ki = 56 nM) with little effects on PKC, protein kinase G, and myosin light-chain kinase. As shown in Fig. 2B, forskolin treatment resulted in ~30% more accumulation of acidic SMN, while KT5720 treatment led to ~40% more accumulation of basic SMN. Transfecting the cells with the catalytic subunit of PKA, but not a dominant negative PKA mutant, resulted in accumulation of acidic SMN, although the effect (~14%) was less dramatic than seen for forskolin (Fig. 2C). Our results with forskolin and KT5720 indicate that at steady state SMN is neither completely phosphorylated nor completely dephosphorylated. This might be expected if phosphorylation regulates SMN function. Other kinases could be affected by non-specific effects of forskolin and KT5720 [13], and some of the in vivo phosphorylation of SMN is likely due to other kinases.
Fig. 1.
Putative phosphorylation sites in SMN. Peptide sequences from human, rat, and mouse SMN were aligned using the ClustalW program. The PhosphoMotif Finder and NetPhosK servers were used to predict serine, threonine, and tyrosine phosphorylation sites in SMN and corresponding kinases for these sites. Putative sites for PKA are highlighted in red, while non-PKA sites are highlighted in grey. Those non-PKA sites that are conserved in all three species, corresponding kinases and kinase motifs, and NetPhosK scores for these sites are listed in Table 1. The three phosphopeptides identified by mass analysis in this study are underlined, and the detected PKA sites are marked by stars.
Table 1.
Prediction of putative phosphorylation sites in human SMN
| Residuea | Putative kinasea | Corresponding motifb (phosphorylated residues in red) | Scorea |
|---|---|---|---|
| S28 | CKII | [pS/pT]XX[E/D/pS/pY] | 0.52 |
| S31 | CKII | pSxx[E/pS/pT] | 0.63 |
| S49 | PKC | [pS/pT]X[R/K] | 0.85 |
| T62 | PKC | N.C.c | 0.80 |
| T69 | PKC | [R/K]X[pS/pT] | 0.66 |
| T128 | CKI | N.C. | 0.53 |
| Y130 | EGFR | N.C. | 0.58 |
| S139 | CKII | N.C. | 0.54 |
| S143 | p38MAPK | pSXXX[pS/pT] | 0.51 |
| S163 | ATM | pSQ | 0.57 |
| S166 | CKII | pSXX[E/D] | 0.70 |
| T167 | CKII | [pS/pT]XX[E/D/pS/pY] | 0.59 |
| Y272 | INSR | N.C. | 0.53 |
| T274 | PKC | N.C. | 0.55 |
Putative phosporylation sites, corresponding kinases, and prediction scores were predicted by NetPhosK 1.0 server (http://www.cbs.dtu.dk/services/NetPhosK/).
Kinase motifs predicted by PhosphoMotif Finder (http://www.hprd.org/PhosphoMotif_finder)
N.C., not a conventional motif for corresponding kinases based on PhosphoMotif Finder prediction.
Fig. 2.
Phosphorylation of SMN in vivo. (A) HEK293 cells were left untreated or treated with calyculin A (threonine and serine phosphatase inhibitor) and sodium vanadate (tyrosine phosphatase inhibitor). Lysates from untreated or treated cells were separated by two-dimensional SDS-PAGE, and the SMN protein (~37 kD) was detected by western blotting using anti-SMN antibodies. (B) HEK293 cells were left untreated or treated with PKA agonist forskolin or PKA inhibitor KT5720. Lysates from untreated or treated cells were analyzed as in A. (C) HEK293 cells were transfected with vector alone, PKA catalytic (PKACA), or dominant negative (PKADN) constructs. Lysates from vector or PKA constructs-transfected cells were analyzed as in A. IF = isoelectrophoresis focusing.
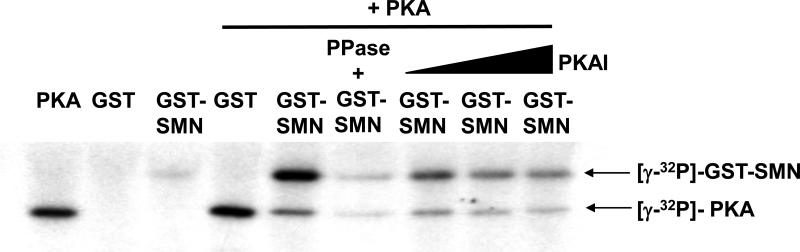
Next we set out to characterize sites of PKA phosphorylation in SMN. GST-tagged full-length SMN expressed in Escherichia coli was mostly insoluble and the majority of the solubilized protein appeared as degraded fragments (data not shown). Only 20 μg of soluble, full-length protein could be obtained from 10 liters of culture. Thus, GST-SMN was expressed in S. frugiperda sf9 cells using a baculovirus expression system to obtain correctly folded protein (Figure S1). Approximately 50 μg of soluble, full-length protein was obtained from one 150-mm dish of infected cells. Purified GST-SMN was used for in vitro phosphorylation of SMN. Fig. 3 shows that PKA efficiently phosphorylated GST-SMN but not GST control; this is consistent with results from a previous study [7]. Specificity of phosphorylation of SMN by PKA was confirmed by reduction of GST-SMN phosphorylation with a PKA inhibitor and by phosphatase treatment of the phosphorylated product. Using optimized conditions, ~200 μg of purified GST-SMN was incubated with or without PKA. Control and PKA-phosphorylated GST-SMN were isolated from SDS-PAGE gels and trypsin-digested. Phosphorylated tryptic fragments were enriched and analyzed by mass spectrometry.
Fig. 3.
Phosphorylation of SMN in vitro by PKA. The purified GST-SMN was incubated with PKA or PKA pre-treated with PKA inhibitor (PKAI) in the presence of γ-32P-ATP at 30°C for 1 hour. Phosphorylation of GST-SMN was then detected by SDS-PAGE followed by film exposure. Some of the phosphorylated GST-SMN protein was subjected to phosphatase (PPase) treatment before gel electrophoresis.
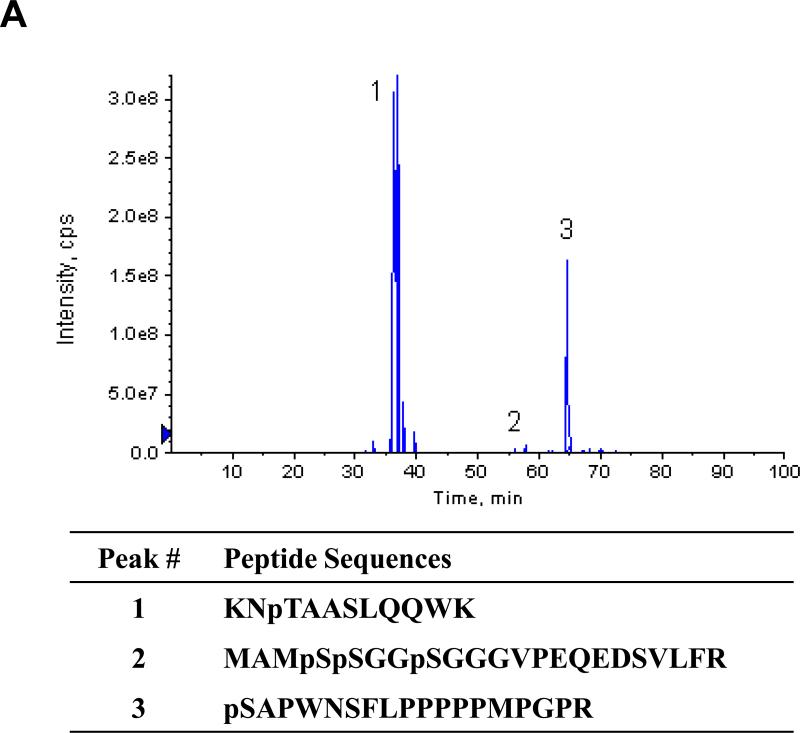
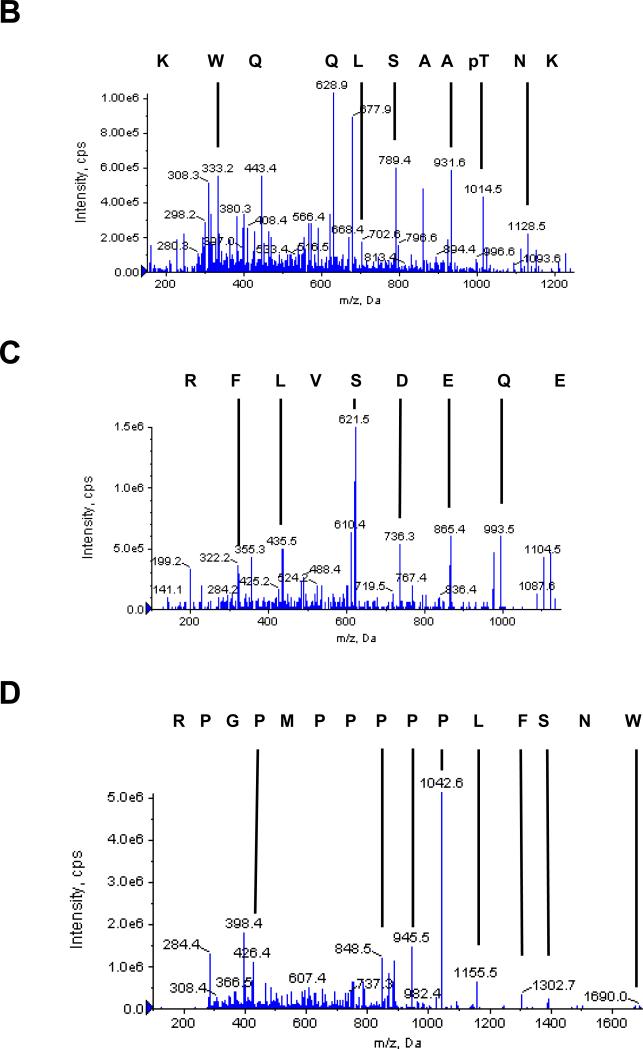
In negative ion mode, three phosphopeptides with characteristic 79 m/z daughter ions were detected (Figs. 4 and S2). MS/MS analysis of corresponding parent ions in positive ion mode identified three peptides KNpTAASLQQWK, MAMpSpSGGpSGGGVPEQEDSV LFR, and pSAPWNSFLPPPPPMPGPR with neutral parent peptides of masses 1353.6442, 2436.877, and 2023.979, respectively (Figs. 4 and S2). These phosphorylation sites correspond to serines 4, 5, 8, 187, and threonine 85 in the native protein. The masses of peaks 1 and 3 correspond to peptides with one phosphorylated amino acid, and the mass of peak 2 corresponds to a peptide with three phosphorylated amino acids. MS/MS analysis clearly identifies the C-terminal amino acid sequence of each peptide. The 1014.5 and 1128.6 masses obtained from peak 1 correspond to a phospho-peptide fragment that has lost a single H3PO4. The fragments of masses 789.4, 860.5, and 931.5 all correspond to peptides predicted to contain an unmodified serine residue, thus identifying threonine as the modified amino acid in this peptide. The most C-terminal serine residues in peptides 2 and 3 were similarly identified as unmodified residues, indicating that the N-terminal serines are the phosphorylated residues. Masses were identified that correspond to a and b series fragments of the N-terminus of the peptide with losses of H3PO4 from the serine residues (Fig. S2). The identification of peak 2 as the N-terminus of SMN was unexpected as the protein was expressed and purified as a fusion protein with GST. We confirmed that the sequence of the expression construct would result in a serine residue of the linker adjacent to the N-terminal methionine of the protein, which is unlikely to be hydrolyzed by trypsin. Although sequencing grade trypsin was used to generate peptides, we conclude that this site must have been partially hydrolyzed by trace contamination of another protease. Note that it is possible that other residues may have been phosphorylated by PKA even though we did not detect them using our MS protocols.
Fig. 4.
MS/MS analysis of phosphopeptides from SMN treated with PKA. (A) Three phosphopeptides were identified by MS of eluted peptides in negative ion mode and the extracted ion chromatogram of the identified peptides is shown. MS analysis of these peptides in positive ion mode identified three phosphopeptides from SMN with a confidence interval ≥ 95%. The corresponding phosphopeptide sequences are shown below. (B-D) Sequences of the three identified phosphopeptides were determined by MS/MS in positive ion mode. Singly charged Y-series peptide ions are identified by masses and vertical lines. The corresponding peptide sequence in C – N direction is shown above the ion spectrum. The ion masses of the parent peptides correspond to mono (peaks 1 and 3) and tri (peak 2) phosphopeptides. Note that the masses of the fragments that contain serine but not threonine correspond to non-phosphorylated peptides, while fragments that contain both the serine and the threonine correspond to modified peptides with loss of one H3PO4. Mascot-based ion assignment is provided in Figure S2.
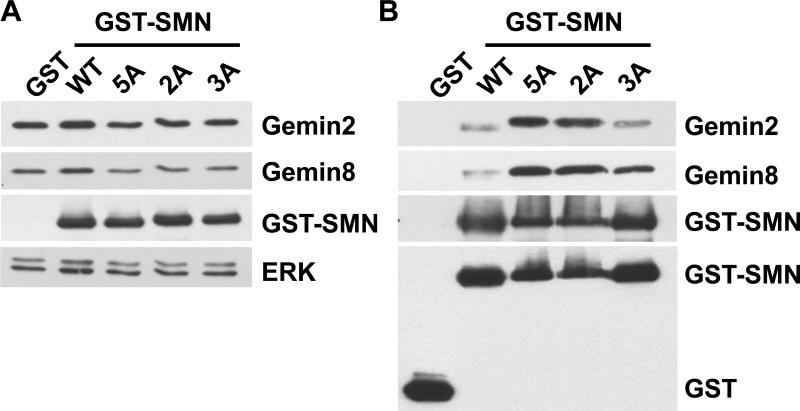
To determine whether the PKA phosphorylation sites have any functional significance, the identified PKA sites were mutated to alanine and transfection experiments performed to determine effects on in vivo binding to Gemin proteins. Wild-type and mutated GST-SMN constructs were transiently expressed in COS-7 cells, and the GST-SMN complexes were isolated using GST pull-down. Mutation of these PKA sites had negligible effect on the steady levels of either GST-SMN or endogenous Gemin2 and Gemin8 protein (Fig 5A). Wild-type GST-SMN was able to bind Gemin2 and Gemin8 in the cells (Fig. 5B). The GST control showed no binding to Gemins. GST-SMN PKA mutants were also able to pull-down the Gemin proteins, and mutation of some of these PKA sites resulted in an increase in association of GST-SMN with Gemin2 and Gemin8. Mutations of all five sites (serines 4, 5, 8, 187, and threonine 85), or two sites (threonine 85 and serine 187) to alanine, showed more enhanced binding than mutations in the N-terminal three serines (serines 4, 5, and 8 to alanines) (Fig. 5B). This result indicates that phosphorylation of threonine 85 and/or serine 187 can regulate binding of SMN to Gemins. This unexpected result requires more extensive studies to determine the role of phosphorylation on SMN complex formation.
Fig. 5.
Phosphorylation of SMN by PKA affects association of SMN with Gemin proteins. (A) COS-7 cells were transfected with each of GST, wild-type GST-SMN (WT), or GST-SMN PKA mutants (2A, T85A/S187A; 3A, S4A/S5A/S8A; and 5A, changing all five PKA sites into alanines). Cell lysates were prepared, 50 μg of lysates were separated by an SDS-PAGE, and levels of GST-SMN, Gemin2, Gemin8, and protein loading control ERK were detected by western blotting. The same membrane was used for each antigen in turn after stripping. (B) Lysates from transfected cells were also subjected to GST pull-down assays. The presence of Gemin2, Gemin8, and GST-SMN (using anti-SMN antibody) or GST control and GST-SMN (using anti-GST antibody) in GST pull-down complexes was detected by western blotting. The same membrane was used for each antigen in turn after stripping.
Survival motor neuron isolated from Hela cells is phosphorylated at serines 28 and 31, and phosphorylation of these sites appears to regulate SMN's activity in snRNP assembly [6]. Global phosphoprotein studies have shown that residues 4, 5, 8, 25, 28, and 31 are phosphorylated in SMN extracted from cells [14]. However, the enzyme(s) that phosphorylates these residues is unknown. Our results indicate that PKA could be responsible for phosphorylation of the 4, 5, and 8 sites in vivo. Direct measurement of phosphorylated residues in cell extracts is compromised by difficulties in isolating sufficient protein, and phosphorylation may not produce detectable steady-state levels of modified residues. Grimmler et al. [6] could only detect phosphorylation of serines 28 and 31 on affinity purified SMN from a 20-liter culture of Hela cells. PKA does not appear to be responsible for phosphorylation of these residues [7]. Using 200 micrograms of recombinant protein, we were able to identify two additional phosphorylation sites at residues 85 and 187. Although additional studies are required to determine whether these residues are phosphorylated in vivo, our GST pull-down analysis indicates that these two sites potentially play a role in regulating in vivo association of SMN with Gemin proteins.
In the nucleus, SMN specifically interacts with the phosphatase PPM1G/PP2Cγ, and dephosphorylation of SMN by this enzyme maintains stability of SMN and controls its localization in Cajal bodies [8]. Together, these studies indicate that phosphorylation may facilitate association of SMN with other proteins, affecting its compartmental localization, stability, and function [7, 8]. We found that GST-SMN expressed in E. coli or S. frugiperda sf9 cells is very unstable, and it has to be eluted from glutathione affinity columns in the presence of protease inhibitors to prevent its degradation, thus supporting the idea that SMN is stabilized either by complex formation or by post-translational modification. Activation of PKA has been shown to enhance incorporation of SMN into the SMN-Gemin complex and to inhibit SMN degradation [7]. In this study, mutation of the potential PKA sites threonines 25 and 122, the sites predicted as the conserved PKA sites by two bioinformatic programs (Fig. 1 and Table 1), and serine 290 failed to affect phosphorylation of SMN by PKA [7] suggesting that SMN is potentially phosphorylated on non-conventional sites. In our study, we have demonstrated that PKA phosphorylates SMN both in vitro and in vivo, and we have identified five phosphorylation sites that are distinct from those examined by others. Mutation of this subset of sites did not appear to influence nuclear localization of SMN in gems/Cajal bodies (data not shown), which indicates that these sites are not important for sub-cellular localization of SMN; however, mutation of these sites did affect association of SMN with Gemin proteins. Further studies are needed to determine how these sites affect binding of SMN with Gemin proteins to regulate SMN complex formation in vivo.
Supplementary Material
Fig. S1. Expression of GST-SMN protein. (A) Sf9 cells were infected with GST-SMN baculoviral stock. After 72 hours incubation, cells were harvested, and GST-SMN was purified from the whole cell lysate. Protein eluted from the affinity column was analyzed by western blotting using anti-SMN and anti-GST antibodies. (B) The purified GST-SMN protein was separated by SDS-PAGE and visualized by Coomassie™ blue staining. Note that the proteins with ~24 Kd in size were identified as degraded GST-SMN and insect GST protein. IB = immunoblotting, L = whole cell lysate, B = proteins retained in the beads after elution, F = flow-through, E1 = first elution, and E2 = second elution.
Fig. S2. MS/MS analysis of phosphopeptides from SMN treated with PKA. Three phosphopeptides were identified by MS of eluted peptides in negative ion mode. MS analysis of these peptides in positive ion mode identified three phosphopeptides from SMN with a confidence interval ≥ 95%. Sequences of the three identified phosphopeptides were determined by MS/MS in positive ion mode (A-C). Singly charged Y-series peptide ions are identified by masses and vertical lines. The corresponding peptide sequence in C – N direction is shown above the ion spectrum. Detailed mass spectrometry data are listed in the corresponding tables (A’-C’).
Acknowledgements
We thank Dr. G. Stanley McKnight at the University of Washington for PKA catalytic and dominant negative plasmids, and we thank Dr. Yusaku Nakabeppu at the Kyushu University for pcDEBdelta plasmid. We are also indebted to Dosh Whye and Leila Choe for technical assistance. This work is supported by Nemours and the National Institute of Health (Delaware INBRE grant 2P20 RR016472-10 and a COBRE grant 5P20 RR020173-04 to support the Center for Pediatric Research at Nemours/Alfred I. duPont Hospital for Children, Wilmington, DE). The authors declare that study sponsor has no role in study design; collection, analysis, and interpretation of data; writing of the report; and/or the decision to submit the paper for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at----.
References
- 1.Lunn MR, Wang CH. Spinal muscular atrophy. Lancet. 2008;371:2120–2133. doi: 10.1016/S0140-6736(08)60921-6. [DOI] [PubMed] [Google Scholar]
- 2.Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossoll W, Bassell GJ. Spinal muscular atrophy and a model for survival of motor neuron protein function in axonal ribonucleoprotein complexes. Results Probl Cell Differ. 2009;48:289–326. doi: 10.1007/400_2009_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cauchi RJ. SMN and Gemins: ‘We are family’ ... or are we?: insights into the partnership between Gemins and the spinal muscular atrophy disease protein SMN. Bioessays. 2010 doi: 10.1002/bies.201000088. [DOI] [PubMed] [Google Scholar]
- 6.Grimmler M, Bauer L, Nousiainen M, Korner R, Meister G, Fischer U. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep. 2005;6:70–76. doi: 10.1038/sj.embor.7400301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnett BG, Munoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petri S, Grimmler M, Over S, Fischer U, Gruss OJ. Dephosphorylation of survival motor neurons (SMN) by PPM1G/PP2Cgamma governs Cajal body localization and stability of the SMN complex. J Cell Biol. 2007;179:451–465. doi: 10.1083/jcb.200704163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Dimatteo D, Funanage VL, Scavina M. Increased susceptibility of spinal muscular atrophy fibroblasts to camptothecin-induced cell death. Mol Genet Metab. 2005;85:38–45. doi: 10.1016/j.ymgme.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Wu CY, Gomez-Curet I, Funanage VL, Scavina M, Wang W. Increased susceptibility of spinal muscular atrophy fibroblasts to camptothecin is p53-independent. BMC Cell Biol. 2009;10:40. doi: 10.1186/1471-2121-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakabeppu Y, Oda S, Sekiguchi M. Proliferative activation of quiescent Rat-1A cells by delta FosB. Mol Cell Biol. 1993;13:4157–4166. doi: 10.1128/mcb.13.7.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu CY, Whye D, Glazewski L, Choe L, Kerr D, Lee KH, Mason RW, Wang W. Proteomic assessment of a cell model of spinal muscular atrophy. BMC Neurosci. 2011;12:25. doi: 10.1186/1471-2202-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Expression of GST-SMN protein. (A) Sf9 cells were infected with GST-SMN baculoviral stock. After 72 hours incubation, cells were harvested, and GST-SMN was purified from the whole cell lysate. Protein eluted from the affinity column was analyzed by western blotting using anti-SMN and anti-GST antibodies. (B) The purified GST-SMN protein was separated by SDS-PAGE and visualized by Coomassie™ blue staining. Note that the proteins with ~24 Kd in size were identified as degraded GST-SMN and insect GST protein. IB = immunoblotting, L = whole cell lysate, B = proteins retained in the beads after elution, F = flow-through, E1 = first elution, and E2 = second elution.
Fig. S2. MS/MS analysis of phosphopeptides from SMN treated with PKA. Three phosphopeptides were identified by MS of eluted peptides in negative ion mode. MS analysis of these peptides in positive ion mode identified three phosphopeptides from SMN with a confidence interval ≥ 95%. Sequences of the three identified phosphopeptides were determined by MS/MS in positive ion mode (A-C). Singly charged Y-series peptide ions are identified by masses and vertical lines. The corresponding peptide sequence in C – N direction is shown above the ion spectrum. Detailed mass spectrometry data are listed in the corresponding tables (A’-C’).