Abstract
A new RNA world has emerged in the past 10 years with the discovery of a plethora of 20- to 30-nucleotide long small RNAs that are involved in various gene silencing mechanisms. These small RNAs have considerably changed our view of the regulation of gene expression in eukaryotic organisms, with a major shift towards epigenetic and post-transcriptional mechanisms. Here we focus on the striking diversity of small silencing RNAs that have been identified in a number of protozoan parasites and their potential biological role.
The small RNA world and RNA silencing
Both prokaryotic and eukaryotic cells are populated with a variety of small RNAs, ranging in size from ~20 up to a few hundred nucleotides (nt). The adjective ‘small’ was initially used to refer to molecules that are shorter than average size, the comparison being made with the large ribosomal RNAs and mRNAs, which are in the size range of hundreds to thousands of nt. Included in the original small RNA category are RNAs involved in translation (tRNAs, 5S RNA), pre-mRNA processing (small nuclear RNAs; snRNAs), processing and modification of pre-rRNA (small nucleolar RNAs; snoRNAs) and protein translocation across the endoplasmic reticulum (7SL RNA), among others. All these small RNAs have in common is that they do not code for proteins, hence the collective name of non-coding RNAs (ncRNAs).
Over the last decade, however, the universe of small ncRNAs has exploded [1] with the discovery of the shortest type of this class, the small silencing RNAs (ssRNAs). They are 20 to 30 nt in length and generally function to downregulate gene expression by a variety of mechanisms, which are collectively referred to as RNA silencing. RNA interference or RNAi, namely the pathway through which long double-stranded RNA (dsRNA) triggers cleavage of homologous transcripts, was the first RNA silencing mechanism discovered [2]. In the space of a few years ssRNAs were recognized as mediators of the RNAi response and related pathways and were shown to correlate with a number of gene silencing phenomena, from post-transcriptional gene silencing (PTGS), first described in plants, to DNA methylation and transcription inhibition, viral demise, heterochromatin formation and chromosome segregation, DNA elimination, transposon and retroposon silencing, and translational control [1,3]. Although the above phenomena display some organism-specific features both at the genetic and mechanistic levels, they are all dependent on ssRNAs.
While the current literature is abounding with acronyms defining ssRNA, making it difficult for the non-cognoscenti to navigate through them, the most well known types of ssRNAs are the small interfering RNAs (siRNAs), the micro RNAs (miRNAs) and the Piwi-associated RNAs (piRNAs). siRNAs and miRNAs are found throughout the eukaryotic kingdom, in single-cell as well as in multicellular organisms, whereas piRNAs are restricted to the germ line of certain metazoans [1,3]. Besides the short length, the second distinctive feature of small silencing RNAs is their association with members of the Argonaute (AGO) protein family [4], which are the central components of the RNA-induced-silencing complex or RISC (Box 1).
Box 1. RNA silencing genes in protozoan parasites.
As extrapolated from model organisms, the minimal RNA silencing machinery consists of Dicer, a Dicer protein partner that binds double-stranded RNA and facilitates Dicer cleavage, an Argonaute ‘slicer’ and a guide siRNA. In addition, in certain organisms, including nematodes and plants, an RNA-dependent-RNA polymerase is required to amplify the RNA silencing response [53]. For the miRNA pathway, Drosha, an RNase III-family enzyme, and a double-stranded RNA binding protein partner are also needed, but these factors do not appear to be conserved in protozoan parasites.
Dicer
Dicer, an RNase III-family enzyme, is classically distinguished by the presence of two RNase III domains, RNase IIIa and RNase IIIb [54], which can be next to each other, like in human Dicer, or at different positions along the polypeptide, as is the case for the two T. brucei Dicer enzymes. In addition, the size and domain structure of Dicers are extremely variable, with human Dicer being over 2,000 amino acids long and G. lamblia Dicer being a polypeptide of a little more than 700 amino acids. Importantly, GlDicer was the first of this class of molecules to be crystallized, and the solution of its structure confirmed that the RNase IIIa and IIIb domains form an internal heterodimer [55]. These studies also led to the model that Dicer acts as a ruler that measures the distance between the 3′ end of the dsRNA, which is bound to the PAZ domain, and the phosphodiester bonds positioned at the active sites.
Argonaute
Argonaute family proteins are at the center of RNAi silencing mechanisms. Several excellent reviews have been published, and the reader is referred to them for a detailed account on the structure and function of this protein family [4,13,56]. Briefly, AGO family proteins are characterized by PAZ, MID and Piwi domains. Only certain AGO proteins, termed ‘slicer’, are endowed with an RNase H-type endonuclease activity, which resides in the C terminal Piwi domain, and functions in transcript cleavage upon target recognition by the guide siRNA. Many AGO proteins, however, including the metazoan AGO family members that are bound to miRNAs, have no detectable endonuclease activity. AGO-miRNA complexes silence gene expression via recruitment of factors involved in mRNA degradation and/or translational repression [1,3]. According to phylogenetic considerations, an AGO-like protein was already present in the last common ancestor of eukaryotes [57]. At present very little is known in terms of slicer activity among the various AGO proteins from RNAi-proficient protozoan parasites. So far, only TbAGO1 has been subjected to detailed functional analysis and shown to be essential for transcript degradation both in the cytoplasm and nucleus [25,58].
RNA-dependent RNA polymerase (RdRp)
RdRp is an enzyme that catalyzes the synthesis of RNA from an RNA template. Originally identified in RNA viruses, many eukaryotic organisms are endowed with RdRps. In relation to the RNAi pathway, in certain organisms, including plants and C. elegans, RdRps are implicated in the amplification of the primary RNAi response through the production of so-called secondary siRNAs. In C. elegans secondary siRNAs bear a di- or tri-phosphate at the 5′ end, the hallmark of newly-synthesized polynucleotides [53]. Among the parasitic protozoa, RdRp-like genes have been identified in G. lamblia, E. histolytica and T. gondii.
Small interfering RNAs and micro RNAs
In reviewing ssRNAs one needs to recall that in 1993 the groups of Ambros [5] and Ruvkun [6] discovered the first example of a miRNA in Caenorhabditis elegans (called at that time small temporal RNA), and they demonstrated that lin-4, a 22-nt RNA species, was involved in regulating the timing of larval development by base pairing with the 3′ untranslated region of lin-14 mRNA. At the time this incredible finding remained largely unnoticed and was even considered a peculiarity of nematodes. However in 2001, sequencing of small 20–30 nt RNAs from Drosophila, HeLa cells and C. elegans [7–9] revealed that miRNAs were plentiful and a universal feature of higher eukaryotes. siRNAs were discovered in 1999, this time in plants, by Baulcombe’s group in a seminal paper published in the journal Science and entitled: “A species of small antisense RNA in post-transcriptional gene silencing in plants” [10]. Following this lead, in 2001 our laboratory reported the identification and sequencing of siRNAs in the protozoan parasite Trypanosoma brucei [11].
Both siRNAs and miRNAs, in a complex with an AGO protein, recognize their targets through base-pair interactions [1,3]. The region of homology between the siRNA and the mRNA can occur at any position, i.e. in the coding region or in the UTRs, and the complementarity is usually perfect along the length of the duplex. Instead, in the case of miRNA/mRNA recognition, the miRNA is only partially complementary to its target, and miRNA binding sites are usually found in the mRNA 3′ UTR. The seed region consisting of nucleotides 2 through 7 of miRNAs determines the specificity of the interaction, although exceptions to this rule have been noted. In particular, plant miRNAs are perfectly complementary to their targets and like siRNAs trigger target cleavage [12].
siRNAs, in a complex with an AGO ‘slicer’, namely an AGO protein endowed with endonuclease activity [13] (Box 1), guide cleavage of target transcripts. In contrast, miRNAs are in general found associated with endonucleolytically-incompetent AGO family members and bring about destabilization of the mRNA target and/or translation inhibition [1,3,14]. However, the exact mechanism of action of miRNAs is still being debated.
The biogenesis of siRNAs and miRNAs
siRNAs are processed from long dsRNAs by the endonuclease Dicer (Box 1), an RNase III-family enzyme, in a complex with a dsRNA-binding protein (dsRBP). Long dsRNA are generated by annealing of sense and antisense transcripts from endogenous genes or transgenes expressing dsRNA in the form of hairpins or rod-like structures. Alternatively, synthetic dsRNA can be introduced into cells by a variety of methods. Dicer cleavage generates short double-stranded fragments (Figure 1), which depending on the organism vary in size between 20 and 30 nt. These primary cleavage products have 5′ phosphate and 3′ OH termini and 2 nt extensions at the 3′ ends, the hallmarks of RNase III-type enzyme cleavage. Only one of the two strands of a duplex siRNA, termed the guide strand, is retained in AGO ‘slicer’, whereas the passenger strand is discarded. For details about the mechanism of guide strand selection and loading into Argonaute, the reader is referred to recent, excellent reviews [3,15]. In certain organisms, like plants, mature siRNAs, which are in a complex with AGO, are methylated at the 3′ end by HEN1, a 2′-O-specific methyltransferase [16].
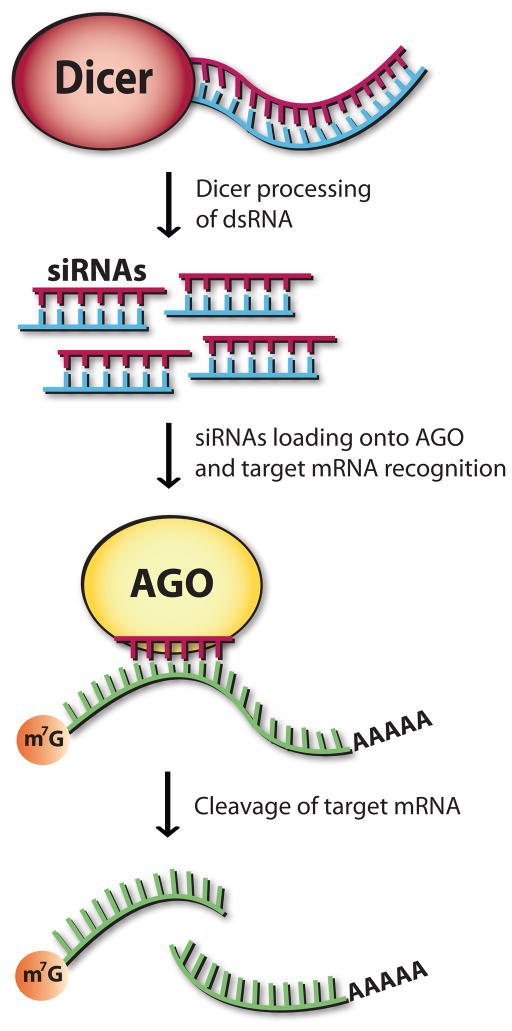
Figure 1. Molecular players of the classical RNAi pathway.
Dicer in association with a dsRNA binding protein (dsRBP) processes long RNA duplexes (dsRNA) into 20–30 nt siRNAs. Next, siRNAs are loaded into Argonaute ‘slicer’ (AGO), the catalytic core of RISC (RNA-induced-silencing complex). The passenger strand is removed and the guide strand interacts directly with AGO to target mRNA for cleavage.
In contrast, the biogenesis of miRNAs is a multistep process that involves two cleavage steps [1,3]. In the nucleus the primary miRNA, which typically is transcribed by RNA polymerase II and codes for several miRNAs, is processed by the microprocessor complex consisting of Drosha, an RNase III-family enzyme, and a dsRNA binding protein (dsRBP; DGCR8 in humans and Pasha in Drosophila) to generate 60–70 nt pre-miRNA hairpins. Pre-miRNAs are then exported from the nucleus to the cytoplasm by the Exportin 5-RAN GTPase complex, and the mature miRNAs are generated by the action of Dicer in a complex with its cytoplasmic dsRBP cofactor. Like siRNAs, miRNAs are in duplex form at this point, with the two strands named miRNA (corresponding to the siRNA guide strand) and miRNA* (corresponding to the siRNA passenger strand). Only the miRNA strand is incorporated into an AGO family member, whereas the miRNA* strand is discarded, although there are exceptions to this rule as well.
RNA silencing pathways and small silencing RNAs in protozoan parasites
In the following sections the current knowledge on RNA silencing pathways and the small silencing RNAs in protozoan parasites will be summarized (Table 2). The focus is on T. brucei, Giardia lamblia, Entamoeba histolytica and Toxoplasma gondii. However, it is important to note that the RNAi pathway is also functional in Trypanosoma congolense [17], Leishmania viannia braziliensis and Crithidia fasciculata [18], although the repertoire of the corresponding small RNAs has not yet been analyzed. In contrast, Trypanosoma cruzi [19], Leishmania major, Leishmania donovani [20] and Plasmodium falciparum are RNAi-deficient [21,22]. At present it can only be speculated what evolutionary force(s) led to the retention or loss of the RNAi pathway.
Table 2.
Classes of small silencing RNAs identified in protozoan parasites
| Organism | Primary siRNAs | Secondary siRNAs | miRNAs |
|---|---|---|---|
| Trypanosoma brucei | yes | ULa | UL |
| Giardia lamblia | likely | UKb | yes |
| Entamoeba histolytica | UK | yes | UK |
| Toxoplasma gondii | yes | UK | yes |
UL, unlikely.
UK, unknown.
Trypanosoma brucei
T. brucei was one of the first organisms where RNAi was discovered [23] and the first protozoan parasite that greatly benefitted from the application of the RNAi tool for analyzing gene function. Over the past thirteen years the T. brucei RNAi mechanism has been investigated in detail through biochemical and genetics approaches. Two Dicer-like proteins, TbDCL1 [24] and TbDCL2 [25], and a single AGO protein, TbAGO1 [26,27], fuel the RNAi mechanism (Table 1). The two Dicer enzymes have two RNase III motifs each (RNase IIIa and IIIb), which are the only recognizable domains, and they occupy different cellular compartments: DCL1 is localized in the cytoplasm, and DCL2 is mostly found in the nucleus. TbAGO1 controls both the nuclear and cytoplasmic RNAi mechanisms and has been shown to be essential for degradation of mRNA in the cytoplasm and of retroposon/repeat transcripts in the nucleus [25,26]. The finding that human AGO2 ‘slicer’ complements, at least in part, TbAGO1 ablation [28] provided further support to the slicer function of trypanosome AGO1. Moreover, comparative genomics indicated the existence of two additional RNAi factors that are specific to trypanosomatid protozoa with functional RNAi machineries (Ullu et al., unpublished data). Based on the current evidence, the following model for the T. brucei RNAi pathway has been proposed [25]. Endogenous dsRNA, derived from annealing of sense and antisense transcripts from retroposons and repeats, is processed to siRNAs in the nucleus by TbDCL2. In the cytoplasm TbDCL1 acts as a sentinel to process dsRNA that either escapes from the nucleus or is formed in the cytoplasm. siRNAs produced from either Dicer are then loaded into TbAGO1 with the assistance of one of the novel RNAi factors recently discovered (Ullu et al., unpublished data).
Table 1.
RNAi genes in protozoan parasites
| Organism | Dicer | Argonaute | RdRp | Refs. |
|---|---|---|---|---|
| Trypanosoma brucei | yes (2a) | yes (1) | no | [24–27] |
| Leishmania braziliensis | yes (2) | yes (1) | no | [18,59] |
| Leishmania major | no | pseudogene | no | [59] |
| Giardia lamblia | yes (1) | yes (1) | yes (1) | [36,55] |
| Entamoeba histolytica | possibly | yes (3) | yes (1) | [43,44,47] |
| Toxoplasma gondii | yes (1) | yes (1) | yes (1) | [21,51] |
indicates the number of genes in each class
To obtain information about the biological role of the endogenous RNAi pathway, we cloned and sequenced size-selected 20–30 nt RNAs isolated from purified polyribosomes more than a decade ago [11]. The choice of the starting material was based on the observation that approximately 10–20% of small RNAs co-sedimented with polyribosomes [29], and that the small RNAs recovered from this fraction had a much lower contamination level with rRNA fragments than those isolated from total RNA. These small RNAs had the features of siRNAs, with a phosphate at the 5′ end, indicative of Dicer cleavage, and interestingly, a blocked 3′ terminus [25]. The latter modification is most likely introduced by a HEN1 family methyltransferase, which is present in the trypanosome genome. The first sequencing effort revealed that the majority of the siRNAs were derived from two retroposon families that inhabit the trypanosome genome, namely the ubiquitous ingi (Kiswahili meaning: many) and the site-specific SLACS (Spliced Leader Associated Conserved Sequence) elements. More recently, a third class of siRNAs derived from 147 bp chromosomal internal repeats were reported, termed CIR147 [25], which were originally identified in putative centromeric regions [30]. High-throughput sequencing of siRNAs associated with TbAGO1 has confirmed the above results, but also indicated that many genomic regions with the potential to generate transcripts in the form of dsRNA, like inverted repeats and convergent transcription units, are the source of siRNAs (Ullu et al., unpublished data). These siRNAs are associated with AGO1, although their abundance is much lower than that of the major siRNA classes. No miRNA-like molecules were detected in these studies, although bioinformatics predictions have suggested the existence of such RNAs [31]. Thus, from the above results one major function of the trypanosome endogenous RNAi pathway could be to free cells of transcripts derived from retroposons, thus decreasing the probability of mobilization of the corresponding elements and promoting genome stability. In support of this possibility, long-term cultures of AGO1 null trypanosomes showed evidence of rearrangements of the SLACS elements [32]. Thus, similarly to what has been described in other organisms, the trypanosome RNAi pathway most likely functions as a nucleic acid-based immune system to defend cells against parasitic nucleic acids. However, the identification of the CIR147 siRNAs is telling us that the function of the RNAi pathway may be more complex than anticipated. These repeats are found in chromosomal internal regions and are surrounded by remnants of ingi retroposons [30]. Not only is this arrangement highly reminiscent of centromeric DNA, but these repeats were identified by Kelly and colleagues as part of putative centromeric regions [30]. However, since only three out of the eleven megabase chromosomes present in T. brucei bear CIR147 repeats [25], it is difficult to imagine that these repeats are markers for centromeric DNA. Nevertheless, they could function as islands for some, yet to be discovered, genome structural purpose. It is important to note that Bastin and colleagues have reported that in Lister 427 trypanosomes ablation of TbAGO1 results in defects in chromosome segregation [27,33], suggesting that the RNAi machinery may function to build pericentric heterochomatin, similar to S. pombe where RNAi is required for proper centromere function [34]. Additionally, retroposons and CIR147 repeats alike may very well be present in heterochromatic regions of the genome, as can be extrapolated from what is known in other organisms. Thus, one important question that needs to be addressed is whether RNAi has a role in heterochromatin formation in T. brucei. The identification of proteins involved in chromatin remodeling and histone modifications specific to heterochromatin will be pivotal towards clarifying whether RNAi plays a role in provoking chromatin silencing in T. brucei.
Giardia lamblia
The G. lamblia genome harbors three RNAi genes (Table 1): an AGO family member, a Dicer enzyme and an RNA-dependent RNA polymerase (RdRP). Although the existence of the RNAi pathway genes was noted many years ago, experimental RNAi as a tool to study gene function in these organisms has not met with much success. Rather, the preferred methods for gene silencing are expression of antisense RNA or transcript cleavage via engineered ribozymes. Nevertheless, a very recent report revealed that long dsRNA can trigger downregulation of gene expression [35], although the system only worked for mRNAs expressed at moderate levels and not for highly abundant mRNAs. Intriguingly, no siRNAs could be detected in these experiments, but this observation needs to be confirmed by further experimentation. Most interesting was the finding that downregulation of GlDicer or GlRdRP, via production of homologous antisense RNA, leads to cells expressing more than one variant-specific surface protein (VSP) gene at one time [36]. Thus, a model has emerged that RNAi is required for antigenic variation in G. lamblia, although the mechanistic details of this phenomenon are not yet understood.
A first insight into the biological role of RNAi in G. lamblia was provided in 2005 by the cloning and sequencing of size-selected 20–30 nt RNAs isolated from trophozoites [37]. This analysis revealed a population of sense and antisense RNAs of approximately 30 nt originating from GilT elements, a non-LTR family of retrotransposons enriched at telomeres [38]. The expression of these GilT small RNAs was confirmed by Northern hybridization and their size was suggestive of siRNA-like molecules. Furthermore, large GilT transcripts of sense and antisense polarity were found in steady-state conditions, suggesting that GilT dsRNA could be formed in vivo, thus providing potential substrates for Dicer cleavage and the generation of siRNAs. However, since the library was constructed using a method that is independent of the phosphate present at the small RNA 5′ end, it is not known whether primary, as well as secondary siRNAs (Box 1), with a polyphosphate 5′ end and possibly made by GlRdRp, were captured. Small RNAs derived from silenced, but not from the expressed VSP gene, have also been documented and implicated in controlling antigenic variation [36]. Lastly, high-throughput sequencing uncovered a class of small RNAs derived from an unusually long tandemly-repeated RNA named Glrep-1, whose sequence is somewhat similar to VSP genes [39]. The authors proposed that these small RNAs represent siRNAs and raised the possibility that they are somehow involved in antigenic variation. In summary, the current evidence suggests that siRNA-like molecules exist in Giardia. However, further experimentation is needed to characterize these RNAs in more detail. Do these molecules have the signature termini of Dicer-dependent cleavage and/or are they synthesized by RdRp? Are they associated with Argonaute? What are the specific silencing functions of these small RNAs?
The potential involvement of the Giardia RNAi machinery in translation repression has been put forward by the work of Saraiya and Wang, who reported the discovery of miRNA-like molecules derived from snoRNAs [40]. They identified a 26-nt RNA (miR2) as a product of Dicer-processed snoRNA GlsR17. Contrary to its nuclear precursor, miR2 was localized to the cytoplasm. Based on reporter gene assays the authors concluded that miR2 likely affects translation, but not stability of target mRNAs. In addition, knock-down of Dicer significantly reduced miR2 levels, implicating the protein in the generation of these molecules. The finding that snoRNAs can enter the RNAi pathway was also reported in human cells where it was directly shown that the corresponding processed products are associated with AGO1 and AGO2 [41]. At present, the mRNA targets of snoRNA-derived miRNAs are not yet known and thus, their functional significance needs to be established.
The presence of miRNA candidates in the Giardia genome has also been explored by high-throughput sequencing [39] and bioinformatics approaches [42]. Collectively, these studies indicate the presence of several tens of potential miRNAs specific to Giardia, and the expression of a few of them was confirmed by RT-PCR [42]. What is needed at this point is to dig in and show that the potential RNA targets, which were only identified bioinformatically, are indeed controlled by specific miRNA candidates. Nevertheless, the results of the above studies are highly suggestive that miRNAs are likely to be important regulators of gene expression in Giardia.
Entamoeba histolytica
In contrast to Giardia, a variety of studies have provided evidence that expression or delivery of dsRNA can be used as a tool to downregulate E. histolytica gene expression [43]. Entamoeba has three predicted AGO-like proteins, one of which (EH_125650) predominates in axenically-grown parasites and is found associated with a special class of ssRNAs [43], as discussed below. Conversely, BLAST searches for RNase III-domain containing proteins in E. histolytica only returned one ORF with a single RNase III domain. In vitro studies indicated that the recombinant protein had detectable RNase III activity and that E. histolytica extracts can degrade dsRNA, although the size of the terminal digestion products was not accurately determined [44]. Thus, at present no firm conclusion can be drawn on the putative Dicer present in this organism. Nevertheless, it is interesting to note that in the fungus Saccharomyces castellii Dicer has a single RNase III domain and most likely dimerizes like its prokaryotic counterparts [45]. Furthermore, the crystal structure and functional studies of a fragment of mouse Dicer indicated that the RNase IIIb domain can homodimerize and is catalytically active [46]. Thus, it is plausible that the RNase III-family protein identified in E. histolytica may function as Dicer, although its role in the generation of ssRNAs needs to be ascertained.
Work carried out in Upinder Singh’s laboratory has revealed a plethora of small RNAs that may well be involved in regulating gene expression by a variety of mechanisms. In order to identify the repertoire of E. histolytica endogenous small RNAs, a 5′-phosphate independent cloning approach aimed at capturing small RNAs with either a 5′-phosphate or a polyphosphate (polyP) structure was used [47]. An abundant population of 27-nt long small RNAs with 5′-polyP termini and a free 3′-OH was identified. Thus, the biogenesis of these molecules involves a biosynthetic pathway, likely carried out by EhRdRP. Importantly, this was the first report of the existence of secondary RNAi effectors in an early divergent protozoan parasite. The 27-nt RNAs mostly mapped to the antisense strand of protein coding genes with a bias toward the 5′ end of predicted mRNAs, associated with the most abundant AGO protein in trophozoites, and their expression inversely correlated with the amount of mRNA present in steady-state in a strain-specific fashion. This latter finding is consistent with a potential regulatory function of the 27-nt RNAs.
Interestingly, the small RNA libraries contained very few sequences derived from non-LTR retrotransposons [47], which have been shown to be abundantly expressed in an E. histolytica virulent strain [48]. This finding was surprising in light of the fact that in most organisms analyzed so far, the repertoire of endogenous siRNAs is largely derived from transposons and retroposons. Although it is possible that the RNAi machinery in E. histolytica does not function in downregulating expression of mobile elements, firm conclusions on this matter cannot be drawn at this time, as the full repertoire of ssRNAs, including primary siRNAs, needs to be investigated.
It should be noted that a significant fraction of the E. histolytica small RNA repertoire was derived from tRNAs, but this population was not considered biologically relevant [47]. However, tandem arrays of tRNA genes make up approximately 10% of the amoeba genome [49], and thus they constitute a highly repeated gene family. Since small RNAs can be generated from repeats, it is possible that tRNA fragments become associated with one of the amoeba AGO proteins and have a regulatory role. Indeed, although little is known about the biological role of small RNAs derived from mature tRNAs, they appear to be part of the RNAi machinery in higher eukaryotes [50].
In summary, studies in E. histolytica have revealed a complex repertoire of small RNAs with potential regulatory functions. The specific role of the 27-nt RNAs needs to be further investigated by determining, for instance, whether they function as siRNAs by triggering target cleavage or as miRNAs by inducing destabilization or translation repression of target mRNAs. Additionally, the identification of secondary siRNAs predicts that primary siRNAs also exists. Lastly, does the Entamoeba genome code for miRNA-like molecules?
Toxoplasma gondii
The RNAi gene repertoire in T. gondii includes one Dicer, one AGO family member and one RdRp (Table 1). TgDicer is a large protein with a structural organization reminiscent of the DCL1 protein of the single cell algae Chlamydomonas reinhardtii [51]. Toxoplasma and Chlamydomonas Dicer-like genes appear to be phylogenetically related, as they form a specific clade. Similarly, TgRdRp is closely related to the Neurospora crassa protein QDE1 and clusters with plant RdRPs. On the other hand, TgAGO has features of metazoan AGOs, localizes predominantly to the cytoplasm, but might also be present in the nuclear compartment [51]. These observations, together with the identification of a complex repertoire of small RNAs (see below) has led to the proposal that in Toxoplasma RNA silencing is brought about by a plant/fungal-like machinery, involved in the generation of ssRNAs, and is effected by a metazoan-like AGO protein. Like TbAGO1 and other AGO-family members [26,52], the N terminus of TgAGO is rich in arginine-glycine-glycine (RGG) residues.
Given that the T. gondii genome has a complete set of RNAi genes, Braun and colleagues considered RNA silencing as a possible strategy employed by the parasite for developmental gene regulation [51]. To test this hypothesis the authors analyzed the repertoire of small RNAs by high-throughput sequencing of a library constructed from size-selected total RNA. Two classes of small RNAs were identified: miRNAs, with metazoan-like features and siRNA-like molecules derived from repeated sequences and satellite DNA. Histones carrying modifications typical of heterochromatin were found associated with these repeated regions, suggesting that a proportion of the corresponding siRNAs may function to mediate heterochromatin formation. Association with TgAGO was confirmed for a number of siRNA and miRNA candidates, underscoring their identification as small silencing RNAs. Interestingly, expression of certain miRNA types varied among the three clonal isolates, which are currently used in laboratory research and have different virulence phenotypes in mice [51]. Whether regulation of miRNA expression affects Toxoplasma virulence is a distinct possibility and an exciting hypothesis for future investigations.
Quite strikingly, immunoprecipitation of epitope-tagged TgAGO followed by mass spectroscopy revealed a rich repertoire of putative interacting factors, which included many of the previously described RISC-interacting proteins [51]. Among these were RNA binding proteins and helicases, factors involved in transcription, translation and chromatin remodeling, and PRMT1, a protein Arginine methyltransferase that may be involved in methylating the RGG-rich N terminus of TgAGO. Furthermore, a proportion of TgAGO co-sedimented with polyribosomes, and this putative association required the RGG domain at the N terminus, as previously described for TbAGO1 [26,52]. Thus, the nature of the small RNAs and the localization of TgAGO suggest that in Toxoplasma RNA silencing operates at the translational level via a miRNA-like pathway and, via a siRNA-mediated mechanism, in heterochromatin formation and possibly in silencing repeats and transposons [51].
Although the paper by Braun and colleagues [51] represents a milestone towards furthering our understanding of the role of RNA silencing in Toxoplasma biology, functional studies remain to be carried out. There are a number of important aspects of Toxoplasma RNA silencing that need to be addressed, including validation of the regulatory function of miRNAs, the identification of mRNA targets and the analysis of the consequences of genetically ablating components of the RNA silencing machinery. Also, it will be very interesting to determine the full repertoire of miRNAs during the various stages of the Toxoplasma life cycle.
Concluding remarks
The analysis of small RNAs is a relatively new area of investigation in the biology of parasitic protozoa. However, from what we know so far it is evident that different organisms express different sets of small RNAs with regulatory potential. The challenge ahead is to analyze their impact on the biology of parasites, with respect to their complex life cycles and pathogenesis. High-throughput sequencing of small RNAs bound to their cognate AGO proteins will provide an invaluable tool to investigate the full repertoire in each organism and perhaps reveal new classes of small silencing RNAs. Lastly, functional studies, in those organisms that have robust genetic tools, like T. gondii, will undoubtedly provide invaluable insights into the small silencing RNA biology.
Acknowledgments
Work in the authors’ laboratory is funded by grants AI28798 and AI56333 from the National Institutes of Health and VDA is the recipient of a James Hudson Brown - Alexander Brown Coxe post-doctoral fellowship. We thank Nikolay Kolev for critical reading of the manuscript and apologize to the many colleagues whose work could not be cited directly due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 4.Ender C, Meister G. Argonaute proteins at a glance. J Cell Sci. 2010;123:1819–1823. doi: 10.1242/jcs.055210. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, et al. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Wightman B, et al. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 9.Lau NC, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 11.Djikeng A, et al. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24–26-nucleotide RNAs. RNA. 2001;7:1522–1530. [PMC free article] [PubMed] [Google Scholar]
- 12.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 13.Tolia NH, Joshua-Tor L. Slicer and the Argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 14.Guo H, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Paroo Z. Biochemical principles of small RNA pathways. Annu Rev Biochem. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- 16.Li J, et al. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue N, et al. Tetracycline-regulated RNA interference in Trypanosoma congolense. Mol Biochem Parasitol. 2002;120:309–313. doi: 10.1016/s0166-6851(02)00015-4. [DOI] [PubMed] [Google Scholar]
- 18.Lye LF, et al. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DaRocha WD, et al. Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol Biochem Parasitol. 2004;133:175–186. doi: 10.1016/j.molbiopara.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol. 2003;128:217–228. doi: 10.1016/s0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 21.Ullu E, et al. RNA interference in protozoan parasites. Cell Microbiol. 2004;6:509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- 22.Baum J, et al. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 2009;37:3788–3798. doi: 10.1093/nar/gkp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngo H, et al. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi H, et al. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA. 2006;12:2063–2072. doi: 10.1261/rna.246906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrick KL, et al. Distinct and overlapping roles for two Dicer-like proteins in the RNA interference pathways of the ancient eukaryote Trypanosoma brucei. Proc Natl Acad Sci USA. 2009;106:17933–17938. doi: 10.1073/pnas.0907766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi H, et al. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol Cell Biol. 2004;24:420–427. doi: 10.1128/MCB.24.1.420-427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durand-Dubief M, Bastin P. TbAGO1, an Argonaute protein required for RNA interference is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol. 2003;1:2. doi: 10.1186/1741-7007-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi H, et al. Functional replacement of Trypanosoma brucei Argonaute by the human slicer Argonaute2. RNA. 2006;12:943–947. doi: 10.1261/rna.20806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Djikeng A, et al. An siRNA ribonucleoprotein is found associated with polyribosomes in Trypanosoma brucei. RNA. 2003;9:802–808. doi: 10.1261/rna.5270203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obado SO, et al. Functional mapping of a trypanosome centromere by chromosome fragmentation identifies a 16-kb GC-rich transcriptional “strand-switch” domain as a major feature. Genome Res. 2005;15:36–43. doi: 10.1101/gr.2895105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallick B, et al. MicroRNA switches in Trypanosoma brucei. Biochem Biophys Res Commun. 2008;372:459–463. doi: 10.1016/j.bbrc.2008.05.084. [DOI] [PubMed] [Google Scholar]
- 32.Patrick KL, et al. Genomic rearrangements and transcriptional analysis of the spliced leader-associated retrotransposon in RNA interference-deficient Trypanosoma brucei. Mol Microbiol. 2008;67:435–447. doi: 10.1111/j.1365-2958.2007.06057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durand-Dubief M, et al. The Argonaute protein TbAGO1 contributes to large and mini-chromosome segregation and is required for control of RIME retroposons and RHS pseudogene-associated transcripts. Mol Biochem Parasitol. 2007;156:144–153. doi: 10.1016/j.molbiopara.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Martienssen RA, et al. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 2005;21:450–456. doi: 10.1016/j.tig.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Rivero MR, et al. Long double-stranded RNA produces specific gene downregulation in Giardia lamblia. J Parasitol. 2010;96:815–819. doi: 10.1645/GE-2406.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prucca CG, et al. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature. 2008;456:750–754. doi: 10.1038/nature07585. [DOI] [PubMed] [Google Scholar]
- 37.Ullu E, et al. Small sense and antisense RNAs derived from a telomeric retroposon family in Giardia intestinalis. Eukaryot Cell. 2005;4:1155–1157. doi: 10.1128/EC.4.6.1155-1157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arkhipova IR, Morrison HG. Three retrotransposon families in the genome of Giardia lamblia: two telomeric, one dead. Proc Natl Acad Sci U S A. 2001;98:14497–14502. doi: 10.1073/pnas.231494798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen XS, et al. High throughput genome-wide survey of small RNAs from the parasitic protists Giardia intestinalis and Trichomonas vaginalis. Genome Biol Evol. 2009;1:165–175. doi: 10.1093/gbe/evp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ender C, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 42.Zhang YQ, et al. Genome-wide computational identification of microRNAs and their targets in the deep-branching eukaryote Giardia lamblia. Comput Biol Chem. 2009;33:391–396. doi: 10.1016/j.compbiolchem.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, et al. RNA interference in Entamoeba histolytica: implications for parasite biology and gene silencing. Future Microbiol. 2011;6:103–117. doi: 10.2217/fmb.10.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abed M, Ankri S. Molecular characterization of Entamoeba histolytica RNase III and AGO2, two RNA interference hallmark proteins. Exp Parasitol. 2005;110:265–269. doi: 10.1016/j.exppara.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 45.Drinnenberg IA, et al. RNAi in Budding Yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du Z, et al. Structural and biochemical insights into the dicing mechanism of mouse Dicer: a conserved lysine is critical for dsRNA cleavage. Proc Natl Acad Sci U S A. 2008;105:2391–2396. doi: 10.1073/pnas.0711506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, et al. Small RNAs with 5′-polyphosphate termini associate with a Piwi-related protein and regulate gene expression in the single-celled eukaryote Entamoeba histolytica. PLoS Pathog. 2008;4:e1000219. doi: 10.1371/journal.ppat.1000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacFarlane RC, Singh U. Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: potential implications for amebic pathogenesis. Infect Immun. 2006;74:340–351. doi: 10.1128/IAI.74.1.340-351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clark CG, et al. Unique organisation of tRNA genes in Entamoeba histolytica. Mol Biochem Parasitol. 2006;146:24–29. doi: 10.1016/j.molbiopara.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 50.Lee YS, et al. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braun L, et al. A complex small RNA repertoire is generated by a plant/fungal-like machinery and effected by a metazoan-like Argonaute in the single-cell human parasite Toxoplasma gondii. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi H, et al. RNA interference in Trypanosoma brucei: the role of the amino-terminal RGG domain and the polyribosome association of Argonaute1. J Biol Chem. 2009;284:36511–36520. doi: 10.1074/jbc.M109.073072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baulcombe DC. Molecular biology. Amplified silencing. Science. 2007;315:199–200. doi: 10.1126/science.1138030. [DOI] [PubMed] [Google Scholar]
- 54.Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- 55.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 56.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 57.Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi H, et al. Function of the trypanosome Argonaute 1 protein in RNA interference requires the N-terminal RGG domain and arginine 735 in the Piwi domain. J Biol Chem. 2004;279:49889–49893. doi: 10.1074/jbc.M409280200. [DOI] [PubMed] [Google Scholar]
- 59.Peacock CS, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]