Summary
Bacteria respond to physical and chemical stresses that affect the integrity of the cell wall and membrane by activating an intricate cell envelope stress response. The ability of cells to regulate the biophysical properties of the membrane by adjusting fatty acid composition is known as homeoviscous adaptation. Here, we identify a homeoviscous adaptation mechanism in Bacillussubtilis regulated by the extracytoplasmic function σ factor σW. Cell envelope active compounds, including detergents, activate a sense-oriented, σW-dependent promoter within the first gene of the fabHa fabF operon. Activation leads to a decrease in the amount of FabHa coupled with an increase in FabF, the initiation and elongation condensing enzymes of fatty acid biosynthesis, respectively. Down-regulation of FabHa results in an increased reliance on the FabHb paralog leading to a greater proportion of straight chain fatty acids in the membrane, and the up-regulation of FabF increases the average fatty acid chain length. The net effect is to reduce membrane fluidity. The inactivation of the σW-dependent promoter within fabHa increased sensitivity to detergents and to antimicrobial compounds produced by other Bacillus spp. Thus, the σW stress response provides a mechanism to conditionally decrease membrane fluidity through the opposed regulation of FabHa and FabF.
Keywords: homeoviscous adaptation, membrane, detergent, ECF σ, fatty acid biosynthesis, condensing enzymes
Introduction
Bacillus subtilis, like other soil microbes, produces a wide variety of secondary metabolites, many of which have antibacterial activity. Many of these compounds affect the integrity of the cell envelope and elicit specific stress responses. B. subtilis is a model system for studying cell envelope stress responses in Gram positive bacteria mediated by a complex network of two-component regulatory systems (TCS) and extracytoplasmic function (ECF) σ factors (Jordan et al., 2008). B. subtilis encodes seven ECF σ factors, several of which are induced by, and confer resistance to, antibiotics targeting the cell envelope (Helmann, 2002). Many cell envelope active compounds are detergents or otherwise affect the biophysical properties of the phospholipid bilayer. As a result, cells have evolved the ability to modify membrane lipid composition to acclimatize to membrane stress (Zhang and Rock, 2008). Some modifications affect the net membrane charge, while others adjust fluidity by changing the fatty acid (FA) composition (Denich et al., 2003; Rivera-Milla et al., 2006). Adjustments in membrane fluidity, known as homeoviscous adaptation, are critical for maintaining the desired biophysical properties such as permeability of the lipid bilayer, protein mobility, protein-protein interactions and active transport processes (Los and Murata, 2004). A clear example of homeoviscus adaptation is the response of B. subtilis to cold-temperature stress (Lu et al., 2004; Aguilar and de Mendoza, 2006; Zhang and Rock, 2008). The decrease in membrane fluidity associated with a rapid decrease in temperature triggers the activation of the DesK histidine kinase that phosphorylates its cognate response regulator (DesR), which activates the expression of des encoding an acyl-desaturase that increases membrane fluidity by introducing a double bond into the FA chains of existing phospholipids (Mansilla et al., 2004; Cybulski et al., 2010).
Several ECF σ factors of B. subtilis are involved in stress responses elicited by compounds that affect membrane integrity and/or fluidity. A strain lacking all seven ECF σ factors displays increased susceptibility to antibiotics and detergents that affect the cell membrane (Luo et al., 2010). The σX regulon includes the phosphatidylethanolamine synthesis genes pssA and psd (Cao and Helmann, 2004) while the σM regulon includes the phosphatidylglycerol hydrolysis enzyme YtpA (Tamehiro et al., 2002; Eiamphungporn and Helmann, 2008) and numerous proteins involved in cell wall synthesis and cell division (Eiamphungporn and Helmann, 2008). Both the σM and σV stress responses are activated when the phosphatidylglycerol content of the membrane is reduced (Hashimoto et al., 2009). The σW regulon includes numerous membrane-localized proteins (Huang et al., 1999) and is activated under conditions of membrane stress, such as the presence of detergents or when membrane proteins are overproduced (Eiamphungporn and Helmann, 2008; Zweers et al., 2009).
Here, we report a σW-dependent pathway that contributes to homeoviscous adaptation in B. subtilis by modifying the membrane phospholipid structure. In contrast with the DesRK pathway, which responds to conditions that decrease membrane fluidity, σW responds to compounds that increase membrane fluidity. Activation of a σW-dependent promoter within the fabHa fabF operon downregulates FabHa and upregulates FabF leading to a higher proportion of straight chain FA and a longer average chain length for membrane phospholipids. These membrane compositional changes reduce bilayer fluidity and increase resistance to detergents and antimicrobial compounds produced by other Bacillus species.
Results and discussion
Identification of an active σW-dependent promoter within fabHa
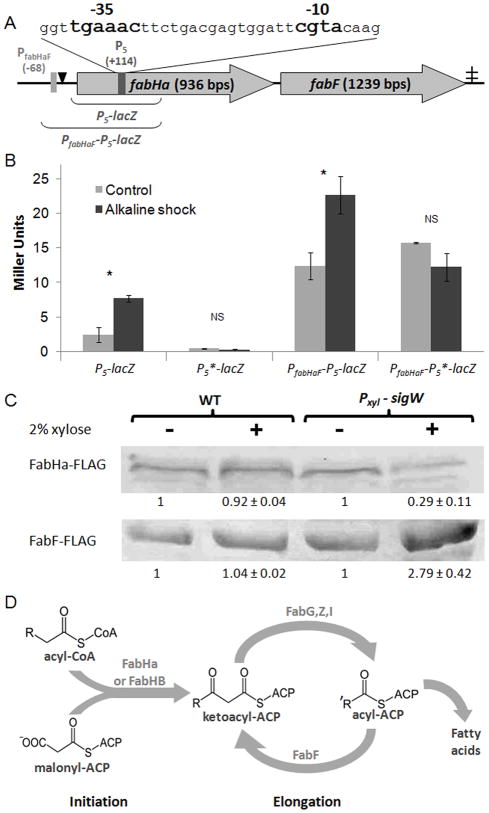
Because ECF σ promoters are, as a class, highly conserved (Helmann, 2002; Koo et al., 2009), computer-based searches have been effective in identifying candidate promoters. The first efforts to identify regulons controlled by ECF σ factors relied on promoter consensus sequence searches directed to intergenic regions (Huang and Helmann, 1998; Huang et al., 1999). Subsequently, the search was expanded to incorporate microarray-based methods to detect mRNAs produced by ECF σ factors in vivo (transcriptional profiling) and in vitro (ROMA; run-off transcription microarray analysis) (Cao et al., 2002a; Cao et al., 2002b; Cao and Helmann, 2004; Eiamphungporn and Helmann, 2008). In the σM regulon, several promoter sites are located within genes (Eiamphungporn and Helmann, 2008), a finding consistent with whole genome chromatin immunoprecipitation studies in other systems (Wade et al., 2006). We therefore searched the B. subtilis genome to determine if other candidate ECF σ promoters were present within annotated genes. Here, we focus on one such element, arbitrarily designated P5, within the fabHa gene. P5 contains a perfect match to the σW consensus in both the -35 (TGAAAC) and -10 (CGTA) elements (Helmann, 2002; Mascher et al., 2007) (Fig. 1A). A 5′-RACE analysis of detergent-treated wild type cells confirmed the presence of a transcript with a 5′ end corresponding to transcription from P5.
Figure 1.
A σW dependent promoter (P5) in fabHa.
A. Schematic map of P5, PfabHaF, FabHa, FabF, and the associated FapR binding site (black triangle). The -35 and -10 elements of P5 are in bold. The locations of the P5 and PfabHaF transcriptional start sites (+114 and −68 respectively) are described relative to the first base of the FabHa translation initiation codon. The ‡ indicates a putative terminator. The DNA regions included in the PfabHaF-P5-lacZ and P5-lacZ fusions are also illustrated.
B. The β-galactosidase activity of the P5-lacZ (HB13069), P5*-lacZ (HB13080), PfabHaF-P5-lacZ (HB13001), and PfabHaF-P5*-lacZ (HB13082) strains grown to mid-log phase in LB with and without alkaline shock. This experiment was performed in biological triplicate and repeated at least three times. Bars represent mean values with error bars indicating standard error. Student’s t tests were performed, and a statistically significant difference (P value ≥ 0.05) between the control and alkaline shocked cells is denoted as * above the bar graph while a non-significant difference is denoted as NS.
C. Detection of FLAG-tagged FabHa and FabF by Western blot with anti-FLAG antibodies in the strains fabHa-FLAG (HB13054), fabF-FLAG (HB13056), fabHa-FLAG Pxyl-sigW (HB13058), and fabF-FLAG Pxyl-sigW (HB13060) with and without xylose treatment. This experiment was performed in biological triplicates and repeated at least three times. The numbers below each band represent the average intensity of that band (± standard error) relative to the non-xylose treated control for that strain. Using Student’s t-tests, a statistically significant difference (P value ≤ 0.05) between the control and xylose treated cells was found in strains containing the Pxyl-sigW construct but not in strains lacking this construct.
D. Overview of FA biosynthesis. Chain initiation requires either FabHa or FabHb. FabHb has a greater ability to accept straight chain precursors than FabHa. Increased abundance of the FabF elongation enzyme can increase the chain length of the resulting FA (Choi et al., 2000; Schujman and de Mendoza, 2008; and this work).
Several of the seven ECF σ factors in B. subtilis overlap in their promoter recognition properties (Mascher et al., 2007; Luo et al., 2010). To determine which ECF σ factors activate the P5 promoter we used a strain with an ectopic P5-lacZ fusion inserted at the amyE locus (HB13069). Using a disk diffusion assay, P5-promoted β-galactosidase activity was observed with activators of the σW-regulon such as polymyxin B, vancomycin, cephalosporin C, D-cycloserine, and triton X-100 (Table 1, Fig. S1) (Cao et al., 2002b). A sigW null mutant (HB13099) exhibited no induction of β-galactosidase. Moreover, a strain deleted for all ECF σ factor genes except for sigW behaved like wild-type. Therefore, σW is both necessary and sufficient for activating P5 in response to antibiotics and detergents.
Table 1.
Summary of disk diffusion assays to monitor induction of P5-lacZ by cell envelope antibiotics
| Stress | P5-lacZ | P5-lacZ ΔσM, V, X, Y, ylaC, Z | P5-lacZ ΔσW |
|---|---|---|---|
| Triton X-100 | +++ | +++ | - |
| Vancomycin | ++ | ++ | - |
| Polymyxin B | + | + | - |
| Cephalosporin | + | + | - |
| D-cycloserine | + | + | - |
The induction ability of the listed antibiotics was measured in the reporter strains HB13069 (P5-lacZ), HB13151 (P5-lacZ ΔsigM ΔsigY ΔsigZ ΔsigV ΔylaC ΔsigX), and HB13099 (P5-lacZ sigW::MLS) using disk diffusion assays (see Fig. S1 for examples). Blue halos were observed after overnight incubation: +++ (strong blue) > ++ (blue) > + (light blue) > - (white after 7 days, no induction). Each disk diffusion assay was performed at least three times with biological triplicates. Data shown is representative of all experiments.
σW regulates transcription of the fabHa-fabF operon
To obtain a more quantitative assessment of P5 activity, we performed β-galactosidase assays on a strain carrying an ectopically located P5-lacZ fusion after induction by alkaline shock as described (Wiegert et al., 2001). P5 displayed a low basal activity that increased ~3-fold in response to alkaline shock (Fig. 1B). To verify that this activity was due to P5, a P5*-lacZ strain was constructed in which the -10 element was changed from CGTA to CGGA (HB13082). No known σW promoters have a G at this position (Cao et al., 2002a; Helmann, 2002), and as predicted this P5*-lacZ strain expressed no detectable β-galactosidase activity either with or without alkaline shock.
The fabHaF operon is expressed from a promoter, PfabHaF, negatively regulated by FapR, a repressor of genes involved in membrane lipid biosynthesis (Schujman et al., 2003; Schujman et al., 2006; Schujman and de Mendoza, 2008). β-galactosidase activity in strain HB13001 containing an ectopically located PfabHaF -P5-lacZ fusion (Fig. 1B) inserted at amyE increased by ~2-fold in response to alkaline shock, whereas no induction was noted for the corresponding PfabHaF -P5*-lacZ fusion strain. Thus, σW-dependent activation of P5 has a measurable impact even in the presence of PfabHaF.
Activating σW increases FabF and reduces FabHa
To determine how P5 affects FabHa and FabF protein levels, strains with ectopic copies of fabHa and fabF containing C-terminal FLAG sequences (HB13054 and HB13056, respectively) were constructed. The fabHa-flag and fabF-flag constructs were expressed under the control of both native promoters (PfabHaF and P5) to ensure that the amount of FabHa-FLAG and FabF-FLAG within the cell approximated wild type levels and was appropriately influenced by P5. P5 was activated using a xylose-inducible copy of sigW. Induction of σW reduced FabHa-FLAG (HB13058) levels by ~4-fold and increased FabF-FLAG (HB13060) by ~3-fold compared to uninduced control cells (Fig. 1C). In contrast, expression was unaffected in control strains lacking the Pxyl-sigW fusion (Fig. 1C) or containing the inactive P5* promoter (Fig. S2A). Analogous experiments using alkaline shock to activate σW resulted in similar effects, albeit of reduced magnitude: FabHa decreased by 53% while FabF increased by 25% (Fig. S2B). Thus, P5 activation directly downregulates FabHa and upregulates FabF protein levels.
Activation of σW alters membrane composition
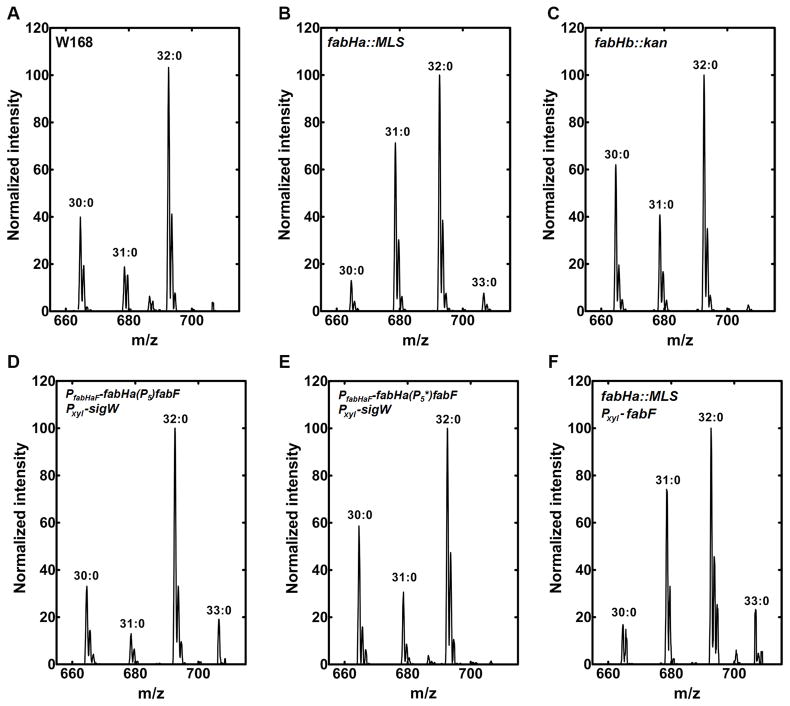
B. subtilis has two initiating condensing enzymes, FabHa and FabHb, that have been biochemically characterized (Fig. 1D): both enzymes utilize branched-chain primers but FabHb is significantly more capable of utilizing the straight chain precursor acetyl-CoA (Choi et al., 2000). The function of the two condensing enzymes in vivo was assessed by determining the FA composition of knockout strains. The inactivation of FabHa (HB 13127) led to a significant increase in the proportion of straight-chain FA (Table 2) with a concomitant increase in 31:0-carbon phosphatidylethanolamine species (Fig. 2A & 2B) compared to W168. The phosphatidylglycerol molecular species profiles were the same as phosphatidylethanolamine (not shown), consistent with phosphatidic acid being a common precursor to both phospholipids. The fabHB deletion strain (HB13115) had a FA composition (Table 2) and molecular species composition (Fig. 2A & 2C) similar to strain W168, although there was a small decrease in the proportion of straight-chain FA. These data are consistent with the biochemical properties of the two enzymes and indicate that FabHa is the principal condensing enzyme responsible for the initiation of FA synthesis in non-stressed B. subtilis cells. A decrease in branched-chain FA synthesis is seen following σW activation when P5 is present (in strain PfabHaF-fabHa(P5)fabF Pxyl-sigW; HB13121), but not in the strain with the mutant P5* promoter (PfabHaF-fabHa(P5*)fabF Pxyl-sigW; HB13122) consistent with the observed downregulation of FabHa expression and an increased reliance on FabHb (Table 2; Fig. 2D & 2E).
Table 2.
The effect of σW on membrane composition and fluidity
| Strain | W168 | PfabHaF- fabHa(P5)fabF Pxyl-sigW | PfabHaF- fabHa(P5*)fabF Pxyl-sigW | fabHa::MLS | fabHb::kan | Pxyl-fabF | fabHa::MLS Pxyl-fabF |
|---|---|---|---|---|---|---|---|
| % C15:0 iso a | 9.6 ± 0.6 | 6.5 ± 0.8 | 10.7 ± 1.4 | 1.8 ± 0.8 | 15.6 ± 0.3 | 6.5 ± 0.9 | 1.9 ± 0.2 |
| % C15:0 anteiso a | 40.0 ± 3.1 | 31.8 ± 3.2 | 42.2 ± 2.4 | 30.3 ± 1.9 | 39.0 ± 2.8 | 32.4 ± 3.1 | 30.0 ± 2.7 |
| % C16:0 iso a | 3.6 ± 0.4 | 2.3 ± 0.5 | 1.7 ± 0.5 | 5.2 ± 0.4 | 4.8 ± 0.8 | 3.5 ± 1.1 | 4.3 ± 0.4 |
| % C16:0 a | 8.3 ± 1.6 | 8.3 ± 0.5 | 7.5 ± 1.1 | 22.2 ± 1.8 | 6.8 ± 1.2 | 8.5 ± 1.6 | 16.8 ± 1.2 |
| % C17:0 iso a | 10.3 ± 2.0 | 9.0 ± 1.7 | 8.1 ± 0.7 | 4.8 ± 0.7 | 10.6 ± 2.2 | 11.9 ± 2.4 | 5.4 ± 1.0 |
| % C17:0 anteiso a | 26.0 ± 1.8 | 32.5 ± 0.8 | 24.8 ± 2.1 | 27.7 ± 0.7 | 20.3 ± 0.4 | 34.0 ± 2.0 | 35.4 ± 2.2 |
| % C18:0 a | 2.2 ± 0.6 | 9.6 ± 1.0 | 2.8 ± 0.8 | 7.8 ± 1.1 | 2.0 ± 0.7 | 3.3 ± 0.4 | 6.0 ± 0.9 |
| % straight FAs a | 10.5 ± 1.0 | 17.9 ± 1.2 | 10.3 ± 1.1 | 29.3 ± 0.7 | 8.8 ± 1.0 | 11.8 ± 1.8 | 22.0 ± 0.7 |
| % iso FAs a | 23.5 ± 2.9 | 17.8 ± 2.9 | 20.6 ± 1.6 | 11.4 ± 1.1 | 31.0 ± 2.0 | 21.9 ± 3.9 | 12.0 ± 1.3 |
| % anteiso FAs a | 66.0 ± 3.5 | 64.3 ± 3.9 | 67.0 ± 2.1 | 58.0 ± 1.5 | 59.3 ± 2.5 | 66.4 ± 4.0 | 65.7 ± 0.9 |
| 17:15 FA ratio a | 0.73 ± 0.07 | 1.08 ± 0.09 | 0.62 ± 0.08 | 1.03 ± 0.11 | 0.57 ± 0.08 | 1.18 ± 0.12 | 1.32 ± 0.22 |
| Fluorescence anisotropy analysisb | 0.185 ± 0.005 | 0.217 ± 0.003 | 0.202 ± 0.004 | ND | ND | ND | ND |
Data derived from FAME analysis of the following strains under xylose inducing conditions: W168, PfabHaF-fabHa(P5)fabF Pxyl-sigW (HB13121), PfabHaF-fabHa(P5*)fabF Pxyl-sigW (HB13122) fabHa::MLS (HB13127), fabHb::kan (HB13115), Pxyl-fabF (HB13128), and fabHa::MLS Pxyl-fabF (HB13132). Data is presented as the average of three trials (± standard error).
Data derived from fluorescence anisotropy analysis of the wild type (W168), PfabHaF-fabHa(P5)fabF amyE::Pxyl-sigW (HB13121), and PfabHaF-fabHa(P5*)fabF amyE::Pxyl-sigW (HB13122) strains under xylose inducing conditions. Data is presented as the average of three trials (± standard error). Student’s t-tests were performed, and all three values were found to be statistically different (P value ≤ 0.05) from each other.
Figure 2.
Phosphatidylethanolamine molecular species of genetically modified strains. The phosphatidylethanolamine molecular species fingerprint was obtained by ESI mass spectrometry of cells grown under inducing conditions (+ xylose). Data shown are representative of least three separate experiments performed in biological triplicates.
A. Wild-type control strain (W168)
B. fabHa::MLS (HB13127)
C. fabHb::kan (HB13115)
D. PfabHaF-fabHa(P5)fabF Pxyl-sigW (HB13121)
E. PfabHaF-fabHa(P5*)fabF Pxyl-sigW (HB13122)
F. fabHa::MLS Pxyl-fabF (HB13132).
Activation of σW (HB13121) also led to an increase in average FA chain length. As measured by the ratio of 17 to 15 carbon length FA (17:15 ratio), activation of σW led to an increase from 0.73 (wild-type) to 1.08. This increase was not observed (ratio of 0.62) in the strain containing the inactive P5* promoter (HB13122, Table 2). An increase in FabF expression is sufficient to account for the increase in 17:15 ratio since overexpression of FabF led to a ratio of 1.18 in strain HB13128. However, an increase in FabF levels alone does not account for all effects of σW on FA chain length: a hallmark of σW activation is the appearance of the C33:0 phosphatidylethanolamine species (Fig. 2D) that reflects a significant increase in 18 carbon FA (Table 2). Overexpression of FabF (HB13128) led to only a modest increase in 18 carbon FA chains which was substantially increased when combined with a fabHa null mutation (HB13132) (Table 2, Fig. 2F). Thus, an increase in the supply of 16 carbon acyl-ACP due to the down-regulation of FabHa provides the substrate for FabF elongation to an 18 carbon acyl-ACP. We conclude that the influence of σW on FA synthesis results from both the down-regulation of FabHa and the up-regulation of FabF protein levels. The net effect is an increase in both the proportion of straight-chain FA and the average FA chain length leading to an altered phospholipid molecular signature indicative of a less fluid bilayer.
The σW-dependent changes in membrane composition reduce membrane fluidity
To determine whether these σW-dependent changes in FA composition were sufficient to significantly alter membrane fluidity we monitored the fluorescence anisotropy of B. subtilis cells using 1,6-diphenyl-1,3,5-hexatriene (DPH). For this analysis we used isogenic strains expressing the intact fabHa-fabF operon containing either P5 (HB13121) or the inactive P5* point mutation (HB13122) following treatment with xylose to induce σW for 30 minutes. Induction of σW led to a substantially higher anisotropy (a lower degree of rotational freedom) consistent with alterations in membrane composition that reduce fluidity (Table 2). The control strain carrying the inactive P5* promoter exhibited a smaller increase in anisotropy indicating that although P5 reduces membrane fluidity, other σW-dependent pathways may also contribute to this reduction. The magnitude of the change in anisotropy dependent on a functional P5 promoter was comparable to that previously reported for a shift in growth temperature from 37°C to 25°C (Beranova et al., 2010), consistent with the hypothesis that P5 can have a physiologically relevant impact on membrane fluidity.
σW contributes to detergent resistance by activating P5
Activation of σW confers resistance to numerous cell envelope active antibiotics and detergents such as vancomycin and triton X-100 (Mascher et al., 2007). The resistance to antibiotics and detergents in strains containing the intact fabHa-fabF operon with either P5 (HB13117) or the inactive P5* promoter (HB13118) was compared to a sigW null mutant (HB6208) using disk diffusion assays. All of the detergents tested (triton X-100, N-lauryl sarcosine, DTAB, amitriptyline, and bile salts) had larger zones of inhibition in the P5* strain than in the strain containing active P5, although the effects were not as pronounced as in the sigW null mutant (Fig. 3). Therefore, σW-dependent alteration of membrane fluidity via activation of P5 contributes to detergent resistance. In contrast, activation of P5 did not affect resistance to either the membrane-active antibiotic daptomycin or the cell wall antibiotic vancomycin (Fig. S3).
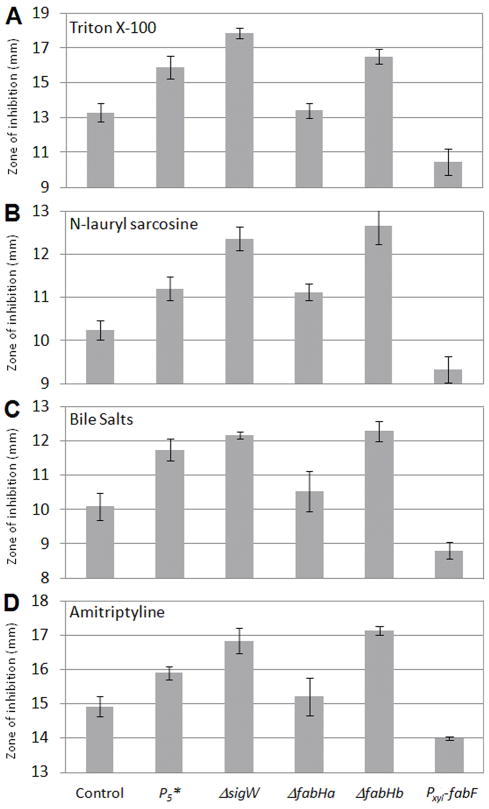
Figure 3.
Disk diffusion assays of detergent sensitivity for the PfabHaF-fabHa(P5)fabF control (HB13117), PfabHaF-fabHa(P5*)fabF (HB13118), sigW::spec (HB6208), fabHa::MLS (HB13127), fabHb::kan (HB13115), and Pxyl-fabF (HB13128) strains. Each bar represents the average zone of inhibition of at least three assays performed with three independent clones of each strain. The y axis shows the zone of inhibition (in millimeters), expressed as total diameter minus diameter of the filter paper disk (5.5 mm). Note that the scales of individual compounds vary for clarity. Error bars represent standard error. Student’s t tests were performed, and P5*, sigW::spec, fabHb::kan, and Pxyl-fabF were found to be significantly different (P value ≤ 0.05) from the wild-type control for all four stresses. The P5* and sigW::spec strains were also significantly different from each other under triton X-100, N-lauryl sarcosine, and amitriptyline treatment.
A. triton X-100
B. N-lauryl sarcosine
C. bile salts
D. amitriptyline
To determine whether detergent resistance is due to downregulation of FabHa, up-regulation of FabF, or both, we monitored detergent resistance in the fabHa::MLS (HB13127), fabHb::kan (HB13115), and xylose-inducible Pxyl-fabF (HB13128) strains. Although inactivating fabHa had little effect on detergent susceptibility, inactivating fabHb increased detergent susceptibility (Fig. 3). The increased susceptibility of the fabHb null strain underscores the importance of FabHb-initiated straight-chain fatty acids in detergent resistance which is driven by FabHa downregulation. The absence of a resistance phenotype in the fabHa::MLS strain could be attributed to the fact that deleting fabHA also removes P5 which renders the strain incapable of upregulating FabF in response to stress. Finally, the FabF-overexpressing strain was more resistant to all detergents tested illustrating the contribution of chain length to resistance (Fig. 3). We conclude that both effects of σW activation contribute to the detergent resistance phenotype.
σW contributes to resistance to antimicrobials made by other Bacillus spp. by activating P5
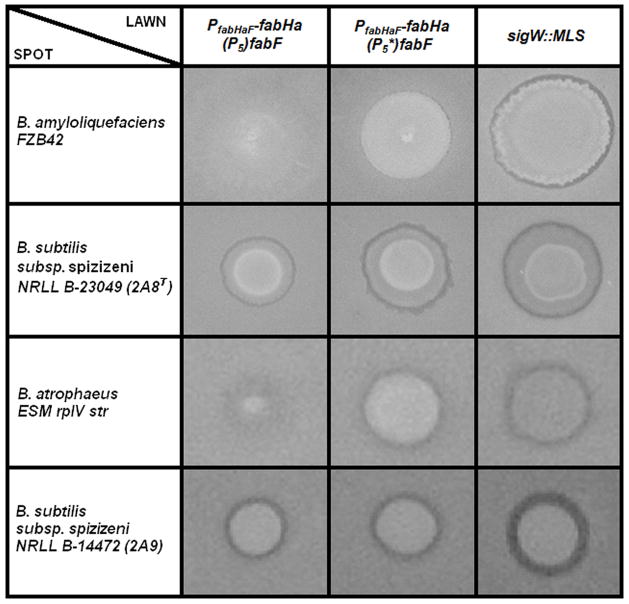
The σW regulon also confers resistance to antimicrobial compounds produced by other Bacillus species (Butcher and Helmann, 2006). We used a spot-on-lawn assay in which an antibiotic producing strain is spotted at high cell density on a lawn of the target strain to test whether σW activation of P5 affects interspecies competition. The six Bacillus species tested are known to produce compounds to which σW confers resistance (Butcher and Helmann, 2006). When the reporter (lawn) strain contained the inactive P5* promoter there was an increased sensitivity to compound(s) produced by three strains: B. amyloliquefaciens, B. subtilis ssp. spizizenii 2A8T, and B. atrophaeus ESM rplV str (Fig. 4). No significant P5-dependent differences in sensitivity were observed with B. subtilis ssp. spizizenii 2A9 (Fig. 4), B. licheniformis, or B. atrophaeus NRS-213 (Fig. S4) as the producer strains. In those strains where P5-dependent modulation of membrane composition may be important, the P5* strain was not as sensitive as the sigW null strain (Fig. 4), consistent with the previous demonstration that other σW-dependent operons contribute to antimicrobial resistance (Butcher and Helmann, 2006).
Figure 4.
Spot on lawn assays depicting the sensitivity of the pMUTIN-P5 (HB13117), pMUTIN-P5* (HB13118), and sigW::spec (HB6208) lawn strains to spots of B. amyloliquefaciens FZB42, B. subtilis subsp. spizizeni 2A8T, B. atrophaeus ESM rplV str, and B. subtilis subsp. spizizenii 2A9. The relative sensitivity of the lawn strains to each spotted strain is reflected by the size of the spot and the zone of inhibition surrounding it after 24 hours growth. A larger spot size and zone of inhibition represents increased sensitivity of the lawn strains to the metabolites produced by the spotted strains. Pictures are representative of at least three assays performed with three independent clones of each strain.
Conclusion
Our study uncovers a novel ECF σ factor dependent pathway that protects against environmental insults to the cell membrane by altering FA synthesis to produce a more rigid phospholipid bilayer. A unique aspect of the system is the presence of an internal, sense-oriented σW promoter that reduces the expression of the gene it is within (fabHa) and elevates expression of the downstream gene (fabF). The coordinate decrease in FabHa and increase in FabF protein levels combine to produce a bilayer with a different constellation of phospholipid species that leads to decreased fluidity. This biophysical response contributes to adaptation to membrane active compounds like detergents and some, yet uncharacterized, antimicrobial compounds produced by other Bacilli. The alterations of membrane phospholipid composition observed upon genetic manipulations of FabHa, FabHb and FabF expression further illustrate the relative importance of each of these enzymes in modulating membrane FA composition and thereby the resistance of cells to environmental insults.
The mechanism by which changes in FabHa and FabF protein levels affect FA composition are understood based on prior enzymological analysis in this and other systems. FabH condenses an acyl-CoA with malonyl-ACP to form a 3-ketoacyl-ACP that initiates new cycles of FA elongation (White et al., 2005). The two B. subtilis FabH isozymes, FabHa and FabHb (Lu et al., 2004), catalyze the same reaction although FabHb has a higher specific activity for straight chain FA substrates than FabHa (Choi et al., 2000). Thus, downregulation of FabHa by σW increases the proportion of straight chain FAs by increasing the cell’s reliance on FabHb (Fig. 1D). The other enzyme affected by P5 is FabF, the elongation condensing enzyme that adds two carbons to the growing FA chain in each round of elongation (White et al., 2005). As the only elongation condensing enzyme in B. subtilis, FabF activity plays a determining role in the final chain length of the acyl-ACP (de Mendoza et al., 2001; Schujman et al., 2008). The acyl-ACPs have 3 possible fates: 1) conversion to acyl-PO4 by PlsX; 2) acylation of lysophosphatidic acid by PlsC; or 3) elongation by another 2 carbons by FabF. Therefore, the σW-dependent upregulation of FabF alters the competitive balance between these 3 fates to increase the average FA chain length, as illustrated in strains where FabF is conditionally induced under xylose control. Together, these increases in the proportion of straight chain FA and average FA chain length reduce membrane fluidity. Sequence comparisons suggest that this regulatory mechanism is likely conserved in those Bacillus spp. containing two FabH paralogs, but conservation of the P5 promoter is not apparent in more distantly related bacteria that contain neither fabHb nor an obvious sigW ortholog (Table S1). In Streptococcus pneumoniae a similar regulatory effect is exerted instead by the essential YycFG TCS which downregulates fabH, upregulates fabF, and increases the average FA chain length of phospholipids in the plasma membrane (Mohedano et al., 2005). However, the physiological role for this homeoviscous adaptation system is not yet clear. Unlike the situation in B. subtilis, Listeria monocytogenes has only a single FabH which displays an altered substrate specificity at different temperatures and thereby contributes to temperature-dependent adjustments in membrane fluidity (Singh et al., 2009).
One of the best characterized mechanisms of homeoviscous adaptation is the DesRK TCS of B. subtilis that regulates expression of a FA desaturase to conditionally increase membrane fluidity (Aguilar & Mendoza, 2006). This system is initiated by the sensor kinase DesK which becomes activated in response to an increase in membrane thickness due to a temperature decrease (Cybulski et al., 2010). In contrast with DesRK, which is activated by conditions that decrease membrane fluidity, the σW-dependent response can be activated, either directly or indirectly, by conditions that increase membrane fluidity. The σW regulon mediates resistance to antimicrobial compounds by activating expression of detoxification enzymes, immunity proteins, and efflux pumps. Here, we extend this suite of mechanisms to include chemical alterations to the membrane that contribute to resistance against the action of membrane-destabilizing compounds. Since the σW regulon is induced by detergents and related membrane-active compounds, this system provides a novel mechanism of homeoviscous adaptation.
Experimental Procedures
Strains, plasmids, and growth conditions
All B. subtilis strains, plasmids, and oligonucleotides (oligos) used in this study are listed Table S2. Bacteria were grown in liquid Luria-Bertani (LB) medium at 37°C with vigorous shaking or on solid LB medium containing 1.5% Bacto agar (Difco) with appropriate selection. Plasmids were amplified in Escherichia coli DH5α before transformation of B. subtilis strains. Ampicillin (amp; 100 μg ml−1) was used to select E. coli transformants. For B. subtilis, antibiotics used for selection were: spectinomycin (spec; 100 μg ml−1), kanamycin (kan; 15μg ml−1), chloramphenicol (cat; 10μg ml−1), and macrolide-lincosoamide-streptogramin B (MLS; contains 1 μg ml−1 erythromycin and 25 μg ml−1 lincomycin).
Genetic techniques
Chromosomal and plasmid DNA transformations were performed as described previously (Harwood and Cutting, 1990). Unless otherwise stated, all PCR products were generated using W168 chromosomal DNA as a template and all strains were verified by sequence analysis (Cornell University Life Sciences Core Laboratories Center).
To create HB13069 (P5-lacZ), a DNA fragment containing P5 was PCR-amplified with primers 4577 and 4852 and cloned into pDG1661 (Guerout-Fleury et al., 1996). The resulting plasmid (pTK022) was linearized by digestion with ScaI and integrated into the amyE locus. To create HB13001 (PfabHaF-P5-lacZ), the same protocol was used except that the DNA fragment was synthesized using primers 4576 and 4577 and the resulting plasmid was pTK001.
To create HB13054 (thrC::PfabHaF-fabHa-FLAG), the fabHa ORF and the PfabHaF promoter were PCR-amplified (primers 4778 and 4780) to generate a product with a BamH1 site upstream of PfabHaF and a flag tag followed by an EcoRI site at the 3′ end of fabHa. The PCR fragment was cloned into pDG1664 (Guerout-Fleury et al., 1996) and integrated into W168 at thrC. To create HB13058 (thrC::PfabHaF-fabHa-fabF-FLAG), the same procedure was used with primers 4778 and 4781.
The Quikchange site-directed mutagenesis kit (Stratagene) and primers 4883 and 4884 were used to change the -10 element of P5 from CGTA in pTK001, pTK013, pTK015, pTK022, and pTK043 to CGGA (designated P5*) in plasmids pTK033, pTK045, pTK046 pTK031, and pTK044, respectively. To incorporate P5* at the genomic locus in B. subtilis, a 525 bp fragment containing P5, PfabHaF, and the FapR binding site was PCR-amplified (primers 4576 and 4577) and cloned into pMUTIN4 (Vagner et al., 1998) to create pTK043. P5* was generated with site-directed mutagenesis as described above to create pTK044. Uncleaved pTK044 was inserted into W168 at the fabHa locus through Campbell integration to create HB13118 in which P5 is changed to P5* at the genomic locus. Note that the upstream regulatory elements remain unchanged. Since this integration vector also incorporated ~8.6 kb of plasmid DNA upstream of PfabHaF, we created a control strain (HB13117) with pMUTIN4 inserted upstream of fabHa while maintaining wild type P5.
Gene deletions were generated using long-flanking homology PCR (LFH-PCR) as described previously (Mascher et al., 2003). To create HB13118 (fabHb::kan), 800 bp DNA fragments flanking fabHb were amplified using primers 5059 and 5060 for the upstream (Kuhry et al.) fragment and 5061 and 5062 for the downstream (DO) fragment. Extensions of ~25 nucleotides were added to the 5′ end of the UP-reverse and the DO-forward primers that were complementary (opposite strand and inverted sequence) to the ends of the kanamycin cassette. 100–150 ng of the UP and DO fragments and 250–300 ng of the resistance cassette were used together with the specific UP-forward and DO-reverse primers in a second joining PCR and the product used to transform B. subtilis. To generate a fabHa null mutant, while retaining fabF under control of PfabHaF (HB13127), an ~800 bp UP fragment was amplified with primers 5164 and 5174. The DO fragment (a PfabHaF-fabF fusion) required the fusion of two PCR product encompassing the PfabHaF promoter (primers 5165 and 5166) and fabF (primers 5167 and 5169). The UP and DO fragments were joined with an MLS cassette by PCR and used to transform B. subtilis. These extra steps were taken to insert the MLS resistance cassette upstream of PfabHaF instead of between PfabHaF and fabF to minimize the effect of deleting fabHa on fabF expression.
To generate HB13128 (Pxyl-fabF), a PCR-amplified fragment (primers 5168 and 5169) containing a ribosome binding site followed by fabF was cloned into pSWEET-bgaB (Bhavsar et al., 2001) and the resulting plasmid (pTK047) was cleaved with PstI and integrated into W168 at amyE.
5′-RACE
The start site of the P5 transcript was determined using 5′ rapid amplification of cDNA ends (5′-RACE) (Schramm et al., 2000). The W168 strain was grown to an OD600 of 0.4 and treated with 0.004% triton X-100 for 30 minutes at 37 °C with aeration. 2 μg of extracted RNA was reverse-transcribed to cDNA using TaqMan reverse transcription reagents (Applied Biosystems) and oligo P5-GSP1 (4520) as a primer. The 3′ end of the cDNA was tailed with poly(dC) using dCTP and terminal deoxynucleotidyl transferase (New England Biolabs). The tailed cDNAs were amplified by PCR with primers AAP (3314) and P5-GSP2 (4521) and sequenced.
Disk diffusion assays
Disk diffusion assays were performed as described (Mascher et al., 2007). Briefly, strains were grown to an OD600 of 0.4. A 100 μl aliquot of these cultures was mixed with 4 ml of 0.7% LB soft agar (kept at 50°C) and directly poured onto LB plates (containing 15 ml of 1.5% LB agar). The plates were dried for 20 min in a laminar airflow hood. Filter paper disks containing the chemicals to be tested were placed on the top of the agar and the plates were incubated at 37°C overnight. The distances between the edge of the inhibition zones and the edge of the filter paper disks were measured. For promoter-lacZ strains, 80 μg ml−1 Xgal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) was added to the agar and the plates were analyzed for the appearance of a blue ring around the edge of the zone of inhibition. The following chemicals and quantities were used in the disk diffusion assays: Triton X-100 10 μl of a 25% solution, amitriptyline 200 μg, polymyxin B 100 μg, vancomycin 100 μg, daptomycin 100 μg, D-cycloserine 1 mg, cephalosporin C 10 μg.
β-galactosidase assays
Strains carrying promoter-lacZ fusions were grown to an OD600 of 0.4 in LB. Cultures were then treated with alkali shock (24 mM NaOH) or a control (H2O) and samples were taken 30 min after treatment. β-galactosidase assays were performed as described by Miller (Miller, 1972).
Western Blots
Strains expressing FLAG-tagged copies of FabHa or FabF were grown to an OD600 of 0.4 and treated with either 2% xylose, 24 mM NaOH, or 200 μL H2O for 30 minutes at 37 °C with aeration. Cells were lysed and subjected to Western blot analysis using anti-FLAG antibodies to detect the fusions as described (Soonsanga et al., 2008). Relative levels of each FLAG-tagged protein were compared using densitometry analysis with ImageJ.
Spot-on-lawn assays
Spot-on-lawn assays were performed as described (Butcher and Helmann, 2006). Briefly, lawn cells were grown to an OD600 of 0.4 in LB. A 100 μl aliquot of these cultures was mixed with 4 ml of 0.7% LB soft agar (kept at 50°C) and directly poured onto LB plates (containing 15 ml of 1.5% LB agar). Plates were dried for 20 min in a laminar flow hood, and 2 μl of the producer strain (OD600 of 0.6) was spotted on top of the agar. Plates were incubated at 37°C overnight (18 h) before observation.
Fluorescence Anisotropy
We analyzed fluorescence anisotropy of B. subtilis strains treated with 1,6-diphenyl-1,3,5-hexatriene (DPH) using a modification of described methods (Svobodová and Svoboda, 1988). Strains were grown to early-log phase (OD600 of 0.2±0.01) in 5 ml LB medium. Cultures were treated with or without 2% xylose and/or 2.5 μM DPH (from a 2.5 mM stock in acetone) and incubated at 37°C with aeration for 30 minutes. For each culture, a 1 ml sample was washed once and suspended in 2 mL phosphate buffer (100 mM, pH 7.0). Fluorescence anisotropy measurements (λex=358 nm, slit width=10 nm; λem=428 nm, slit width=15 nm) were performed with a PerkinElmer LS55 luminescence spectrometer. The correction for the fluorescence intensity of non-labeled cells was calculated according to (Kuhry et al., 1985).
Lipid analysis
Cells were grown in M9 minimal medium supplemented with 0.05% casamino acids, 10 μg ml−1 tryptophan, 0.1% glutamate, 1 mM MgSO4, 0.1 mM CaCl2, 500 nM MnCl2, 10μM FeCl3, 0.5% glucose and 2% xylose. Cells were harvested and lipids extracted as described by Bligh and Dyer (Bligh and Dyer, 1959). FA methyl esters were prepared using methanol-HCl and were identified and quantified using a Hewlett-Packard model 5890 gas chromatograph equipped with a flame ionization detector as described (Choi et al., 2000). FA compositions are expressed as weight percent. Samples for molecular species analysis were dried and resuspended in chloroform:methanol:formic acid (50:50:1). Mass spectra were obtained using a Thermo Finnigan TSQ Quantum Ultra triple quadrupole mass spectrometer (Thermo Fisher Scientific) operated in the positive ESI mode using neutral loss scanning for 141 m/z corresponding to the loss of the phosphoryl headgroup of phosphatidylethanolamine (Leonardi et al., 2009). Ion source parameters were: spray voltage 3000 V, capillary temperature 270°C, capillary offset 35 V, and tube lens offset set by infusion of the polytyrosine tuning and calibration solution (Thermo Electron, San Jose, CA) in electrospray mode. Acquisition parameters for phosphatidylethanolamine were: scan range 600–900 m/z, scan time 0.5 s, neutral loss mass 141.0 m/z, collision energy 30 V, peak width Q1 and Q3 0.7 FWHM, and Q2 CID gas 0.5 mTorr. The sample was injected into the loop using a syringe pump at a flow rate of 5 μl min−1 and the data were collected for 3 min. and analyzed using QuantumTune software version 1.2 (Thermo Fisher Scientific).
Statistical analysis
All experiments were performed with a minimum of three biological replicates. Unless otherwise noted, data is presented as mean ± standard error. Statistical evaluation of the data was performedby the use of unpaired Student’s t tests. A value of P ≤ 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM47446 to JDH; GM034496 to COR), Cancer Center (CORE) Support Grant CA21765 and the American Lebanese Syrian Associated Charities.
Contributor Information
Anthony W. Kingston, Email: awk57@cornell.edu.
Chitra Subramanian, Email: Chitra.subramanian@stjude.org.
Charles O. Rock, Email: Charles.rock@stjude.org.
References
- Aguilar PS, de Mendoza D. Control of fatty acid desaturation: a mechanism conserved from bacteria to humans. Mol Microbiol. 2006;62:1507–1514. doi: 10.1111/j.1365-2958.2006.05484.x. [DOI] [PubMed] [Google Scholar]
- Beranova J, Mansilla MC, de Mendoza D, Elhottova D, Konopasek I. Differences in cold adaptation of Bacillus subtilis under anaerobic and aerobic conditions. J Bacteriol. 2010;192:4164–4171. doi: 10.1128/JB.00384-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar AP, Zhao X, Brown ED. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: Conditional complementation of a teichoic acid mutant. Appl Environ Microbiol. 2001;67:403–410. doi: 10.1128/AEM.67.1.403-410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Butcher BG, Helmann JD. Identification of Bacillus subtilis σW-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol Microbiol. 2006;60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- Cao M, Kobel PA, Morshedi MM, Fang M, Wu W. Defining the Bacillus subtilis σW regulon: A comparative analsis of promoter consensus search, run-off transcription/macroarrray analysis (ROMA), and transcriptional profiling approaches. J Mol Biol. 2002a;315:443–457. doi: 10.1006/jmbi.2001.5372. [DOI] [PubMed] [Google Scholar]
- Cao M, Wang T, Ye R, Helmann JD. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol Microbiol. 2002b;45:1267–1276. doi: 10.1046/j.1365-2958.2002.03050.x. [DOI] [PubMed] [Google Scholar]
- Cao M, Helmann JD. The Bacillus subtilis extracytoplasmic-function σX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J Bacteriol. 2004;186:1136–1146. doi: 10.1128/JB.186.4.1136-1146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Heath RJ, Rock CO. β-ketyacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J Bacteriol. 2000;182:365–370. doi: 10.1128/jb.182.2.365-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski LE, Martin M, Mansilla MC, Fernandez A, de Mendoza D. Membrane thickness cue for cold sensing in a bacterium. Curr Biol. 2010;20:1539–1544. doi: 10.1016/j.cub.2010.06.074. [DOI] [PubMed] [Google Scholar]
- de Mendoza D, Aguilar P, Schujman G. Biosynthesis and function of membrane lipids. In: Hoch JA, Sonenshein AL, Losick R, editors. Bacillus subtilis and Its Closest Relatives: From Genes to Cells. Washington D.C: American Society for Microbiology; 2001. pp. 43–55. [Google Scholar]
- Denich TJ, Beaudette LA, Lee H, Trevors JT. Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J Microbiol Meth. 2003;52:149–182. doi: 10.1016/s0167-7012(02)00155-0. [DOI] [PubMed] [Google Scholar]
- Eiamphungporn W, Helmann JD. The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol Microbiol. 2008;67:830–848. doi: 10.1111/j.1365-2958.2007.06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons Ltd; 1990. [Google Scholar]
- Hashimoto M, Takahashi H, Hara Y, Hara H, Asai K, Sadie Y, Matsumoto K. Induction of extracytoplasmic function sigma factors in Bacillus subtilis cells with membranes of reduced phosphatidylglycerol content. Genes Genet Syst. 2009;84:191–198. doi: 10.1266/ggs.84.191. [DOI] [PubMed] [Google Scholar]
- Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- Huang X, Helmann JD. Identification of target promoters for the Bacillus subtilis sigma X factor using a consensus-directed search. J Mol Biol. 1998;279:165–173. doi: 10.1006/jmbi.1998.1765. [DOI] [PubMed] [Google Scholar]
- Huang X, Gaballa A, Cao M, Helmann JD. Identification of target promoters for the Bacillus subtilis extracytoplasmic function σ factor, σW. Mol Microbiol. 1999;31:361–371. doi: 10.1046/j.1365-2958.1999.01180.x. [DOI] [PubMed] [Google Scholar]
- Jordan S, Hutchings MI, Mascher T. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol Rev. 2008;32:107–146. doi: 10.1111/j.1574-6976.2007.00091.x. [DOI] [PubMed] [Google Scholar]
- Koo BM, Rhodius VA, Nonaka G, deHaseth PL, Gross CA. Reduced capacity of alternative sigmas to melt promoters ensures stringent promoter recognition. Genes Dev. 2009;23:2426–2436. doi: 10.1101/gad.1843709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhry JG, Duportail G, Bronner C, Laustriat G. Plasma membrane fluidity measurements on whole living cells by fluorescence anisotropy of trimethylammoniumdiphenylhexatriene. Biochim Biophys Acta. 1985;845:60–67. doi: 10.1016/0167-4889(85)90055-2. [DOI] [PubMed] [Google Scholar]
- Leonardi R, Frank MW, Jackson PD, Rock CO, Jackowski S. Elimination of the CDP-ethanolamine pathway disrupts hepatic lipid homeostasis. J Biol Chem. 2009;284:27077–27089. doi: 10.1074/jbc.M109.031336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los DA, Murata N. Membrane fluidity and its roles in the perception of environmental signals. Biochim Biophys Acta. 2004;1666:142–157. doi: 10.1016/j.bbamem.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Lu YJ, Zhang YM, Rock CO. Product diversity and regulation of type II fatty acid synthases. Cell Biol. 2004;82:145–155. doi: 10.1139/o03-076. [DOI] [PubMed] [Google Scholar]
- Luo Y, Asai K, Sadaie Y, Helmann JD. Transcriptomic and phenotypic characterization of a Bacillus subtilis strain without extractyoplasmic function σ factors. J Bacteriol. 2010;192:5736–5745. doi: 10.1128/JB.00826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansilla MC, Cybulski LE, albanesi D, de Mendoza D. Control of membrane lipid fluidity by molecular thermosensors. J Bacteriol. 2004;186:6681–6688. doi: 10.1128/JB.186.20.6681-6688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol. 2003;50:1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x. [DOI] [PubMed] [Google Scholar]
- Mascher T, Hachmann AB, Helmann JD. Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors. J Bacteriol. 2007;189:6919–6927. doi: 10.1128/JB.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Mohedano ML, Overweg K, Fuente Adl, Reuter M, Altabe S, Mulholland F, et al. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J Bacteriol. 2005;187:2357–2367. doi: 10.1128/JB.187.7.2357-2367.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Milla E, Stuermer CAO, Málaga-Trillo E. Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cell Mol Life Sci. 2006;63:343–357. doi: 10.1007/s00018-005-5434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm G, Bruchhaus I, Roeder T. A simple and reliable 5′-RACE approach. Nuc Acids Res. 2000;28:e96. doi: 10.1093/nar/28.22.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schujman GE, Paoletti L, Grossman AD, de Mendoza D. FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev Cell. 2003;4:663–672. doi: 10.1016/s1534-5807(03)00123-0. [DOI] [PubMed] [Google Scholar]
- Schujman GE, Guerin M, Buschiazzo A, Schaeffer F, Llarrull LI, Reh G, et al. Structural basis of lipid biosynthesis regulation in Gram-positive bacteria. EMBO J. 2006;25:4074–4083. doi: 10.1038/sj.emboj.7601284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schujman GE, Altabe S, de Mendoza D. A malonyl-CoA-dependent switch in the bacterial response to a dysfunction of lipid metabolism. Mol Microbiol. 2008;68:987–996. doi: 10.1111/j.1365-2958.2008.06202.x. [DOI] [PubMed] [Google Scholar]
- Schujman GE, de Mendoza D. Regulation of type II fatty acid synthase in Gram-positive bacteria. Curr Opin Microbiol. 2008;11:148–152. doi: 10.1016/j.mib.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Singh AK, Zhang YM, Zhu K, Subramanian C, Li Z, Jayaswal RK, et al. FabH selectivity for anteiso branched-chain fatty acid precursors in low-temperature adaptation in Listeria monocytogenes. FEMS Microbiol Lett. 2009;301:188–192. doi: 10.1111/j.1574-6968.2009.01814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soonsanga S, Lee JW, Helmann JD. Conversion of Bacillus subtilis OhrR from a 1-Cys to a 2-Cys peroxide sensor. J Bacteriol. 2008;190:5738–5745. doi: 10.1128/JB.00576-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svobodová J, Svoboda P. Cytoplasmic membrane fluidity measurements on intact living cells of Bacillus subtilis by fluorescence anisotropy of 1,6-diphenyl 1,3,5-hexatriene. Folia Microbiol (Praha) 1988;33:1–9. doi: 10.1007/BF02928006. [DOI] [PubMed] [Google Scholar]
- Tamehiro N, Okamoto-Hosoya Y, Okamoto S, Ubukata M, Hamada M, Naganawa H, Ochi K. Bacilysocin, a novel phospholipid antibiotic produced by Bacillus subtilis 168. Antimicrob Agents Ch. 2002;46:315–320. doi: 10.1128/AAC.46.2.315-320.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner V, Dervyn E, Ehrlich SD. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- Wade JT, Roa DC, Grainger DC, Hurd D, Busby SJW, Struhl K, Nudler E. Extensive functional overlap between σ factors in Escherichia coli. Nat Struct Mol Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- White SW, Zheng J, Zhang YM, Rock CO. The structural biology of type II fatty acid biosynthesis. Annu Rev Microbiol. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- Wiegert T, Homuth G, Versteeg S, Schumann W. Alkaline shock induces the Bacillus subtilis σW regulon. Mol Microbiol. 2001;41:59–71. doi: 10.1046/j.1365-2958.2001.02489.x. [DOI] [PubMed] [Google Scholar]
- Zhang YM, Rock CO. Membrane lipid homesostasis in bacteria. Nat Rev Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- Zweers JC, Wiegert T, Dijl JMV. Stress-responsive systems set specific limits to the overproduction of membrane proteins in Bacillus subtilis. Appl Environ Microbiol. 2009;75:7356–7364. doi: 10.1128/AEM.01560-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.