Abstract
tRNA-guanine transglycosylases (TGTs) are responsible for incorporating 7-deazaguanine-modified bases into certain tRNAs in eubacteria (preQ1), eukarya (queuine) and archaea (preQ0). In each kingdom, the specific modified base is different. We have found that the eubacterial and eukaryal TGTs have evolved to be quite specific for their cognate heterocyclic base and that Cys145 (E. coli) is important in recognizing the amino methyl side chain of preQ1 (Chen et al. (2011) Nuc. Acids Res.39, 2834). A series of mutants of the E. coli TGT have been constructed to probe the role of three other active site amino acids in the differential recognition of heterocyclic substrates. These mutants have also been used to probe the differential inhibition of E. coli versus human TGTs by pteridines. The results indicate that mutation of these active site amino acids can “open up” the active site, allowing for the binding of competitive pteridine inhibitors. However, even the “best” of these mutants still does not recognize queuine at concentrations up to 50 μM, suggesting that other changes are necessary to adapt the eubacterial TGT to incorporate queuine into RNA. The pteridine inhibition results are consistent with an earlier hypothesis that pteridines may regulate eukaryal TGT activity
Keywords: RNA modification, queuine, biopterin
Introduction1
The tRNA-modifying enzyme tRNA-guanine transglycosylase (TGT) is found to be utilized across all three kingdoms of life, eukarya, eubacteria and archaea, with differing heterocyclic substrate and tRNA recognition [1]. In eukarya and eubacteria, TGT incorporates queuine or preQ1 (7-aminomethyl-7-deazaguanine), respectively, into position 34, the anticodon wobble position of tRNAs [2, 3], while the archaeal TGT exchanges the 7-deazaguanine derivative, preQ0 (7-cyano-7-deazaguanine), into position 15 of the D loop of tRNAs [4]. In all cases, the heterocyclic substrate is exchanged for a guanine at the third position of an RNA hairpin loop.
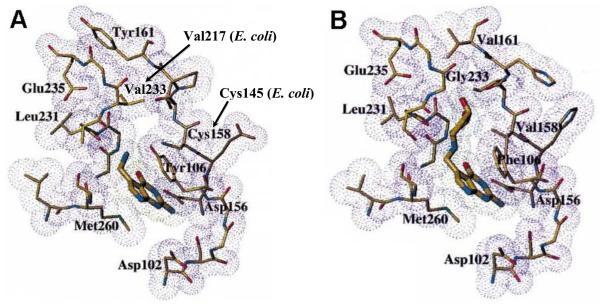
Although the crystal structure of the eukaryal TGT has yet to be solved, the determination of the Zymomonas mobilis TGT structure along with sequence homology analysis have provided insight into the major structural characteristics of all TGTs, including a (β/α)8 TIM barrel and a zinc-binding motif [5]. A follow-up study reports a comparison of the active sites of eubacterial and eukaryal TGTs based on the structure of Z. mobilis TGT and a homology model of the C. elegans enzyme [6] (Figure 1). Several differences in amino acid composition were observed in the active sites of eubacterial versus eukaryal TGTs. For instance, valine 217 (E. coli), which is replaced by a glycine in the eukaryal TGT (corresponding to G232 in human), is thought to form the ceiling of the heterocyclic binding pocket and sterically prevent the enzyme from accommodating queuine, due to the larger cyclopentenediol side chain that preQ1 lacks. Additionally, replacement of the eubacterial C145 by valine allows binding of queuine in the active site of the eukaryal enzymes. Queuine binding to the eukaryal TGT is postulated to be enhanced (with respect to preQ1 binding to the eubacterial enzyme) through additional hydrophobic interactions between the cyclopentenediol moiety and V145 [6]. Furthermore, the two hydroxyls from this moiety are predicted to be involved in hydrogen bonding with main chain carbonyls within the binding pocket. Additional examination of the crystal structure and homology model, as well as sequence alignments, have led us to postulate that additional amino acid substitutions may be involved in dictating differential substrate specificity (Table 1). In particular, T146 and P147 are replaced in the eukaryal TGT with Val and Ser, respectively.
Figure 1. Eubacterial and Eukaryal TGT Active Sites.
A) PreQ1-Bound Z. mobilis TGT Active Site (Crystal Structure) and (B) Predicted Queuine-Bound C. elegans QTRT1 Active Site (Homology Model). Amino acids are labeled based on Z. mobilis TGT numbering. C158 corresponds to C145 in E. coli and V158 corresponds to V161 in human. V233 corresponds to V217 in E. coli and G232 in human. Adapted with permission from Romier et al., (1997) FEBS Letters 416, 93-98.
Table 1.
TGT Active Site Amino Acids From Three Kingdoms
| Amino Acidsa | Eukarya (9b) | Eubacteria (85b) | Archaea (19b) |
|---|---|---|---|
| 145 | Valine 9/9c | Cysteine 83/85 Alanine 1/85 Leucine 1/85 |
Proline 19/19 |
| 146 | Valine 7/9 Isoleucine 1/9 Leucine 1/9 |
Threonine 42/85 Proline 26/85 Valine 12/85 Isoleucine 4/85 Alanine 1/85 |
Threonine18/19 Phenylalanine 1/19 |
| 147 | Serine 3/9 Lysine 2/9 Histidine 2/9 Asparagine 1/9 Valine 1/9 |
Proline 65/85 Alanine 8/85 Glutamine4/85 Glutamate3/85 Arginine 3/85 Valine 1/85 Glycine 1/85 |
Proline 12/19 Glycine 3/19 Threonine 2/19 Aspartate 1/19 Lysine 1/19 |
| 217 | Glycine 9/9 | Valine 84/85 Isoleucine 1/85 |
Proline 15/19 Valine 3/19 Threonine1/19 |
E. coli TGT numbering
The numbers of species chosen for sequence alignments in each kingdom.
The number of instances of the specific mutation relative to the total number of sequences examined.
In addition to preQ1 and queuine, a variety of guanine/preQ1/queuine structural analogues were also previously utilized to study the substrate specificity of the eukaryal TGT [3, 7]. Of all the derivatives, pteridines are particularly interesting as they are not only involved in several enzymatic reactions (serving as cofactors), but also serve as precursors to other biological molecules (such as folates). Although not substrates for TGT, pteridines have been shown to reduce/block formation of Q-tRNA in vivo and this appears to be due to the inhibition of TGT [7, 8]. Pteridine-containing Drosophila extracts have been shown to inhibit the incorporation of queuine into tRNA in L-M mouse fibroblasts [8]. Furthermore, biopterin was found to inhibit the TGT from mouse fibroblasts with a Ki of approximately 1 μM [7]. Moreover, upon induction of erythroid differentiation of Friend murine erythroleukemia cells, a 3-fold increase (to a maximum of 30-40 μM) in the level of cell tetrahydrobiopterin was observed [9, 10]. Concurrent with this increase in tetrahydrobiopterin were 5- to 10-fold increases in non-modified, queuine cognate tRNAs. Also interestingly, early studies have suggested that the queuosine biosynthesis pathway utilizes the same precursor (GTP) as the tetrahydrobiopterin (BH4) and tetrahydrofolate (THF) biosynthesis pathways [11]. It was later shown that those biosynthesis pathways appear to share the same first enzymatic step, which is mediated by GTP cyclohydrolase I [12, 13]. In addition, a very recent study has demonstrated that queuosine deficiency in eukaryotes leads to a loss of ability to generate tyrosine from phenylalanine, which results from a down regulation of the cofactor BH4 during the biotransformation [14]. All of these observations suggest a physiological link between cellular pteridines and queuine modification in vivo.
We have recently reported a study of the E. coli and human TGTs, utilizing radiolabeled preQ1 and queuine along with site-directed mutagenesis to probe the role of C145 in the E. coli enzyme [15]. We concluded that C145 and the corresponding valine in the human TGT are partly responsible for the differential recognition of preQ1 between these enzymes. In order to further probe the structural requirements for the differential recognition of the cyclopentenyldiol side chain of queuine, we have generated additional mutants of the E. coli TGT, focusing on amino acids 145, 146, 147 & 217. We have also used these mutants to probe the inhibition of the E. coli TGT by pteridines. To confirm these results, inhibition studies were conducted to probe the interaction of biopterin with the human TGT.
Materials and Methods
Reagents
Unless otherwise specified, reagents were from Sigma-Aldrich. Bradford reagent and bovine serum albumin (BSA) were from Bio-Rad. Yeast extract and bactotryptone were from Difco or Fisher. Restriction enzymes and buffers were from New England BioLabs. Glycerol, HEPES, NTPs, and phenol were from Pharmacia. RNase inhibitor, pyrophosphatase, and urea were from Roche. Biopterin and pterin were from Schircks Laboratories. DNA oligonucleotides, agarose, dithiothreitol (DTT), isopropyl β-D-thiogalactopyranoside (IPTG) and DNA ladders were ordered from Invitrogen. All restriction enzymes and Vent® DNA polymerase were ordered from New England Biolabs. The ribonucleic acid triphosphates (NTPs), pyrophosphatase and kanamycin sulfate were ordered from Roche Applied Sciences. The deoxyribonucleic acid triphosphates (dNTPs) were ordered from Promega. Scriptguard™ RNase Inhibitor was ordered from Epicentre. Epicurian coli® XL2-Blue ultracompetent cells were ordered from Stratagene. TG2 cells, K12 (DE3, Δtgt)/pLysS cells and K12 (DE3, Δtgt)/pRIPL cells were from laboratory stocks. His•Bind resin and lysonase bioprocessing reagent were also purchased from Novagen. The QIAPrep® Spin Miniprep and Maxiprep Kits were ordered from Qiagen. Precast PhastGels and SDS buffer strips were from VWR. Whatman GF/C Glass Microfibre Filters and Amicon Ultra Centrifugal Filter Devices, carbenicillin were ordered from Fisher. Radiolabeled guanine was from Moravek. [3H]-Queuine (7.3 Ci/mmol) was obtained via custom synthesis from Moravek. The human TGT and tRNATyr (human and E. coli) were prepared as previously reported [16, 17].
E. coli TGT Mutants
The active site mutants of the E. coli TGT were generated via QuikChange mutagenesis following standard procedures. The resulting clones were then screened for the presence of the desired mutation through restriction digests of the prepared plasmids contained therein and were subsequently sequenced at the University of Michigan Biomedical Research Core Facilities. Wild-type and mutant enzymes were prepared as previously described [18].
Guanine Incorporation Assays with the E. coli TGT
Reaction mixtures consisting of 100 mM HEPES (pH 7.3), 20 mM MgCl2, 5 mM DTT, saturating concentrations (20 μM) of tRNA and varying concentrations of [14C]-guanine (55-56 mCi/mmol, Moravek), were treated with 100 - 200 nM TGT and incubated at 37 °C. The total volume of a typical assay mixture was 400 μL. During a 10-15 min time interval, 70 μL aliquots were removed at a specified time and quenched in 2 mL 5% TCA. Glass microfibre filters (Whatman, GF/C) were rinsed with approximately 1 mL of TCA prior to application of the quenched samples to the filters. The test tube, containing the quenched sample was rinsed three times with TCA and the filter unit was rinsed with 100% EtOH. Filters were then dried under a heat lamp for approximately 15 min. Liquid scintillation counting was used to determine the amount of radioactivity (incorporated guanine) in each sample. Disintegrations per min (dpm) were converted to pmol guanine by multiplying by the specific activity of the radiolabeled substrate (mCi / mmol) and then by the conversion factor 1 Ci / 2.2 × 1012 dpm. These data were analyzed using Kaleidagraph, where initial velocities were calculated from linear fits of incorporated guanine versus time plots. Plots of initial velocity versus substrate concentration were generated and fit to the Michaelis-Menten equation by non-linear regression to determine kcat and KM.
Pteridine Inhibition Studies
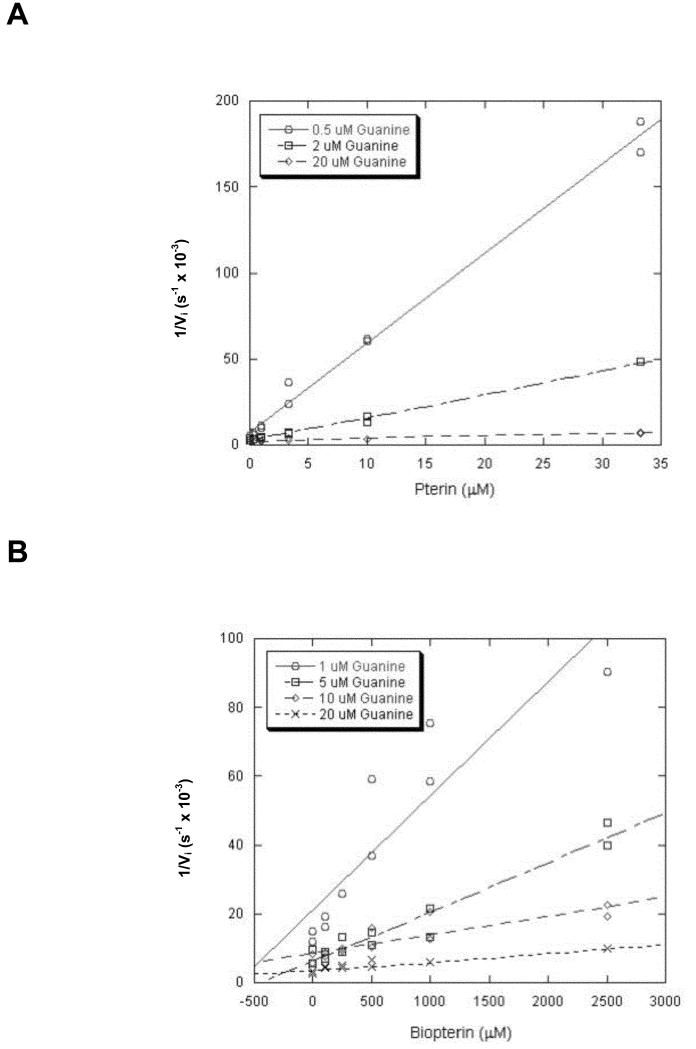
As with the kinetic parameter determinations, Kis for the mutant TGTs were determined at 20 μM tRNA, 100 mM HEPES (pH 7.3), 20 mM MgCl2, 5 mM DTT. The inhibitor concentrations were varied (4-6 concentrations) for each of 3-4 fixed concentrations of guanine. These experiments were performed in duplicate. Initial velocities were obtained for each experiment, and these data were analyzed via Dixon plots as 1/vi values were plotted versus inhibitor concentration. A linear fit was obtained for each guanine concentration (Figure 2 and Supporting Information). Each line was then used to determine Ki according to the equation:
Figure 2. Inhibition of E. coli TGT (V217G).
A) Inhibition by Pterin and B) Inhibition by Biopterin. These curves were obtained from two independent determinations of initial velocity data as described in Methods. The tRNA concentration was held constant at 20 μM while guanine and inhibitor concentrations were varied over the ranges shown.
The average of these Ki values is reported (Table 3), along with the standard deviation of the individual Ki values. A three dimensional fit of the data (using Grafit) further validated that the pteridines act as competitive inhibitors of TGT, with respect to guanine [7]. While experiments were not designed to generate data to be fit in this manner, these results confirmed the competitive inhibition model and were consistent with the Dixon analyses.
Table 3.
Pterin Inhibition Constants for Wild-type and Mutant E. coli TGTs
| E. coli TGT |
Ki Pterin a (μM) |
Ki Biopterin a (μM) |
|---|---|---|
| Wild-Type | 0.42 ( ±0.1) | > 3 mM b |
| V217G | 0.90 ( ±0.35) | 550 ( ±92) |
| C145V/T146V | 2.2 ( ±0.18) | 550 ( ±69) |
| C145V/T146V/V217G | 1.3 ( ±0.61) | 160 ( ±25) |
| C145V/T146V/P147N/V217G | 37 ( ±5.2) | 1050 ( ±100) |
Standard errors are in parentheses. Inhibition constants were calculated from the average of duplicate determinations of initial velocity data as described in the Methods section.
KI was above the biopterin solubility limit.
Biopterin Inhibition of the Human TGT
The biopterin stock solution (10 mM) was dissolved in water with the addition of 1% (v/v) 14.8 N ammonium hydroxide. Due to the light sensitivity of biopterin, the diluted biopterin solutions were prepared without direct light exposure and protected from light while stored. A serial dilution of biopterin samples (0.025-0.2 mM, pH 7.3) was prepared and the absorbance of each dilution was determined at 354 nm. The extinction coefficient obtained (8.12 mM−1cm−1) is comparable with the reported value in the literature (6.3 mM−1cm−1, pH 7.6) [19]. The queuine exchange assays were performed by varying the concentrations of [3H]-queuine (1, 2.5 and 5 μM) and biopterin (10-500 μM), while holding tRNA concentrations at 10 μM (saturating condition) in HEPES reaction buffer (100 mM HEPES, pH 7.3; 20 mM MgCl2; 5 mM DTT) at a final volume of 400 μL. All reaction mixtures were incubated at 37°C for purposes of equilibration before initiating the reaction with the addition of the human TGT (100 nM), and the studies were performed in duplicate. The initial velocities of queuine incorporation were determined by converting the slopes of these plots (pmol/min) to units of sec−1, taking into account the concentration of the enzyme and aliquot size. The data were then analyzed graphically by plotting either (A) 1/Vini vs. [Biopterin] or (B) [queuine]/Vini vs. [Biopterin] to obtain Dixon plot or Cornish-Bowden plot. The average data points (with error bars representing their standard deviations) were plotted. However, all of the individual data points were fit via linear regression using Kaleidagraph (Abelbeck Software). To obtain the Ki for the inhibitor, the following equation was used: slope (obtained from the Dixon plot) = KM / (kcat)(Ki)[S], where KM and kcat with respect to queuine have been determined previously.
Results
Kinetic parameters (KM and kcat), with respect to guanine, were determined for he E. coli TGT mutants V217G, C145V/T146V, C145V/T146V/V217G, and C145V/T146V/P147N/V217G (will subsequently be referred to as single, double, triple, and quadruple mutants, respectively) (Table 2, Supporting Information). Following initial characterization of these mutants, studies were conducted to examine their inhibition by pterin and biopterin (Figure 2, Table 3 and Supporting Information). Wild-type enzyme was inhibited by pterin; however, no inhibition of wild-type TGT was observed with 3 mM biopterin, a maximum concentration dictated by solubility of the compound. Similarly, the mutant enzymes were also inhibited by pterin. Significant differences in inhibition were observed between pterin and biopterin. For biopterin, the Ki for the triple mutant was the lowest of the four enzymes. Among these enzymes, the quadruple mutant was affected least by these pteridine inhibitors (similar to the initial kinetic studies which suggest that this mutant recognizes guanine poorly).
Table 2.
Kinetic Parameters wrt Guanine for Wild-type and Mutant E. coli TGTs
| E. coli TGT |
kcata (10−3•s−1) |
KMa (μM) |
kcat/KMa (10−3•s−1•μM−1) |
|---|---|---|---|
| Wild-type | 6.29 (0.12) | 0.35 (0.03) | 18 (1.6) |
| V217G | 0.259 (0.003) | 1.00 (0.10) | 0.26 (0.03) |
| C145V/T146V | 0.53 (0.03) | 0.87 (0.18) | 0.61 (0.13) |
| C145V/T146V/V217G | 1.22 (0.03) | 2.09 (0.24) | 0.58 (0.07) |
| C145V/T146V/P147N/V217G | 0.62 (0.09) | 35.0 (13.5) | 0.017 (0.007) |
Standard errors are in parentheses. Kinetic parameters were calculated from the average of three replicate determinations of initial velocity data as described in the Methods section.
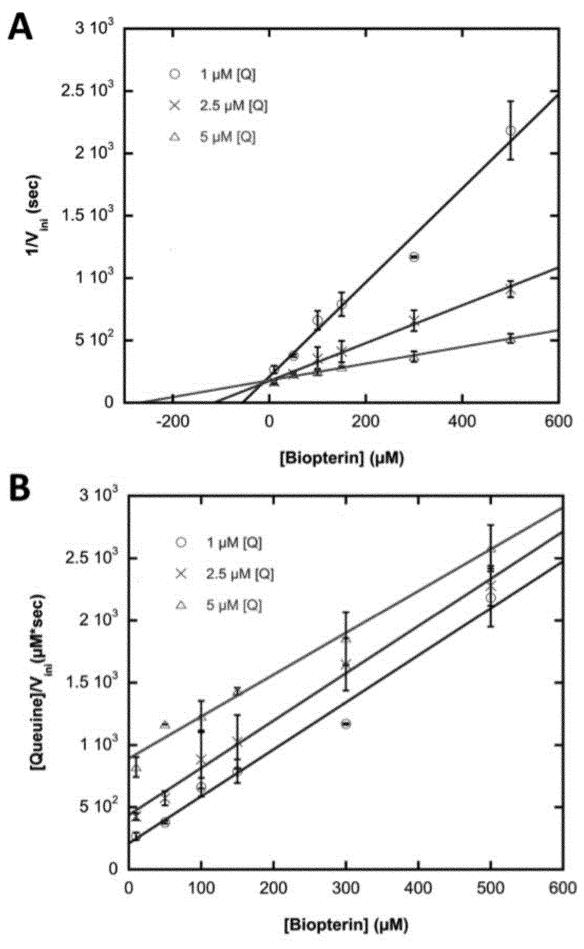
With respect to queuine, biopterin was found to be a competitive inhibitor of the human TGT according to intersecting lines in a Dixon plot (Figure 4A), and parallel lines in a Cornish-Bowden plot (Figure 4B). Derived from the Dixon plot, the inhibition constant (Ki) of biopterin was determined to be 8.72 ± 0.62 μM, which is within two orders of magnitude of the KM for queuine (0.26 ± 0.03 μM). This result suggests that biopterin is not a particularly potent inhibitor of the human TGT, consistent with previous reports regarding the eukaryal TGT [7].
Figure 4. Inhibition of Human TGT-catalyzed Queuine Incorporation by Biopterin.
A) Dixon and B) Cornish-Bowden Plots. Curves were obtained from the average of two independent determinations of initial velocity data. Error bars were generated from the standard deviation within each point.
Discussion
Despite their high levels of sequence homology, bacterial and eukaryal TGTs are known to utilize different physiological substrates (preQ1 and queuine, respectively). Romier et al. suggested, based upon their homology model (Figure 1), that C145 and V217 (E. coli numbering) could serve important roles in excluding queuine from the active sites of bacterial TGTs [6]. Subsequent examination of bacterial and eukaryal TGT sequence homology (Table 1), in addition to this structural homology model, prompted us to consider the contributions of T146 and P147 as well. In order to probe the roles of these four active site residues in differential substrate recognition, these amino acids in the E. coli TGT were mutated to residues found in eukaryal TGTs.
The V217G single mutant and C145V/T146V double mutant were created separately, as these mutants appear to address opposite sides of the TGT active site. C145 and V217 are conserved among nearly all bacterial TGTs. In contrast, all known sequences for eukaryal TGTs contain a valine and a glycine at positions corresponding to 145 and 217, respectively. While the threonine at position 146 in E. coli is less stringently conserved, this residue is excluded from the corresponding position in all known eukaryal TGTs. In order to assess the cumulative effects of these mutations, a triple mutant (C145V/T146V/V217G) was also created. Finally, the proline found at position 147 was mutated to asparagine in the C145V/T146V/P147N/V217G mutant. This proline is present in nearly 2/3 of known bacterial TGT sequences and is excluded from all known eukaryal TGT sequences. All four of these mutants were constructed using QuikChange mutagenesis, and were expressed in E. coli K12 (DE3, Δtgt)/pLysS, yielding the mutant enzymes without any contaminating wild-type. No significant changes from wild-type enzyme were observed in these proteins' chromatographic or electrophoretic behavior during purification and analysis (data not shown).
All four mutants exhibit substantially reduced, 6-25 fold, kcat values relative to the wild-type, using guanine as a substrate (Table 2). However the KM values are much closer (within an order of magnitude), with the exception of the quadruple mutant, which is 100-fold higher than wild-type. Previously, we have shown that some mutants of C145 exhibit increased kcat and KM values [15], consistent with our determination that product release is rate-limiting [20], i.e., poorer binding due to a faster off-rate would result in faster kcat and larger KM. Additional mutations of the other residues cause decreases in kcat and increases in KM to relatively smaller extents (Table 2). Additional mutation of the proline at position 147 to asparagine increases KM nearly 20-fold over that observed for the triple mutant and 100-fold over the wild-type value. Consequently, kcat/KM is decreased about 1000-fold. While this mutation is obviously deleterious (at least in conjunction with the three other mutations) in the context of the E. coli TGT scaffold, proline is not found at this position in all known eukaryal TGTs. Furthermore, proline is not absolutely conserved at this position among eubacterial TGTs [15]. Thus, it appears that mutation of this proline to asparagine disrupts binding of substrates in the active site of the E. coli enzyme; however, complementary changes elsewhere in the protein scaffold may be responsible for allowing substitution of this proline in eukaryal TGTs.
Precedent, both biochemical and physiological, exists for pteridine inhibition of TGTs. Farkas et al. reported that pteridines were inhibitors of the TGT from rabbit reticulocytes competitive with respect to guanine [7]; furthermore, a concentration of 0.48 μM pterin was reported to result in 44% inhibition. In light of these findings, we performed Dixon analyses on our inhibition data. Dixon plots (plots of 1/vi versus [I] at a fixed [S]) are an efficient approach to analyze data resulting from studies in which the mode of inhibition is known to be competitive. For each of the pteridine and mutant pairs (with one exception), the Dixon analyses (in replicate, at 3-4 different guanine concentrations) converged upon well-defined values for Ki. To further substantiate the Dixon analyses and confirm competitive inhibition, three-dimensional analysis of the data (using an equation for a competitive mode of inhibition) was performed using GraFit (not shown). These results confirmed these pteridines to be competitive inhibitors of TGT and replicated well the Ki values determined by the Dixon analyses
Pterin was observed to inhibit all mutants other than the quadruple mutant with Kis ranging from 0.90 μM to 2.2 μM. Interestingly, these values seem to mirror the guanine KMs for these enzymes (even the quadruple mutant, with its poorer binding, follows this trend). As with data reported by Hoops et al. [21], this trend is consistent with KMs for TGT being reflective of Kd values for the enzyme.
Biopterin inhibition of this series of enzymes revealed well-defined differences in binding. While the wild-type enzyme exhibited no detectable inhibition at biopterin concentrations up to 3 mM, the mutant enzymes were all inhibited by biopterin. Furthermore, the 160 μM Ki reported for the triple mutant enzyme is comparable to the biopterin inhibition reported for the rabbit reticulocyte enzyme (39% inhibition at 20 μM) [7]. The effects of mutations on either side of the active site (V217G and C145V/T146V) appear to be additive, as the single and double mutants exhibit Ki values for biopterin that are significantly higher than that observed for the triple mutant (Table 3).
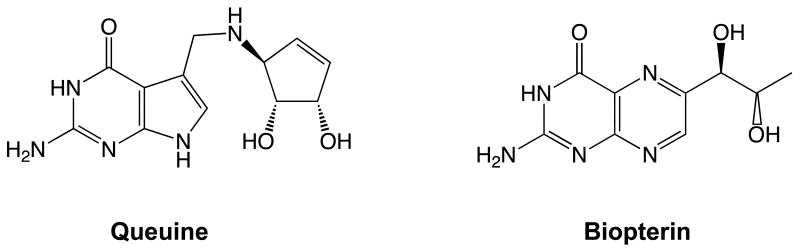
To put the above results in context, we directly examined biopterin inhibition of the human TGT. Biopterin was confirmed to be a competitive inhibitor of the human TGT with a Ki value (8.7 μM) ca. 30-fold higher than the KM for queuine (Table 4). The inhibition observed by biopterin was expected, as the diol side chain of biopterin seems to be capable of partially fulfilling the interactions that the cyclopentenediol moiety of queuine provides (Figure 3). The kcat values for the E. coli TGT mutants are lower than those for the E. coli wild-type and human TGTs. The triple mutant, TGT (C145V/T146V/V217G), appears to be the “best” mutant as it has only about a 5-fold reduction in kcat, less than a 10-fold increase in KM for guanine relative to wild-type and exhibits the lowest Ki for biopterin. We then tested the triple mutant to see if it would recognize and utilize queuine as a substrate. Unfortunately, we observed no incorporation of queuine at concentrations up to 50 μM (data not shown). It seems likely that the greater flexibility of the acyclic diol side chain of biopterin allows it to adapt to the incompletely enlarged active sites of the E. coli TGT mutants, whereas the more restricted and larger side chain of queuine may not be able to do so.
Table 4.
Queuine Kinetic Parameters and Biopterin Inhibition Constant for Human TGT
|
kcata (10−3•s−1) |
KMa (μM) |
kcat/KMa (10−3•s−1•μM−1) |
|
|---|---|---|---|
| Queuine b | 8.2 (±0.2) | 0.26 (±0.03) | 31.6 (±3.4) |
| Biopterin |
KI a (μM) 8.7 (±0.6) |
||
Standard errors are in parentheses.
From Chen et al. (2011) Nuc. Acids Res. 39, 2834.
Figure 3.
Structures of Queuine and Biopterin.
In conclusion, we have shown that mutation of key residues in the E. coli TGT active site can “open up” the site to allow for the binding of competitive pteridine inhibitors. The fact that the “best” mutant still does not incorporate queuine suggests that other changes are necessary for efficient recognition of queuine. This postulate seems plausible considering that the cyclopentenediol moiety of queuine, compared to the side of biopterin, is not only structurally bulkier, but most likely also more rigid. We have also confirmed the inhibition of the human TGT by biopterin, consistent with earlier reports on other eukaryal TGTs, and supportive of the concept that pteridines may regulate eukaryal TGT activity in vivo.
Supplementary Material
Acknowledgements
This work was supported by the University of Michigan, College of Pharmacy, Vahlteich and UpJohn Research Funds (GAG) and the NIH-NIGMS GM007767 (CET trainee).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was supported in part by NIH GM07765 (CET, trainee) and GM065489 (GAG) and the University of Michigan, College of Pharmacy, Vahlteich and UpJohn Research Funds (GAG).
Abbreviations used: Q, queuine; preQ1, 7-aminomethyl-7-deazaguanine; TGT, tRNA-guanine transglycosylase; BSA, bovine serum albumin; DTT, dithiothreitiol; IPTG, isopropyl β-D-thiogalactopyranoside; HEPES, hydroxyethylpiperazine-ethylsulfonate; Tris-HCl, tris(hydroxymethyl) aminomethane hydrochloride; SDS, sodium dodecyl sulfate; PAGE, polyacrylamide gel electrophoresis; TCA, trichloroacetic acid.
Supplementary Material
Plots for guanine incorporation kinetics and pterin and biopterin inhibition of wild-type and mutant E. coli TGTs can be found in the Supplementary Material.
References
- 1.Garcia GA, Kittendorf JD. Transglycosylation: A mechanism for RNA modification (and editing?) Bioorganic Chemistry. 2005;33:229–251. doi: 10.1016/j.bioorg.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okada N, Nishimura S. Isolation and Characterization of a Guanine Insertion Enzyme, a Specific tRNA Transglycosylase, from Escherichia coli. Journal of Biological Chemistry. 1979;254:3061–3066. [PubMed] [Google Scholar]
- 3.Shindo-Okada N, Okada N, Ohgi T, Goto T, Nishimura S. Transfer Ribonucleic Acid Guanine Transglycosylase Isolated from Rat Liver. Biochemistry. 1980;19:395–400. doi: 10.1021/bi00543a023. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe M, Matsuo M, Tanaka S, Akimoto H, Asahi S, Nishimura S, Katze JR, Hashizume T, Crain PF, McCloskey JA, Okada N. Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to archaeal tRNA, proceeds via a pathway involving base replacement on the tRNA polynucleotide chain. Journal of Biological Chemistry. 1997;272:20146–20151. doi: 10.1074/jbc.272.32.20146. [DOI] [PubMed] [Google Scholar]
- 5.Romier C, Reuter K, Suck D, Ficner R. Crystal structure of tRNA-guanine transglycosylase: RNA modification by base exchange. EMBO Journal. 1996;15:2850–2857. [PMC free article] [PubMed] [Google Scholar]
- 6.Romier C, Meyer JEW, Suck D. Slight sequence variations of a common fold explain the substrate specificities of tRNA-guanine transglycosylases from the three kingdoms. FEBS Letters. 1997;416:93–98. doi: 10.1016/s0014-5793(97)01175-7. [DOI] [PubMed] [Google Scholar]
- 7.Farkas WR, Jacobson KB, Katze JR. Substrate and Inhibitor Specificity of tRNA-Guanine Ribosyltransferase. Biochimica et Biophysica Acta. 1984;781:64–75. doi: 10.1016/0167-4781(84)90124-6. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson KB, Farkas WR, Katze JR. Presence of Queuine in Drosophilia melanogaster. Correlation of Free Pool with Queuosine Content of tRNA and Effect of Mutations in Pteridine Metabolism. Nuc. Acids Res. 1981;9:2351. doi: 10.1093/nar/9.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin VK, Farkas WR, Agris PF. Specific Changes in Q-Ribonucleoside Containing Transfer RNA Species During Friend Leukemia Cell Erythroid Differintiation. Nucleic Acids Res. 1980;8:3481–3489. doi: 10.1093/nar/8.15.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parniak MA, Andrejchyshyn S, Marx S, Kleiman L. Alterations in cell tetrahydrobiopterin levels may regulate queuine hypomodification of tRNA during differentiation of murine erythroleukemia cells. Experimental Cell Research. 1991;195:114–118. doi: 10.1016/0014-4827(91)90506-p. [DOI] [PubMed] [Google Scholar]
- 11.Kuchino Y, Kasai H, Nihei K, Nishimura S. Biosynthesis of the Modified Nucleoside Q in Transfer RNA. Nucleic Acids Res. 1976;3:393–398. doi: 10.1093/nar/3.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips G, El-Yacoubi B, Lyons B, Alvarez S, Iwata-Reuyl D, de Crecy-Lagard V. Biosynthesis of 7-Deazaguanosine-Modified tRNA Nucleosides: a New Role for GTP Cyclohydrolase I. Journal of Bacteriology. 2008;190:7876–7884. doi: 10.1128/JB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarty RM, Bandarian V. Deciphering deazapurine biosynthesis: Pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chemistry & Biology. 2008;15:790–798. doi: 10.1016/j.chembiol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rakovich T, Boland C, Bernstein I, Chikwana VM, Iwata-Reuyl D, Kelly VP. Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation. Journal of Biological Chemistry. 2011 doi: 10.1074/jbc.M111.219576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YC, Brooks AF, Goodenough-Lashua DM, Kittendorf JD, Showwalter HD, Garcia GA. Evolution of eukaryal tRNA-guanine transglycosylase: insight gained from the heterocyclic substrate recognition by the wild-type and mutant human and Escherichia coli tRNA-guanine transglycosylases. Nucleic Acids Res. 2011;39:2834–2844. doi: 10.1093/nar/gkq1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen YC, Kelly VP, Stachura SV, Garcia GA. Characterization of the human tRNA-guanine transglycosylase: Confirmation of the heterodimeric subunit structure. RNA-a Publication of the RNA Society. 2010;16:958–968. doi: 10.1261/rna.1997610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kittendorf JD, Barcomb LM, Nonekowski ST, Garcia GA. tRNA-guanine transglycosylase from Escherichia coli: Molecular mechanism and role of aspartate 89. Biochemistry. 2001;40:14123–14133. doi: 10.1021/bi0110589. [DOI] [PubMed] [Google Scholar]
- 18.Goodenough-Lashua DM, Garcia GA. tRNA-Guanine Transglycosylase from Escherichia coli: a Ping-Pong Kinetic Mechanism is Consistent with Nucleophilic Catalysis. Bioorganic Chemistry. 2003;31:331–344. doi: 10.1016/S0045-2068(03)00069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viscontini M, Loeser E, Karrer P. Fluoreszierende Stoffe aus Drosophila melanogaster .8. Isolierung und Eigenschaften des Pteridins HB2. Helvetica Chimica Acta. 1958;41:440–446. [Google Scholar]
- 20.Garcia GA, Chervin SM, Kittendorf JD. Identification of the Rate-Determining Step of tRNA-Guanine Transglycosylase from Escherichia coli. Biochemistry. 2009;48:11243–11251. doi: 10.1021/bi901501a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoops GC, Townsend LB, Garcia GA. tRNA-guanine transglycosylase from Escherichia coli: Structure-activity studies investigating the role of the aminomethyl substituent of the heterocyclic substrate preQ1. Biochemistry. 1995;34:15381–15387. doi: 10.1021/bi00046a047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.