Abstract
Matrix metalloproteinases (MMPs) are implicated in cancer development and progression and are associated with prognosis. Single-nucleotide polymorphisms (SNPs) of MMPs, most frequently located in the promoter region of the genes, have been shown to influence cancer susceptibility and/or progression. SNPs of MMP-1, -2, -3, -7, -8, -9, -12, -13 and -21 and of the tissue inhibitor of metalloproteinases (TIMPs) TIMP-1 and TIMP-2 have been studied in digestive tract tumors. The contribution of these polymorphisms to the cancer risk and prognosis of gastrointestinal tumors are reviewed in this paper.
Keywords: Matrix metalloproteinase, Tissue inhibitor of metalloproteinase, Single nucleotide polymorphism, Promoter region, Digestive tract, cancer
INTRODUCTION
The matrix metalloproteinases (MMPs) belong to a metzincin superfamily of zinc-containing proteinases. Other members of this superfamily are the ADAM (a disintegrin and metalloproteinase) family and the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family, which have also been reported to be implicated in cancer progression[1,2]. MMP as well as ADAM/ADAMTS family members are inhibited by tissue inhibitors of metalloproteinases(TIMP)s. RECK (reversion-inducing, cysteine-rich protein with Kazal motifs) is a membrane-anchored inhibitor of MMPs and ADAMs[3]. Even though ADAM, ADAMTS, and RECK are closely related to the MMPs and TIMPs, their single-nucleotide polymorphisms (SNPs) in gastrointestinal cancer have not yet been studied and are therefore not included in this review.
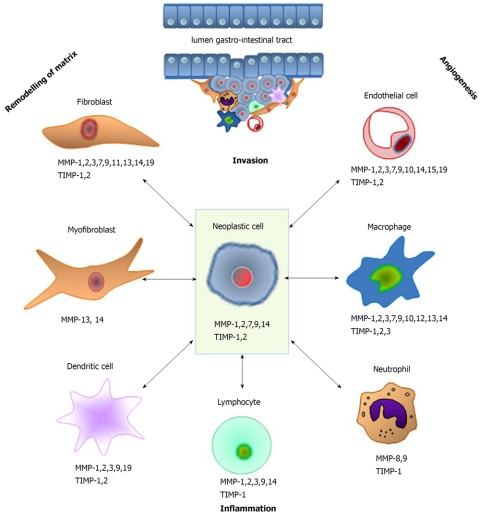
MMPs have proven to be of relevance for cancer development and prognosis in various organ systems. The 23 members of this family of endopeptidases all share a catalytic domain, a pro-peptide and a hemopexin-like C-terminal domain. According to their structure and major function or substrates, the MMPs are subdivided in the following subgroups: collagenases (MMP-1, -8 and -13), stromelysins (MMP-3, -10 and -11), matrilysins (MMP-7 and -26), gelatinases (MMP-2 and -9), membrane-type MT-MMPs (MMP-14, -15, -16, -17, -24 and -25), and others[4-6]. The most firmly established function of MMPs is the degradation/remodeling of extracellular matrix. By cleavage of receptors and their ligands they also influence various growth and signaling pathways in normal and pathological conditions[6]. In cancer, MMPs are involved in angiogenesis by regulating the bio-availability of vascular endothelial growth factor (VEGF) (e.g. MMP-9) and the cleavage of matrix-bound VEGF (MMPs -3, -7, -9 and -16)[7]. On the other hand, cleavage of plasminogen by MMP-2, -9 and -12 leads to the production of angiostatin, an inhibitor of angiogenesis[8,9]. Furthermore, MMPs have been suggested to interfere in the balance between growth signals and growth-inhibiting signals [e.g. by modulating the transforming growth factor-β (TGF-β) pathway and activation of the epidermal growth factor (EGF) receptor], to regulate the induction of apoptosis by cleavage of Fas ligand (by MMP-7), to play a role in the creation of a metastatic niche (MMP-3, -9 and -10), and to control inflammation (MMP-2, -3, -7, -8, -9, -12) and invasive processes (MMP-1, -2, -3, -7, -13 and -14), see Figure 1[6,10]. It has become increasingly clear that MMPs are not always detrimental since they also show anti-tumor effects, as illustrated by the inhibiting effects on angiogenesis described before[11]. MMPs are secreted as inactive pro-enzymes that need activation to exert their proteolytic properties. Their actions can be counteracted by specific inhibitors, i.e. the TIMPs.
Figure 1.
Schematic overview of the various types of matrix metalloproteinase-producing cells that are involved in the different processes during the various stages of cancer. MMP: Matrix metalloproteinase; TIMP: Tissue inhibitor of metalloproteinase.
Single-Nucleotide Polymorphism is the most common type of genetic variation. The estimated number of SNPs in the human genome is 10 million, but only a small part of these polymorphisms are functionally relevant. Most of the functional SNPs are located in the promoter region of the gene and are therefore expected to influence gene expression (Table 1)[12-30]. In this paper, we review the association of SNPs in the MMP and TIMP genes with the risk, the phenotype, and prognosis of gastrointestinal tumors.
Table 1.
Effects of matrix metalloproteinases single nucleotide polymorphisms on promoter activity
| MMP | SNP | Effect of mutation | Influence on promoter activity in vitro | Reference |
| MMP-1 | -1607 1G/2G | Extra Guanine (2G) creates a binding site for transcription factor Ets-1 | Increased in 2G allele (4-fold) | Rutter et al[18] |
| MMP-2 | -1306 C/T | C to T substitution disrupts Sp-1 binding site | Decreased in T-allele | Price et al[22] |
| MMP-2 | -735 C/T | C to T substitution influences Sp-1 binding site | Decreased in T-allele | Yu et al[15] |
| MMP-2 | -790 T/G | Three transcription factors1 bind to T (but not G) allele sequence | Decreased in G allele2 | Vasku et al[28] |
| MMP-2 | -955 A/C | Unknown | Effect unclear | Price et al[22] |
| MMP-2 | -1575 G/A | G to T substitution decreases estrogen receptor α binding | Decreased in A allele | Harendza et al[27] |
| MMP-3 | -1171 5A/6A3 | Transcription suppressor binds with higher affinity to 6A allele | Decreased in 6A allele (2-fold) | Ye et al[19] |
| MMP-3 | Lys45Glu | Lys to Glu substitution in exon 2 of gene | Effect unclear | Ouyang et al[16] |
| MMP-7 | -181 A/G | Nuclear proteins bind with higher affinity to G allele | Increased in G allele4 (2- to 3- fold) | Jormsjö et al[17] |
| MMP-7 | -153 C/T | T allele binds additional nuclear proteins compared with C allele | Increased in T allele4 (2- to 3- fold) | Jormsjö et al[17] |
| affinity for proteins that bind to both alleles higher in C allele | ||||
| MMP-8 | -799 C/T | Influences binding of transcription factor? | Increased in T allele5 | Wang et al[23] |
| MMP-8 | 17 C/G | Influences binding of transcription factor? | Increased in G allele5 | Wang et al[23] |
| MMP-9 | -90 CA(n) | Number of repeats influences strength of nuclear binding | Increased in n = 21 vs n = 14, n = 18 | Shimajiri et al[12] |
| MMP-9 | -1562 C/T | C to T substitution disrupts nuclear protein binding site | Increased in T allele | Zhang et al[20] |
| MMP-9 | R279Q | Arg to Gln substitution in fibronectin type II domains | Effect unclear6 | Wu et al[13] |
| MMP-9 | P574R | Pro to Arg substitution in hemopexin domain | Effect unclear6 | Wu et al[13] |
| MMP-9 | R668Q | Arg to Gln substitution in hemopexin domain | Effect unclear6 | Wu et al[13] |
| MMP-12 | -82 A/G | A to G substitution results in decreased affinity for transcription factor AP-1 | Decreased in G allele | Jormsjö et al[21] |
| MMP-12 | 1082 A/G | Asn to Ser substitution at coding region of hemopexin domain | Effect unclear | Joos et al[25] |
| MMP-13 | -77 A/G | A to G substitution results in decreased affinity for transcription factor AP-1 | Decreased in G allele (2-fold) | Yoon et al[24] |
| MMP-21 | C572T | Ala to Val substitution in enzymes catalytic domain | Effect unclear | Shagisultanova et al[29] |
| TIMP-1 | 372 C/T | Unknown, located in exon 5, no effect on transcription or amino-acid sequence | Effect unclear | Hinterseher et al[26] |
| TIMP-2 | -418 G/C | G to C substitution results in disruption of Sp-1 binding site | Decreased in C allele2 | Hirano et al[30] |
| TIMP-2 | 303C/T | Unknown, located in exon 3, no effect on transcription or amino-acid sequence | Effect unclear | Kubben et al[14] |
The three transcription factors are: GKLF (Gut-enriched Krueppel-like factor), S8 and Evi1 (ectopic viral integration site 1 encoded factor);
Not confirmed;
Formerly known as -1612 5A/6A;
Only in combination MMP-7 -181G/-153T;
Only in combination MMP-8 -799T/-381G/+17G, in cells resembling chorion cytotrophoblasts;
Probably influences substrate binding and inhibitor binding. MMP: Matrix metalloproteinase; SNP: Single-nucleotide polymorphisms; TIMP: Tissue inhibitor of metalloproteinase.
LITERATURE SEARCHE
Data sources
Electronic literature searches using PubMed, Embase and Web of Science were used to identify published papers concerning SNPs of MMPs, TIMPs, ADAMs, ADAMTS and RECK in gastrointestinal cancer up to September 2010. Search terms used included the MeSH heading “digestive system neoplasm” as well as all different types of gastrointestinal tumors mentioned separately, in combination with the MeSH heading “matrix metalloproteinases”, as well as all individual MMPs mentioned separately, combined with the MeSH heading “polymorphism, genetic” or synonyms of the term SNP. Papers were included when written in English or in any other language, provided that an English abstract was available. Full papers, as well as letters and abstracts, were included in this review. Publications concerning in vitro or animal studies only were excluded. Results are arranged by tumor type.
Software
To generate the forest plot, IBM SPSS statistics 17.0 was used.
ESOPHAGEAL CANCER
The incidence of esophageal cancer shows great geographical variation. This tumor is more common in Southern Africa and Eastern Asia than in Europe and Northern America (source: GLOBOCAN; http://globocan.iarc.fr). In the Asian population almost all cases of esophageal cancer are squamous cell cancers, whereas in the Western world adenocarcinoma occurs more often and its incidence has risen over recent decades. Because the pathophysiology and risk factors of squamous cell cancer and adenocarcinoma are different, we will discuss these two tumor types separately. An overview of the studies included in this paragraph is shown in Table 2[31].
Table 2.
Polymorphisms of tissue inhibitor of matrix metalloproteinases in esophageal cancer
| Gene | SNP | Reference | Cancer type | Ethnicity | Case/control | Outcome parameter | Results | Parameter OR | OR | 95% CI | P value |
| MMP-1 | -1607 1G/2G | Bradbury[33] | EA | Caucasian | 313/455 | Cancer risk | Increased cancer risk in 1G/2G and 2G/2G | 2G/2G vs 1G/1G | 1.83 | 1.2-2.8 | 0.005 |
| Overall survival | No difference in overall survival | ||||||||||
| Fruh[32] | EA | Caucasian | 101/101 | Cancer risk | No difference in cancer risk | ||||||
| Jin[42] | ESCC | Chinese | 234/350 | Cancer risk | No difference in cancer risk | ||||||
| LN metastases | No difference in LN metastases | ||||||||||
| MMP-2 | -735 C/T | Li[39] | ESCC | Chinese | 335/624 | Cancer risk | No difference in cancer risk | ||||
| Yu[15] | ESCC | Chinese | 527/777 | Cancer risk | Increased cancer risk in CC (trend) | CC vs CT+TT | 1.30 | 1.04-1.63 | 0.056 | ||
| Sun[31] | ESCC | Chinese | 335/624 | Cancer risk | No difference in cancer risk | ||||||
| Chen[40] | ESCC | Mongolian | 188/324 | Cancer risk | Increased cancer risk in TT | TT vs CC | 4.82 | 1.59-14.60 | |||
| MMP-2 | -1306 C/T | Fruh[32] | EA | Caucasian | 101/101 | Cancer risk | No difference in cancer risk | ||||
| Cancer risk and HP | HP infection protects against EA in CC | CC vs CT+TT | 0.29 | 0.1-0.7 | |||||||
| Chen[40] | ESCC | Mongolian | 188/324 | Cancer risk | No difference in cancer risk | ||||||
| Li[39] | ESCC | Chinese | 335/624 | Cancer risk | Increased cancer risk in CC | CC vs CT+TT | 1.57 | 1.10-2.23 | 0.010 | ||
| Yu[15] | ESCC | Chinese | 527/777 | Cancer risk | Increased cancer risk in CC | CC vs CT+TT | 1.52 | 1.17-1.96 | 0.001 | ||
| Sun[31] | ESCC | Chinese | 335/624 | Cancer risk | Increased cancer risk in CC | CC vs CT+TT | 1.57 | 1.10-2.23 | |||
| MMP-3 | -1171 6A/5A | Bradbury[33] | EA | Caucasian | 313/455 | Cancer risk | Increased cancer risk in 6A/5A and 5A/5A | 5A/5A vs 6A/6A | 1.61 | 1.0-2.5 | 0.030 |
| Overall survival | No difference in overall survival | ||||||||||
| Fruh[32] | EA | Caucasian | 101/101 | Cancer risk | No difference in cancer risk | ||||||
| Cancer risk and HP | HP infection protects against EA in 6A/6A | 6A/6A vs 5A/5A + 5A/6A | 0.04 | 0.002-0.9 | 0.040 | ||||||
| Zhang[44] | ESCC | Chinese | 234/350 | Cancer risk | No difference in cancer risk | ||||||
| Cancer in smoker | Increased cancer risk in 5A allele in smokers | 5A/5A + 5A/6A vs 6A/6A | 1.95 | 1.08-3.53 | |||||||
| LN metastases | Increased risk of LN metastases in 5A allele | 5A/5A vs 6A/6A | 2.24 | 1.07-4.69 | 0.030 | ||||||
| Infiltration depth | No difference in infiltration depth | ||||||||||
| MMP-3 | Lys45Glu | Ouyang [16] | ESCC | Chinese | 227/378 | Cancer risk | No difference in cancer risk | ||||
| MMP-7 | -181 A/G | Zhang et al[45] | ESCC | Chinese | 258/350 | Cancer risk | Increased cancer risk in AG and GG | AG+GG vs AA | 1.83 | 1.12-2.99 | |
| LN metastases | No difference in LN metastases | ||||||||||
| MMP-9 | R279Q | Wu [13] | ESCC | Chinese | 132/132 | Cancer risk | No difference in cancer risk | ||||
| MMP-9 | P574R | Wu et al[13] | ESCC | Chinese | 132/132 | Cancer risk | Increased cancer risk in RR | RR vs PP | 4.08 | 1.58-10.52 | 0.000 |
| MMP-9 | R668Q | Wu et al[13] | ESCC | Chinese | 132/132 | Cancer risk | No difference in cancer risk | ||||
| MMP-9 | -1562 C/T | Xia et al[43] | ESCC | Chinese | Cancer risk | No difference in cancer risk | |||||
| MMP-12 | -82 A/G | Bradbury et al[33] | EA | Caucasian | 313/455 | Cancer risk | No difference in cancer risk | ||||
| Overall survival | No difference in overall survival | ||||||||||
| Fruh et al[32] | EA | Caucasian | 101/101 | Cancer risk | No difference in cancer risk | ||||||
| Cancer risk and HP | HP infection protects against EA in AA | AA vs AG/GG | 0.44 | 0.2-0.8 | 0.020 | ||||||
| MMP-12 | 1082 A/G | Bradbury et al[33] | EA | Caucasian | 313/455 | Cancer risk | No difference in cancer risk | ||||
| Overall survival | No difference in overall survival | ||||||||||
| MMP-12 | -82 A/G | Li et al[39] | ESCC | Chinese | 335/624 | Cancer risk | No difference in cancer risk | ||||
| Cancer risk | No difference in cancer risk | ||||||||||
| MMP-13 | -77 A/G | Zhang et al[41] | ESCC | Chinese | 316/609 | Cancer risk | No difference in cancer risk |
SNP: Single nucleotide polymorphism; EA: Esophageal adenocarcinoma; ESCC: Esophageal squamous cell carcinoma; LN: Lymph node; HP: Helicobacter pylori; OR: Odds Ratio; 95% CI: 95% confidence interval.
ESOPHAGEAL ADENOCARCINOMA
Only two papers describe the relationship between polymorphisms of MMPs and esophageal adenocarcinoma (EA). One of these studies focused on the protective effect of Helicobacter pylori (H. pylori) infection in patients with different genotypes of MMP-1 (-1607 1G/2G), MMP-2 (-1306 C/T), MMP-3 (-1171 6A/5A) and MMP-12 (-82 A/G)[32]. In individuals with an MMP-2-1306 CC (wild-type) genotype, H. pylori infection (at any time during life) strongly protects against EA [adjusted odds ratio (OR) 0.29, 95% confidence interval (CI) 0.1-0.7]. In persons with a CT or TT genotype, the esophageal cancer risk was not influenced by H. pylori infection. To a lesser extent, the protective effect of H. pylori infection on the development of EA was also seen in carriers of the MMP-3 wild-type (6A/6A) and MMP-12 wild-type (-82 AA). However, no association between any of the studied MMP polymorphisms and overall risk of EA was found. The second paper, published by the same group, investigated the polymorphisms of MMP-1 (-1607 1G/2G), MMP-3 (6A/5A), and MMP-12 (-82 A/G) in relation to the risk and overall survival of EA[33]. In a cohort of 313 cancer patients and 455 controls, they found an increased cancer risk in 2G-allele carriers of the MMP-1 -1607 1G/2G polymorphism [2G/2G vs 1G/1G: adjusted OR 1.83 (95% CI: 1.2-2.8), P = 0.005]. 5A-allele carriers of the MMP-3 polymorphism also had an increased risk of developing EA in the same patient population (5A/5A vs 6A/6A, OR 1.61, 95% CI: 1.0-2.5, P = 0.03). The various genotypes of the MMP-12-82 A/G polymorphism were not associated with an EA risk. There was no difference in survival of the patients in relation to any of the above mentioned polymorphisms.
ESOPHAGEAL SQUAMOUS CELL CARCINOMA
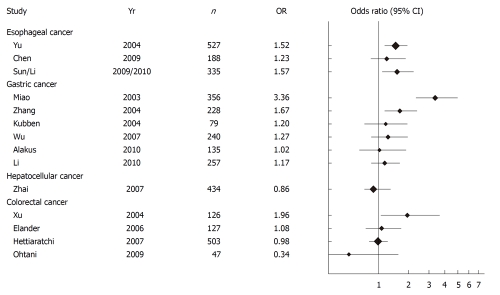
Matrix metalloproteinase-2 is overexpressed in esophageal squamous cell cancer (ESCC)[34-37]. There are two known functionally important SNPs in the promoter region of the MMP-2 gene, MMP-2-1306 C/T and MMP-2-735 C/T. The C to T transition at the -1306 position disrupts a Sp-1 transcription factor binding site and thereby reduces promoter activity[22] (Table 1). The allele frequency of the minor (T) allele is significantly lower in the Asian population (13.6%) than in the European population (23.3%)[38]. Increased risk of developing ESCC in individuals with -1306 CC genotype (compared to CT+TT genotype) has been reported in two large cohorts of Chinese patients (Figure 2)[15,39]. In Mongolian patients, the association between the different genotypes and incidence of ESCC did not reach statistical significance[40]. A recent meta-analysis showed that the -1306 CC genotype, which is the genotype with the highest transcriptional activity[22], is associated with an increased overall cancer risk and this association was maintained in the subgroup analysis of ESCC patients[38]. These findings suggest an important role for the MMP-2 -1306 C/T polymorphism in cancer development, which led us to the idea of plotting the results for this MMP-2 polymorphism derived from all the publications included in this review. Figure 2 illustrates that in gastrointestinal cancers the association between MMP-2 -1306 C/T polymorphism and cancer risk is not unidirectional.
Figure 2.
Forest plot of gastrointestinal cancer risk associated with the MMP-2 -1306 C/T polymorphism. Results are expressed as Odds ratios ± 95% confidence interval (CI) for CC vs CT+TT. The size of the diamonds indicates the size of the study cohort.
The reports on the association of the MMP-2 -735 C/T polymorphism are more dispersed. T allele carriers of this polymorphism show a lower transcriptional activity[15], which could explain the trend towards increased cancer risk in CC carriers compared to CT+TT carriers, which was reported in a Chinese population (OR 1.30, 95% CI: 1.04-1.63, P =0.056)[15]. However, these results were not confirmed in another large Chinese cohort[39] and a Mongolian study even found the opposite, higher ESCC cancer risk in TT carriers compared to CC, OR = 4.82, 95% CI: 1.59-14.60[40].
No association between the different promoter polymorphisms of MMP-1 (-1607 C/T), MMP- 9 (-1562 C/T), MMP-12 (-82 A/G), MMP-13 (-77 A/G) or between a polymorphism in the catalytic domain of MMP-9 (R279Q) or R668Q and the occurrence of ESCC has been found[13,39,41-43]. A polymorphism of MMP-3, located in the promoter region at position -1171 (5A/6A) was not associated with the overall risk of developing ESCC. However, the cancer risk was lower in smokers with a 6A/6A genotype (cancer risk 5A/6A vs 6A/6A: OR = 2.12, 95% CI: 1.16-3.90) and the risk of lymph node metastases was lower in 6A allele carriers (risk of lymph node metastases 5A/6A vs 6A/6A: OR = 2.24, 95% CI: 1.07-4.69)[44]. An increase in ESCC was also observed in AG and GG carriers of the MMP-7-181 A/G polymorphism (AG+GG vs AA: OR = 1.83, 95% CI: 1.12-2.99)[45]. MMP-7 is one of the smallest MMPs and has the capability of degrading a variety of extracellular matrix components, including elastin, type IV collagen, fibronectin, vitronectin, aggrecan and proteoglycans[46]. In various cancer types, including esophageal cancer, MMP-7 is over-expressed and associated with worse prognosis[47-49]. The G-allele of -181A/G is associated with higher basal transcriptional activity in vitro[17], which could explain the contribution of MMP-7 over-expression to prognosis.
In summary, the association between genotype and esophageal cancer susceptibility is most prominent for the MMP-2 -1306 C/T polymorphism with an increased risk of squamous cell cancer and an H. pylori protection against adenocarcinoma in the CC genotype carriers. In an Asian population, ESCC risk is increased in G-allele carriers of the MMP-7 -181 A/G polymorphism. No clear association was found between any of the investigated SNPs and disease progression or prognosis.
GASTRIC CANCER
Gastric cancer is the second largest cause of global cancer related mortality. The World Health Organization reported 803.000 deaths worldwide in 2004. There is a male preponderance and known risk factors are H. pylori infection and tobacco smoking[50]. Most of the data concerning polymorphisms of MMPs in gastric cancer concern the Asian population, reflecting the much higher incidence of gastric tumors in the Eastern world compared to Western Europe and the United States. Table 3 gives an overview of the studies discussed in this paragraph.
Table 3.
Polymorphisms of matrix metalloproteinases in hepatocellular carcinoma
| Gene | SNP | Reference | Cancer type | Ethnicity | Case/control | Outcome parameter | Results | Parameter OR | OR | 95% CI | P value |
| MMP-1 | -1607 1G/2G | Matsumura[65] | Gastric | Japanese | 215/166 | Cancer risk | No difference in cancer risk | ||||
| Clinicopathol. par. | No correlation with any clinicopath. par. | ||||||||||
| Jin[42] | GCA | Chinese | 183/350 | Cancer risk | No difference in cancer risk | ||||||
| LN metastases | No difference in LN metastases | ||||||||||
| MMP-2 | -1306 C/T | Zhang[53] | Gastric | Chinese | 228/774 | Cancer risk | Increased cancer risk in CC | CC vs CT/TT | 1.67 | 1.17-2.38 | |
| Wu[55] | Gastric | Taiwanese | 240/283 | Cancer risk | No difference in cancer risk | ||||||
| LN metastases | Increased risk of lymphatic invasion in CC | CC vs CT+TT | 2.77 | 1.27-6.04 | 0.01 | ||||||
| Venous invasion | Increased risk of venous invasion in CC | CC vs CT+TT | 2.93 | 1.27-6.78 | 0.012 | ||||||
| Survival | No difference in survival | ||||||||||
| Kubben[14] | Gastric | Caucasian | 79/169 | Cancer risk | No difference in cancer risk | ||||||
| Tumor-related survival | No difference in tumor-related survival | ||||||||||
| Alakus[56] | Gastric | Caucasian | 135/58 | Cancer risk | No difference in cancer risk | ||||||
| MMP-2 protein expression | No correlation with protein expression | ||||||||||
| Overall survival | No difference in overall survival | ||||||||||
| Li[39] | GCA | Chinese | 257/624 | Cancer risk | No difference in cancer risk | ||||||
| Miao[54] | GCA | Chinese | 356/789 | Cancer risk | Increased cancer risk in CC | CC vs CT+TT | 3.36 | 2.34-4.97 | |||
| Distant metastases | No difference in metastases | ||||||||||
| MMP-2 | -735 C/T | Li[39] | GCA | Chinese | 257/624 | Cancer risk | Trend towards increased cancer risk in CC | CC vs CT+TT | 1.36 | 0.99-1.87 | 0.06 |
| Cancer risk in non-smoker | Increased cancer risk in CC in non-smoker | CC vs CT+TT | 1.7 | 1.07-2.68 | 0.02 | ||||||
| MMP-3 | -1171 6A/5A | Zhang[44] | GCA | Chinese | 183/350 | Cancer risk | No difference in cancer risk | ||||
| Cancer risk in smoker | No difference in cancer risk in smoker | ||||||||||
| LN metastases | No difference in LN metastases | ||||||||||
| Infiltration depth | No difference in infiltration depth | ||||||||||
| MMP-8 | -799 C/T | Kubben[14] | Gastric | Caucasian | 79/169 | Cancer risk | No difference in cancer risk | ||||
| Tumor-related survival | No difference in tumor-related survival | ||||||||||
| MMP-7 | -181 A/G | Sugimoto[63] | Gastric | Japanese | 160/434 | Cancer risk | Increased cancer risk in G-allele carriers | AG+GG vs AA | 2.32 | 1.24-4.35 | 0.009 |
| Cancer stage | Increased cancer stage in G allele carriers | AG+GG vs AA | 3.66 | 1.54-8.73 | 0.003 | ||||||
| Kubben[14] | Gastric | Caucasian | 79/169 | Cancer risk | More AA and less AG in cancer group | AG+GG vs AA | 0.50 | 0.28-0.87 | < 0.04 | ||
| Tumor-related survival | No difference in tumor-related survival | ||||||||||
| Li[64] | Gastric | Chinese | 338/380 | Cancer risk | Increased cancer risk in G allele carriers | AG+GG vs AA | 1.95 | 1.24-3.05 | 0.004 | ||
| LN metastases | Increased risk of LN metastases in G allele | AG+GG vs AA | 0.040 | ||||||||
| Cancer stage | More advanced cancer stage in G allele | AG+GG vs AA | 0.007 | ||||||||
| Alakus[56] | Gastric | Caucasian | 135/58 | Cancer risk | No difference in cancer risk | AG+GG vs AA | 1.06 | 0.69-1.64 | 0.79 | ||
| Overall survival | No difference in overall survival | ||||||||||
| Zhang[45] | GCA | Chinese | 201/350 | Cancer risk | Increased cancer risk in G allele carriers | AG+GG vs AA | 1.96 | 1.17-3.29 | |||
| LN metastases | No difference in LN metastases | ||||||||||
| MMP-7 | -153 C/T | Kubben[14] | Gastric | Caucasian | 79/169 | Cancer risk | No difference in cancer risk | ||||
| Tumor-related survival | No difference in tumor-related survival | ||||||||||
| MMP-8 | 17 C/G | Kubben[14] | Gastric | Caucasian | 79/169 | Cancer risk | No difference in cancer risk | ||||
| Tumor-related survival | No difference in tumor-related survival | ||||||||||
| MMP-9 | R279Q | Tang[59] | Gastric | Chinese | 74/100 | Cancer risk | No difference in cancer risk | ||||
| LN metastases | Increased risk of LN metastases in RR | RR vs QQ+RQ | 5.74 | 1.59-13.43 | |||||||
| MMP-9 | P574R | Tang[59] | Gastric | Chinese | 74/100 | Cancer risk | No difference in cancer risk | ||||
| LN metastases | Increased risk of LN metastases in PP | PP vs RR+PR | 4.17 | 1.39-11.78 | |||||||
| MMP-9 | -1562 C/T | Kubben[14] | Gastric | Caucasian | 79/169 | Cancer risk | No difference in cancer risk | ||||
| Tumor-related survival | No difference in tumor-related survival | ||||||||||
| Matsumura[57] | Gastric | Japanese | 177/224 | Cancer risk | No difference in cancer risk | ||||||
| Infiltration depth | Deeper submucosal infiltration in T allele | CT+TT vs CC | 2.61 | 1.07-6.34 | 0.034 | ||||||
| Lymphatic invasion | Increased lymphatic invasion in T allele | CT+TT vs CC | 2.27 | 1.09-4.74 | 0.028 | ||||||
| TNM classification | More advanced stage cancer in T allele | CT+TT vs CC | 2.26 | 1.12-4.55 | 0.022 | ||||||
| Venous invasion | No difference in venous invasion | CT+TT vs CC | 1.98 | 0.99-3.97 | 0.053 | ||||||
| Zhang[53] | Gastric | Chinese | 228/774 | Cancer risk | No difference in cancer risk | ||||||
| Wu[58] | Gastric | Taiwanese | 263/354 | Cancer risk | No difference in cancer risk | ||||||
| MMP-12 | -82 A/G | Li[39] | GCA | Chinese | 335/624 | Cancer risk | No difference in cancer risk | ||||
| Cancer risk in smoker | No difference in cancer risk in smoker | ||||||||||
| MMP-13 | -77 A/G | Li[39] | GCA | Chinese | 257/624 | Cancer risk | No difference in cancer risk | ||||
| Cancer risk in smoker | Decreased cancer risk in AG in smoker | AG vs AA/AG | 0.47 | 0.28-0.8 | 0.01 | ||||||
| Zhang[41] | GCA | Chinese | 243/609 | Cancer risk | No difference in cancer risk | ||||||
| Cancer risk in smoker | Decreased cancer risk in AG in smoker | ||||||||||
| TIMP-1 | 372 C/T | Kubben[14] | Gastric | Caucasian | 79/169 | Cancer risk | No difference in cancer risk | ||||
| Tumor-related survival | No difference in tumor-related survival | ||||||||||
| TIMP-2 | -418 G/C | Kubben[14] | Gastric | Caucasian | 79/169 | Cancer risk | No difference in cancer risk | ||||
| Tumor-related survival | No difference in tumor-related survival | ||||||||||
| Wu[55] | Gastric | Taiwanese | 240/283 | Cancer risk | No difference in cancer risk | ||||||
| LN metastases | Increased risk of LN metastases in GG | GG vs CG+GG | 2.87 | 1.22-6.76 | 0.16 | ||||||
| Venous invasion | Increased venous invasion in GG | GG vs CG+GG | 2.65 | 1.08-6.49 | 0.033 | ||||||
| Survival | No difference in survival | ||||||||||
| Yang[66] | Gastric | China | 206/206 | Cancer risk | More C-alleles in cancer patients | CC+CG vs GG | 1.51 | 1.00-2.26 | 0.049 | ||
| Clinicopathol. par. | No correlation with any clinicopath. par. | ||||||||||
| TIMP-2 | 303 C/T | Kubben[14] | Gastric | Caucasian | 79/169 | Cancer risk | No difference in cancer risk | ||||
| Tumor-related survival | Better survival in CC patients | CC vs CT/TT | 4.45 | 1.81-10.9 | 0.001 | ||||||
| Alakus[56] | Gastric | Caucasian | 135/58 | Cancer risk | No difference in cancer risk | ||||||
| LN metastases | More LN metastases in CC | 0.01 | |||||||||
| Distant metastases | Increased risk of distant metastases in CC | 0.022 | |||||||||
| Survival | No difference in survival |
SNP: Single nucleotide polymorphism; GCA: Gastric Cardia Adenocarcinoma; LN: Lymph node; OR: Odds ratio; 95% CI: 95% confidence interval; Clinicopath. par.: Clinicopathological parameters. 1Except histological subtype.
The gelatinases MMP-2 and MMP-9 are upregulated in gastric cancer and increased MMP-2 and MMP-9 protein levels in tumor tissue of gastric cancer patients are associated with poor prognosis[51,52]. Two papers reported a significant increase in gastric cancer risk in -1306 CC carriers of the MMP-2 SNP[53,54], while four other studies did not find such a correlation (Figure 2)[14,39,55,56]. In a recent meta-analysis, the MMP-2 -1306 CC genotype was associated with a significant increase in gastric cancer susceptibility[38], but this meta-analysis did not include two studies that reported no difference in cancer risk between the different genotypes[14,56]. Survival was not influenced by the -1306 C/T polymorphism in any of these studies. However, Wu et al detected an increase in lymphatic and venous invasion in individuals with a CC genotype[55]. A second functional polymorphism of MMP-2 is a C to T transition at position -735 in the promoter region of the gene. This substitution influences a Sp1 transcription factor binding site, resulting in lower promoter activity in T allele carriers[15], similar to the C to T transition at position -1306. There is a trend towards increased cancer risk in MMP-2-735 CC individuals, and this correlation is particularly significant in smokers.
The C to T substitution at position -1562 of the promoter region of MMP-9 results in the loss of binding to this region of a repressor nuclear protein, resulting in an increase in transcriptional activity in macrophages[20]. T-allele carriers of this polymorphism had deeper submucosal infiltration, more frequent lymphatic invasion and more advanced stage cancer compared to non-T allele carriers[57]. Nevertheless, none of the four publications describing the MMP-9 -1562 C/T polymorphism in gastric cancer found an association between the various genotypes and cancer risk[14,53,57,58]. In addition, Kubben et al did not find an association between the MMP-9 polymorphisms and tumor-related survival[14]. Two non-synonymous SNPs located in an exon of MMP-9, R279Q and P574R, were both associated with the risk of lymph node metastases in gastric cancer (higher risk in the RR and PP genotype, respectively), but did not show a relationship with gastric cancer risk[59].
MMP-7 over-expression has been demonstrated in various forms of cancer. In gastric cancer, MMP-7 expression has been linked to cancer progression and survival[60-62]. The genotype distribution of the -181 A/G polymorphism of MMP-7 is significantly different in various parts of the world; the frequency of the minor G-allele being 8.8% in the Asian population and 42.0% in the European population[38]. An increased risk of gastric cancer in G-allele carriers of the MMP-7 -181A/G polymorphism, who have a higher transcriptional activity, was reported in three studies[45,63,64]. These findings are in line with the observations in esophageal squamous cell cancers, as described above. Patients with the GG and AG genotype had a more advanced cancer stage. Interestingly, these findings are in contrast with two other papers, where either no correlation between the MMP-7 polymorphisms and gastric cancer risk was found[56] or there was even an inverse correlation, i.e. a higher percentage of MMP-7 -181 AA genotype in the gastric cancer group compared to the control group[14]. The discrepancy between these findings might be explained by a difference in ethnicity: in the first three papers all patients had an Asian background, whereas the latter two papers concern Caucasian patients.
The SNPs of MMP-1 (-1607 1G/2G), MMP-3 (-1171 5A/6A), MMP-7 (-153 C/T), MMP-8 (17 C/G) and MMP-8 (-799 C/T) are reported not to be associated with gastric cancer risk or prognosis[14,42,44,65]. Smokers with the AG genotype of the MMP-13 -77A/G polymorphism were reported to have a decreased risk of developing gastric cancer[39,41]. A trend towards increased cancer risk was observed in individuals with the AG genotype of the MMP-12 -82 A/G polymorphism[39,41].
One of the mechanisms that regulates MMP activity, in addition to promoter polymorphisms, is the interaction with TIMPs. The contribution of gene polymorphisms of TIMP-1 and TIMP-2 has only been studied sporadically in gastric cancer. The 372 C/T polymorphism of TIMP-1 did not correlate with cancer risk or cancer-related survival[14]. The G to C substitution at position -418 in the promoter region of the TIMP-2 gene has been suggested to disrupt a Sp-1 binding site, presumably leading to decreased TIMP-2 transcription[30]. Yang et al[66] studied this TIMP-2 polymorphism in a group of 206 gastric cancer patients and 206 controls. The gastric cancer risk was elevated in C-allele carriers (CC + GC vs GG, adjusted OR = 1.51, 95% CI: 1.00-2.26, P = 0.049). Two other papers that described the contribution of this polymorphism on gastric cancer occurrence, did not find an association between the different genotypes and cancer risk[14,55]. In a cohort of 240 Taiwanese gastric cancer patients, Wu et al[55] found increased lymph node metastases, increased serosal invasion and increased venous invasion in patients with the TIMP-2 GG genotype. Despite these results, neither in the Taiwanese study, nor in a Dutch study, was an association with survival reported[14,55]. The function of the 303C/T polymorphism of TIMP-2, located in exon 3 of the gene, is unclear. Both Kubben et al[14] and Alakus et al[56] found no correlation between genotype and gastric cancer risk. The finding in the study of Alakus et al that patients with the TIMP-2 303CC genotype more often have lymphatic and distant metastases seems to contradict with the findings of Kubben et al, who showed a significantly better tumor-related survival in patients with the CC genotype. This discrepancy could possibly be due to the low number of patients with a CT or TT genotype in both studies.
To conclude, an increased gastric cancer susceptibility seems to be present in Asian (but not in Caucasian) G-allele carriers of the MMP-7 -181 A/G polymorphism, an association also seen with ESCC. While some studies reported an association between genotype and clinicopathological parameters or prognosis, these results were not confirmed by others and are thus not consistent, except for the finding that MMP-7-181 AG or GG genotype patients in the Asian population seem to have a more advanced tumor stage than patients with the AA genotype[63,64].
SMALL INTESTINAL CANCER
Tumors of the duodenum, jejunum and ileum are rare and there are in fact no data on the effect of functional polymorphisms of matrix metalloproteinases in these tumors. Only one paper describes MMP-2, -7, -9, -11 and -13 protein levels in 25 patients with a carcinoid tumor localized in the ileum. Except for MMP-2, none of the MMP protein levels were associated with survival[67]. Surprisingly, low MMP-2 expression in the primary carcinoid tumor is correlated with an unfavorable outcome of the disease. This finding, which contrasts with observations made in many other gastrointestinal tumors, might indicate that these neuro-endocrine tumors have a different proteolytic phenotype compared to the other tumors which are adenocarcinomas or (in case of proximal or mid-esophageal cancers) squamous cell carcinomas.
PANCREATIC CANCER
Over-expression of MMP-1, MMP-2, MMP-7 and MMP-9 protein in pancreatic cancer is associated with more advanced tumor stage and poor prognosis[68-74], whereas high glandular TIMP-2 expression is associated with better survival in pancreatic ductal adenocarcinoma[75]. However, until now, there are no reports on the functional polymorphisms of MMPs and TIMPs in malignant tumors of the pancreas.
CHOLANGIOCARCINOMA
Bile duct tumors are rare in the general population. Patients with primary sclerosing cholangitis (PSC) have an increased risk of developing cholangiocarcinoma. Wiencke et al[76] investigated the association of MMP-1 and MMP-3 promoter polymorphisms in 165 PSC patients. Fifteen of these patients developed cholangiocarcinoma; all of these were 1G-allele carriers of the MMP-1 -1607 1G/2G polymorphism, compared to 72% of the whole PSC population. This finding is somewhat surprising since the 2G allele of this SNP is associated with a higher level of transcription and in most cancers, as for example esophageal adenocarcinomas, associated with increased cancer risk or worse prognosis. The number of PSC patients in this study was too small to draw definite conclusions about the role of this promoter-SNP in PSC-associated cholangiocarcinoma.
HEPATOCELLULAR CARCINOMA
Hepatocellular carcinoma (HCC) is the fifth most common cancer in men and the eighth in women worldwide[77]. The geographic distribution of this most common form of primary liver cancer follows the distribution of hepatitis B and C infection, as most of the patients have a background of liver cirrhosis or hepatitis B-infection. Several studies have looked into the effect of single nucleotide polymorphisms of MMPs on the incidence of HCC and its relation to survival of the patients (Table 4). None of the MMP gene polymorphisms that have been studied in HCC patients is correlated with cancer risk. In one paper, the number of 2G/2G homozygotes of the MMP-1 -1607 1G/2G polymorphism was slightly increased in HCC patients with a background of chronic hepatitis C virus (HCV)-related liver disease compared to patients with HCV-related chronic liver disease without HCC[78]. However, when the same patient group was compared with healthy controls, this relationship was no longer present[79]. In hepatitis B virus (HBV) patients with or without HCC, the genotype distribution of the MMP-1 -1607 polymorphism was similar[80]. No association was found between the MMP-2 -1306 C/T polymorphism and the risk of hepatocellular carcinoma[80], although patients with the CC genotype did experience an increase in HCC recurrence after liver transplantation compared to patients with the CT genotype (CT vs CC, OR = 0.42, 95% CI: 0.18-0.99, P < 0.05; there were no TT patients in the cohort). When compared with healthy controls, HCV-infected patients with HCC were more often 5A-allele carriers of the -1171 5A/6A polymorphism[79]. However, when compared to HCV infected patients without HCC, no difference in genotype distribution was found[78], which might suggest that the 5A-allele interferes with the development of the underlying disease instead of with the development of HCC. 5A allele carriers did have larger tumor diameters at the time of diagnosis and a poorer prognosis[78,79]. The SNPs MMP-2 -735 C/T, MMP-7 -181 A/G, MMP-8 -799C/T, MMP-9 -1562 C/T, MMP-12 -82 A/G, MMP-13 -77 A/G and MMP-21 C572T were found not to be associated with an increased risk of developing HCC[78-82]. One paper which describes the impact of the TIMP-2 -418 G/C polymorphism in a group of 92 HCC patients and 70 patients with chronic liver disease without signs of HCC found no association with HCC occurrence or prognosis[83].
Table 4.
Polymorphisms of matrix metalloproteinases in hepatocellular carcinoma
| Gene | SNP | Reference | Ethnicity | Case/control | Parameter | Results | Parameter OR | OR | 95% CI | P value |
| MMP-1 | -1607 1G/2G | Zhai[80] | Chinese | 434/480 | Cancer risk | No difference in cancer risk | ||||
| Okamoto[78] | Japanese | 95/83 | Cancer risk | Increased risk of HCC in 2G/2G | 0.002 | |||||
| Clinicopath. par. | No correlation with any clinicopath. par. | |||||||||
| Survival | No difference in survival | |||||||||
| Okamoto[79] | Japanese | 92/170 | Cancer risk | No difference in cancer risk | ||||||
| Survival/clinicopath. par. | No difference in survival/clinicopath. par. | |||||||||
| MMP-2 | -735 C/T | Zhai[80] | Chinese | 434/480 | Cancer risk | No difference in cancer risk | ||||
| Cancer risk | No difference in cancer risk | |||||||||
| MMP-2 | -1306 C/T | Zhai[80] | Chinese | 434/480 | Cancer risk | No difference in cancer risk | ||||
| Wu[82] | Chinese | 93/0 | HCC recurrence after LTx | More CC in recurrence group | CT vs CC | 0.42 | 0.18-0.99 | < 0.05 | ||
| MMP-3 | -1171 5A/6A | Zhai[80] | Chinese | 434/480 | Cancer risk | No difference in cancer risk | ||||
| Okamoto[78] | Japanese | 95/83 | Cancer risk | No difference in cancer risk | ||||||
| HCC diameter at diagnosis | Larger diameter in 5A allele carriers | |||||||||
| Survival | Decreased survival in 5A allele carriers | |||||||||
| Okamoto[79] | Japanese | 92/170 | Cancer risk | Increased cancer risk in 5A allele carriers | ||||||
| Survival/clinicopath. par. | Decreased survival in 5A allele carriers | 0.035 | ||||||||
| MMP-7 | -181 A/G | Qiu[81] | Chinese | 434/480 | Cancer risk | No difference in cancer risk | ||||
| MMP-8 | -799 C/T | Qiu[81] | Chinese | 434/480 | Cancer risk | No difference in cancer risk | ||||
| MMP-9 | -1562 C/T | Zhai[80] | Chinese | 434/480 | Cancer risk | No difference in cancer risk | ||||
| Wu[82] | Chinese | 93/0 | HCC recurrence after LTx | No difference in HCC recurrence | ||||||
| Okamoto[78] | Japanese | 95/83 | Cancer risk | No difference in cancer risk | ||||||
| Differentiation grade | Differentiation worse in T allele carriers | 0.03 | ||||||||
| Survival | No difference in survival | |||||||||
| Okamoto[79] | Japanese | 92/170 | Cancer risk | No difference in cancer risk | ||||||
| Survival/clinicopath. par. | No difference in survival/clinicopath. par. | |||||||||
| MMP-12 | -82 A/G | Zhai[80] | Chinese | 434/480 | Cancer risk | No difference in cancer risk | ||||
| MMP-13 | -77 A/G | Zhai[80] | Chinese | 434/480 | Cancer risk | No difference in cancer risk | ||||
| MMP-21 | C572T | Qiu[81] | Chinese | 434/480 | Cancer risk | No difference in cancer risk | ||||
| TIMP-2 | -418 G/C | Okamoto[83] | Japanese | 92/70 | Cancer risk HCC | No difference in cancer risk | ||||
| Survival | No difference in survival |
SNP: Single nucleotide polymorphism; LN: Lymph node; Clinicopath. par.: Clinicopathological parameters; OR: Odds ratio; 95% CI: 95% confidence interval; HCC: Hepatocellular carcinoma; LTx: Liver transplantation.
In conclusion, polymorphisms of MMPs are not associated with HCC susceptibility. MMP-genotype may possibly influence the course of the disease in HCC patients, as HCV-infected HCC patients carrying the 5A allele of the MMP-3 -1171 5A/6A polymorphism appear to have worse survival rates[78,79]. In addition, it has been reported that HCC recurrence after liver transplantation is increased in MMP-2 -1306 CC genotype carriers when compared to CT patients[82].
COLORECTAL CANCER
Worldwide, colorectal cancer (CRC) is the fourth most common cancer in men and the third most common cancer in women[84]. Continents with a high incidence of colorectal cancer include Europe and North America. The lowest incidence is found in Asia, Africa and South America. In Eastern Europe and Japan, CRC incidence has increased over recent years, probably due to a “Westernization” of lifestyle[84]. The effect of MMP polymorphisms on lung, breast and colorectal cancer has been reviewed previously by Decock et al[85]. The studies that are included in the present review are shown in Table 5.
Table 5.
Polymorphisms of matrix metalloproteinases in colorectal cancer
| Gene | SNP | Reference | Ethnicity | Case/control | Parameter | Results | Parameter OR | OR | 95% CI | P value |
| MMP-1 | -1607 1G/2G | Ghilardi[92] | Caucasian | 60/164 | Cancer risk | Increased cancer risk in 2G/2G | 2G/2G vs 1G/1G + 1G/2G | 2.21 | 1.17-4.16 | 0.014 |
| Distant metastases | Increased risk of metastases in 2G/2G | 2G/2G vs 1G/1G + 1G/2G | 4.73 | 1.46-15.26 | 0.008 | |||||
| Zinzindohoue[99] | Caucasian | 201/0 | Survival | Overall survival worse in 2G/2G | 2G/2G vs 1G/1G | 5.4 | 2.0-14.7 | 0.001 | ||
| Hettiaratchi[96] | Australian | 503/471 | Cancer risk | No difference in cancer risk | ||||||
| Survival | Increased survival in 2G/2G | 2G/2G vs 1G/2G + 1G/1G | 0.43 | 0.19-0.96 | 0.040 | |||||
| Clinicopath. par. | No correlation with any clinicopath. par. | |||||||||
| Woo[89] | Korean | 185/304 | Cancer risk | Increased cancer risk in 2G/2G and G-allele | 2G/2G in patients vs controls | 1.8 | 1.23-2.64 | 0.044 | ||
| LN metastases | More often >10 LN in 2G/2G | |||||||||
| Fang[94] | Chinese | 237/252 | Cancer risk | No difference in cancer risk | ||||||
| Xu[97] | Chinese | 126/126 | Cancer risk | No difference in cancer risk | ||||||
| Clinicopath. par. | No correlation with any clinicopath. par. | |||||||||
| Przybylowska[98] | Caucasian | 33/52 | Cancer risk | No difference in cancer risk | ||||||
| Hinoda[91] | Japanese | 101/127 | Cancer risk | Increased cancer risk in 2G/2G | 2G/2G vs 1G/1G + 1G/2G | 2.08 | 1.22-3.53 | 0.007 | ||
| Clinicopath. par. | No correlation with any clinicopath. par. | |||||||||
| Biondi[93] | Caucasian | 63/164 | Cancer risk | More 2G allele in cancer patients | < 0.08 | |||||
| de Lima[95] | Brasilian | 130/130 | Cancer risk | No difference in cancer risk | ||||||
| Distant metastases | Increased risk of metastases in 1G allele (trend) | |||||||||
| LN metastases | No difference in LN metastases | |||||||||
| Elander[90] | Caucasian | 127/208 | Cancer risk | Increased cancer risk in 2G allele carriers | 2G allele vs 1G allele | 1.41 | 1.02-1.96 | 0.037 | ||
| Clinicopath. par. | No correlation with any clinicopath. par. | |||||||||
| Kouhkan[88] | Iranian | 150/100 | Cancer risk | Increased cancer risk in 2G/2G and G-allele | ||||||
| Distant metastases | Earlier metastases in 2G/2G | |||||||||
| MMP-2 | -1306 C/T | Xu[102] | Chinese | 126/126 | Cancer risk | Increased cancer risk in CC | CC vs CT+TT | 1.96 | 1.06-3.64 | < 0.05 |
| Infiltration depth | More serosa/adventitia involvement in CC | CC vs CT+TT | 0.042 | |||||||
| Hettiaratchi[96] | Australian | 503/471 | Cancer risk | No difference in cancer risk | ||||||
| Survival | No difference in survival | |||||||||
| Clinicopath. par. | No correlation with any clinicopath. par. | |||||||||
| Langers[105] | Caucasian | 215/0 | Survival | 10 year survival worse in TT | CC/CT vs TT | 1.4 | 1.02-1.91 | 0.038 | ||
| Ohtani[103] | Japanese | 47/67 | Cancer risk | No difference in cancer risk | ||||||
| Elander[90] | Caucasian | 127/208 | Cancer risk | No difference in cancer risk | ||||||
| Clinicopath. par. | No correlation with any clinicopath. par. | |||||||||
| MMP-2 | -790 T/G | Xu[104] | Chinese | 126/126 | Cancer risk | No difference in cancer risk | ||||
| Infiltration depth | No difference in infiltration depth | |||||||||
| MMP-2 | -955 A/C | Xu[104] | Chinese | 126/126 | Cancer risk | No difference in cancer risk | ||||
| Infiltration depth | No difference in infiltration depth | |||||||||
| MMP-2 | -1575 G/A | Xu[104] | Chinese | 126/126 | Cancer risk | Increased cancer risk in GG and G allele | GG vs GA+AA | 1.96 | 1.06-3.64 | 0.04 |
| Infiltration depth | More serosa/adventitia infiltration in GG | GG vs GA+AA | < 0.05 | |||||||
| MMP-3 | -1171 5A/6A | Hinoda[91] | Japanese | 101/127 | Cancer risk | Increased cancer risk in 6A/6A | 6A/6A vs 5A/5A + 5A/6A | 2.11 | 1.17-3.82 | 0.01 |
| Clinicopath. par. | No correlation with any clinicopath. par. | |||||||||
| Biondi[93] | Caucasian | 63/164 | Cancer risk | No difference in cancer risk | ||||||
| Ghilardi[92] | Caucasian | 60/164 | Cancer risk | No difference in cancer risk | ||||||
| Distant metastases | No difference in metastases | |||||||||
| Woo[89] | Korean | 185/304 | Cancer risk | No difference in cancer risk | ||||||
| LN metastases | No difference in LN metastases | |||||||||
| Ohtani[103] | Japanese | 47/67 | Cancer risk | No difference in cancer risk | ||||||
| Elander[90] | Caucasian | 127/208 | Cancer risk | No difference in cancer risk | ||||||
| Clinicopath var. | No correlation with any clinicopath. par. | |||||||||
| Zinzindohoue[99] | Caucasian | 201/0 | Survival | No difference in overall survival | ||||||
| Hettiaratchi[96] | Australian | 503/471 | Cancer risk | No difference in cancer risk | ||||||
| Survival | No difference in survival | |||||||||
| Clinicopath. par. | No correlation with any clinicopath. par. | |||||||||
| Xu[97] | Chinese | 126/126 | Cancer risk | No difference in cancer risk | ||||||
| Clinicopath. par. | No correlation with any clinicopath. par. | |||||||||
| MMP-7 | -153 C/T | Ghilardi[111] | Caucasian | 58/111 | Cancer risk | Increased cancer risk in T allele carriers | T allele in patients vs controls | 2.2 | 0.89-5.48 | 0.05 |
| Clinicopath. par. | No correlation with any clinicopath. par. | |||||||||
| Langers[110] | Caucasian | 174/0 | Survival | Better survival in CC patients | CC vs CT+TT (LR) | 14 | 0.001 | |||
| MMP-7 | -181 A/G | Woo[89] | Korean | 185/304 | Cancer risk | No difference in cancer risk | ||||
| LN metastases | No difference in LN metastases | |||||||||
| Fang[94] | Chinese | 237/252 | Cancer risk | No difference in cancer risk | ||||||
| Ghilardi[111] | Caucasian | 58/111 | Cancer risk | Increased cancer risk in GG | GG in patients vs controls | 2.41 | 0.98-5.89 | 0.03 | ||
| Distant metastases | GG more often distant metastases | G in M+ vs M- | 7.5 | 2.07-27.19 | 0.001 | |||||
| LN metastases | GG more frequent LN metastases | |||||||||
| Ohtani[103] | Japanese | 47/67 | Cancer risk | No difference in cancer risk | ||||||
| Langers[110] | Caucasian | 174/0 | Survival | No difference in survival | ||||||
| de Lima[95] | Brasilian | 130/130 | Cancer risk | No difference in cancer risk | ||||||
| Distant metastases | No difference in metastases | |||||||||
| LN metastases | No difference in LN metastases | |||||||||
| MMP-9 | R279Q | Woo[89] | Korean | 185/304 | Cancer risk | No difference in cancer risk | ||||
| LN metastases | No difference in LN metastases | |||||||||
| Xing[106] | Chinese | 137/199 | Cancer risk | No difference in cancer risk | ||||||
| LN metastases | No difference in LN metastases | |||||||||
| Fang[94] | Chinese | 237/252 | Cancer incidence | Increased cancer risk in RR | RR vs QQ | 2.21 | 1.25-3.93 | 0.006 | ||
| MMP-9 | -90(CA)n | Woo[89] | Korean | 185/304 | Cancer risk | No difference in cancer risk | ||||
| LN metastases | No difference in LN metastases | |||||||||
| MMP-9 | -1562 C/T | Xu[107] | Chinese | 126/126 | Cancer risk | No difference in cancer risk | ||||
| Infiltration depth | No difference in infiltration depth | |||||||||
| Woo[89] | Korean | 185/304 | Cancer risk | Increased cancer risk in CC patients | GG in patients vs controls | 1.7 | 1.04-2.66 | 0.03 | ||
| LN metastases | No difference in LN metastases | |||||||||
| Xing[106] | Chinese | 137/199 | Cancer risk | No difference in cancer risk | ||||||
| LN metastases | Increased risk of LN metastases in CT+TT | CT+TT vs CC | 0.02 | |||||||
| Langers[105] | Caucasian | 215/0 | Survival | No difference in survival | ||||||
| Ohtani[103] | Japanese | 47/67 | Cancer risk | No difference in cancer risk | ||||||
| Elander[90] | Caucasian | 127/208 | Cancer risk | No difference in cancer risk | ||||||
| Clinicopath. par. | No relationschip with clinicopath. par. | |||||||||
| MMP-12 | -82A/G | Woo[89] | Korean | 185/304 | Cancer risk | No difference in cancer risk | ||||
| LN metastases | No difference in LN metastases |
SNP: Single nucleotide polymorphism; LN: Lymph node; Clinicopath. par.: Clinicopathological parameters; OR: Odds ratio; aOR: Adjusted odds ratio; 95% CI: 95% confidence interval.
MMP-1, an interstitial collagenase, degrades fibrillar collagens type I, II, III, V, IX, and X, that form the most abundant class of extracellular matrix proteins in the interstitium[86]. The MMP-1 gene is located on chromosome 11q22. Insertion of an extra guanine (G) at the -1607 promoter position creates an Ets-1 transcription factor binding site (5’-GGA-3’) leading to a significant increase in transcription activity in normal fibroblasts[18]. Both alleles are common in the general population; the allele frequency of the 2G allele is 64% in the Asian population and 52% in the European population[87]. In the Caucasian population, the frequency of the homozygote -1607 2G/2G polymorphism is about 30%. Several papers reported an increased colorectal cancer susceptibility in either 2G/2G homozygotes or 2G-allele carriers of the MMP-1 -1607 polymorphism[88-93]. However, a number of other studies found no association between cancer risk and MMP-1 genotype[94-98]. With exception of the studies of Fang et al and Hettiaratchi et al, all studies included a relatively small number of patients, which could explain the differences in results. Hettiaratchi et al included the largest cohort (503 Australian CRC patients, 471 controls) of all studies so far[96]. Besides the lack of association between the genotype and CRC susceptibility, in this cohort the 5-year survival was increased in 2G/2G homozygotes. All other studies, which have either looked at survival or correlation with clinicopathological parameters, showed that the 2G/2G genotype is either associated with worse survival[99], with unfavourable clinicopathological parameters, like increased risk of metastases at time of diagnosis[92], a higher number of affected lymph nodes[89], or with earlier distant metastases[88]. Patient selection could possibly account for this discrepancy, since Hettiaratchi et al only included patients who did not have synchronous metastases at the time of diagnosis, and the influence of MMP-1 on the cancer process may change during different stages of cancer progression. De Lima et al reported a higher risk of lymph node metastases in patients carrying a 1G-allele, although this association was not significant with a P value of 0.09[95]. In all the other abovementioned papers, no association of MMP-1 gene polymorphisms and clinicopathological parameters was found. In two meta-analyses, 2G-allele carriers showed a significantly increased risk of developing colorectal cancer when compared with homozygous 1G allele carriers[87,100]. However, the large cohort of Hettiaratchi et al[96] was not included in these meta-analyses and inclusion of this study might lead to loss of significance.
Lièvre et al[101] studied the influence of genetic polymorphisms in the MMP-1 (-1607 1G/2G) gene in 295 patients with large adenomas and 302 patients with small adenomas, the premalignant condition to colorectal cancer, and in 568 polyp-free controls. No difference was found in the genotype distribution between patients with large adenomas and patients with small adenomas or healthy controls.
In a population of 126 CRC patients and 126 healthy controls, Xu et al[102] found an increase in CRC susceptibility in patients with the CC genotype of the MMP-2 -1306 C/T polymorphism. These findings were not supported by Hettiaratchi et al[96], Elander et al[90] and Ohtani et al[103], who found no influence of the MMP-2 genotype on the colorectal cancer risk (Figure 2). Difference in ethnicity (Australian vs European vs Japanese) or sample size might be the underlying cause of this discrepancy. Two meta-analyses, both including the study of Xu et al, showed no association between the MMP-2 -1306 C/T polymorphism and colorectal cancer susceptibility[87,100]. Xu et al[104] also reported that patients with the CC genotype had more frequent serosa/adventitia involvement, while none of the other studies described any correlation with clinicopathological parameters or survival, except for the study of Langers et al, where the TT genotype was shown to be an indicator of poor 10-year survival[89-93,96,97,99,103,105]. In the Xu et al[104] cohort of 126 CRC patients and 126 control patients, two other polymorphisms of MMP-2 (-790 T/G, -955 A/C) were not associated with cancer susceptibility or infiltration depth, while GG genotype carriers of the MMP-2 -1575 G/A polymorphism had an increased risk of developing CRC and more frequent serosa or adventitia invasion compared to the other genotypes, similar to that with the -1306 CC genotype[102]. The similarity in these observations are probably because the MMP-2 -1575 G/A, -1306 C/T, -790 G/T and -735 C/T polymorphisms have been found to be in almost complete pair-wise linkage (dis)equilibrium[28].
The most frequently studied MMP-9 polymorphism is the C to T substitution at position -1562 of the promoter region, which increases transcriptional activity. In a population of 185 Korean colorectal cancer patients and 304 controls, individuals with the CC genotype had an increased risk for developing CRC (OR = 1.7, 95% CI: 1.04-2.66, P = 0.033)[89]. None of the other studies found similar results[90,103-107]. A meta-analysis that included the studies of Elander et al[90], Xu et al[104,107], Woo et al[89] and Xing et al[106] showed no significant association of the -1562 C/T MMP-9 polymorphism and colorectal cancer[100]. The same conclusion was reached in a second meta-analysis[87]. Xing et al[106] reported a decrease of lymph node metastases in 137 Chinese CRC patients with the CC genotype of the MMP-9 -1562 SNP, whereas the other studies did not find an association with lymph node metastases, survival, infiltration depth or any other clinicopathological variable. The mechanism of action of MMP-9 in cancer is intriguing and not as straightforward as some of the other MMPs. In colorectal cancer, both very high and very low levels of MMP-9 in tumor tissue seem to be associated with poor prognosis compared to intermediate MMP-levels[105]. Similarly, in ovarian cancer, the presence of MMP-9 within the ovarian cells is associated with better survival, whereas higher stromal expression is a marker of worse prognosis[108]. The -90(CA)14-27 polymorphism, in which the number of CA repeats influences expression of MMP-9, is not associated with cancer risk or the risk of lymph node metastases[89]. Thus, no consistent relation emerges between MMP-9 genotypes and CRC expression. In a single study of 185 Taiwanese colorectal cancer patients, no association of the MMP-12 -82A/G polymorphism and colorectal cancer risk of development or lymph node metastases was found[89].
Insertion of an extra Adenosine (A) at position -1171 of the MMP-3 promoter generates a 6A allele with lower promoter activity compared to the 5A allele[19]. This polymorphism has been studied quite extensively in colorectal cancer, and in all but one paper, no contribution to cancer risk, clinicopathological parameters or survival was demonstrated[89-93,96,97,99,103]. Only Hinoda et al found a two-fold increase in CRC risk in the 6A/6A homozygotes (OR = 2.11, 95% CI: 1.16-3.82, P = 0.013) in the previously mentioned study of a Japanese cohort of 101 CRC patients and 127 controls. In 302 patients with small adenomas and 568 polyp-free controls, the 6A/6A genotype of the -1171 MMP-3 polymorphism was associated with a significant risk of small adenomas (OR = 1.50, 95%CI: 0.99-2.28, P = 0.008) and this association was even stronger in individuals with the combined genotype MMP-3 -1171 6A/6A + MMP-1 -1607 2G/2G (OR =1.88, 95%CI: 1.08-3.28, P = 0.001)[101]. When the MMP-3 genotype of 295 patients of that study with large adenomas was compared to either the patients with small adenomas or the polyp-free controls, no difference in genotype distribution was found. These findings suggest that this MMP-3 5A/6A polymorphism (and the -1607 1G/2G polymorphism) might be of importance early in the process of adenoma formation. The 6A/6A genotype of the MMP-3 -1171 5A/6A polymorphism has a lower transcriptional activity and higher plasma levels of MMP-3 were measured in 5A/5A homozygote patients with acute coronary syndrome compared to 6A/6A homozygotes[109]. Apparently, the association between this polymorphism and increased susceptibility for developing early colorectal adenomas does not provide an insight into the functional activity of the protein.
No clear association between MMP-7 -181 A/G polymorphism and colorectal cancer incidence, lymph node metastases or survival was found in most of the publications[89,94,95,103,110]. The only exception is by Ghilardi et al[111] who showed that the GG genotype increases the colorectal cancer risk. Furthermore, in the 58 patients with colorectal cancer included in this study, the CC genotype predisposed for lymph node metastases and distant metastases at the time of diagnosis[111]. Ghilardi et al[111] also studied the C/T polymorphism at position -153 of the MMP-7 promoter and found an increase in colorectal cancer risk in T allele carriers, but no association with any of the clinicopathological variables. In a study of 174 colorectal cancer patients, Langers et al[110] reported that patients with the CC genotype had a better 10-year survival than the patients with the CT or TT genotype (CC vs CT+TT: Log Rank 14.0, P = 0.0009). The study of Ghilardi et al[111] included 58 patients, a relatively small number for studying the influence of gene polymorphisms on cancer susceptibility and prognosis. This may explain the discordant results between the different studies and illustrates the need for larger sample sizes. Peng et al tried to solve this problem by performing meta-analyses of case control studies investigating the role of gene polymorphisms of MMP-1, -2, -3, -7 and -9 on cancer susceptibility in lung, head and neck, esophageal, gastric, colorectal, hepatocellular, breast, renal, bladder, cervical, ovarian, endometrial, prostate and skin cancer[38,87]. In these meta-analyses, a consistent positive association with colorectal cancer risk was observed for the MMP-1 -1607 1G/2G polymorphism, but not for MMP-2 -735C/T, MMP-2 -1306 C/T, MMP-7 -181A/G and MMP-9 -1562 C/T.
In summary, although data are still emerging there appears to be evidence for associations between the MMP-1 -1607 1G/2G, MMP-2 -1306 C/T, MMP-7 -181 A/G and MMP-9 -1562 C/T polymorphisms and CRC susceptibility. In affected individuals, an association of the MMP polymorphism with the course of the disease or prognostic parameters was reported in some studies, as shown in Table 3, although these results await further confirmation.
DISCUSSION
The three major regulatory mechanisms that eventually determine the function of MMPs are transcription, activation of latent MMPs and inhibition by specific inhibitors. Along with local activation and inhibition, regulation of transcription seems to be of major importance for the function of MMPs[112]. Most of the promoter polymorphisms that are described in this review have been shown to influence promoter activity and to increase or decrease transcription in vitro, as shown in Table 1. Some SNPs are associated with gastrointestinal cancer susceptibility and in some cases, a correlation with clinicopathological parameters and outcome of the disease was observed. Surprisingly, only a few studies have actually looked at the correlation between the promoter polymorphism of MMPs and the corresponding tumor protein levels. Two studies reported no association between the different genotypes and MMP protein expression in the tumor[56,105]. It would be interesting to correlate the values in normal tissues from these patients with their genotypes to further elucidate their contribution to the phenotypic expression of the MMPs in cancer patients. Besides the regulation of expression by transcription, the presence of MMPs in the (tumor) microenvironment depends on the inactivation/clearing, which is regulated by the inhibitors. High clearance could lead to low protein levels despite high levels of expression. Furthermore, a specific genotype can have different (and even opposite) effects in different cell types. Wang et al showed that the -799T/-381G/+17G haplotype of MMP-8 increased promoter activity in cells resembling chorion cytotrophoblasts, but the same haplotype decreased promoter activity in a leukocyte cell line and had no effect on promoter activity in a macrophage cell line[23]. Cell-specific functional effects of SNPs have been described for several cancer-associated proteins[113,114]. This phenomenon makes the translation of the effect of a promoter polymorphism on gene transcription to the in vivo situation even more complex, especially as many different stromal cell types as well as tumor cells are involved in the production of MMPs (Figure 1). A correlation between a particular polymorphism and cancer susceptibility does not necessarily demonstrate the implication of the corresponding gene in the process of cancer development or progression. It could also be the result of a linkage (dis)equilibrium between the examined (potentially functionally neutral) SNP and another (potentially functionally important) SNP[85].
Although for some MMP-polymorphisms the results between different studies are unanimous, there is often a discrepancy between the results of different studies on the same polymorphism. However, some trends can be observed. An increased incidence of esophageal cancer in CC carriers of the MMP-2 -1306 C/T polymorphism was reported in 2 studies[15,39] and this association was corroborated in a meta-analysis[38]. In the Asian population, G-allele carriers of the MMP-7 -181 A/G polymorphism have an increased risk of developing both esophageal cancer and gastric cancer[45,63,64]. In hepatocellular cancer, no association was found between any of the MMP SNPs and cancer risk, although 5A-allele carriers of the MMP-3 5A/6A polymorphism might have a worse prognosis. Although some studies concerning CRC report a correlation between cancer incidence and the MMP-1 1G/2G, MMP-2 -1306 C/T, MMP-7 -181A/G and MMP-9 -1562 C/T polymorphism, the only association that was found to be significant in a meta-analysis was a higher cancer risk in 2G allele carriers of the MMP-1 1G/2G polymorphism[87]. Sometimes, as for the MMP-7 -181 A/G polymorphism in gastric cancer, the variability in results between the different studies is likely to be explained by ethnic differences between the study groups. Different genotype distributions of MMP-2 and MMP-9 SNPs have been reported in Caucasians and African-Americans, which seem to be associated with differences in prevalence of cancer and cardiovascular disease[115]. The diverse results in the publications described in this review emphasize the need for studies on larger numbers of patients before definite associations between genetic polymorphism and susceptibility to cancer or with the course of the disease in affected individuals can be established. There is a need for large cohorts of patients who are genotyped, and information about disease progression, lymph node metastases, distant metastases and prognosis needs gathered. In the meantime meta-analyses rather than single studies are the best indicators of the practical value of single SNPs. The recent meta-analysis of Zhou et al[116] including almost 3000 breast carcinoma patients, suggested MMP-2 -1306 C/T as a potential indicator, whereas the SNPs of MMP-1, MMP-3 and MMP-9 were not indicative.
Genome-wide association studies (GWAS) may further highlight the genes that are important in identifying people at high risk for the development of cancer or patients who are likely to have an unfavorable outcome of their disease.To date, fourteen loci identified by GWAS analysis have been shown to influence the risk of developing colorectal cancer[117]. None of them is located in any of the MMP genes. However, in an extensive mutation analysis of the human genome in which 13.023 genes were involved, Sjöblom et al[118] identified MMP-2, ADAM29 and three ADAMTS family members among the 69 CAN genes that are often mutated in colorectal cancer.
CONCLUSION
To predict the cancer risk in a population and the outcome of the disease in affected individuals, a genomic profile including functional SNPs of several genes would probably be a better tool than the use of a single SNP. Being key players in the process of cancer development and progression, SNPs of selected MMPs or TIMPs could be included in such a profile to predict disease susceptibility and/or the course of a disease. Because of the heterogeneity of previous studies that have included a relatively small number of patients, further research on large cohorts of cancer patients and healthy controls is needed before a definite conclusion can be drawn about the impact of these genes on gastro-intestinal cancer risk and prognosis.
Footnotes
Peer reviewer: Asahi Hishida, MD, PhD, Department of Preventive Medicine, Nagoya University Graduate School of Medicine, Tsurumai-cho 65, Showa-ku, Nagoya 466-8550, Japan
S- Editor Wang JL L- Editor Hughes D E- Editor Ma WH
References
- 1.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 2.Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer. 2008;8:929–941. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- 3.Shiomi T, Lemaître V, D'Armiento J, Okada Y. Matrix metalloproteinases, a disintegrin and metalloproteinases, and a disintegrin and metalloproteinases with thrombospondin motifs in non-neoplastic diseases. Pathol Int. 2010;60:477–496. doi: 10.1111/j.1440-1827.2010.02547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborti S, Mandal M, Das S, Mandal A, Chakraborti T. Regulation of matrix metalloproteinases: an overview. Mol Cell Biochem. 2003;253:269–285. doi: 10.1023/a:1026028303196. [DOI] [PubMed] [Google Scholar]
- 5.Mook OR, Frederiks WM, Van Noorden CJ. The role of gelatinases in colorectal cancer progression and metastasis. Biochim Biophys Acta. 2004;1705:69–89. doi: 10.1016/j.bbcan.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawinkels LJ, Zuidwijk K, Verspaget HW, de Jonge-Muller ES, van Duijn W, Ferreira V, Fontijn RD, David G, Hommes DW, Lamers CB, et al. VEGF release by MMP-9 mediated heparan sulphate cleavage induces colorectal cancer angiogenesis. Eur J Cancer. 2008;44:1904–1913. doi: 10.1016/j.ejca.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 8.Cornelius LA, Nehring LC, Harding E, Bolanowski M, Welgus HG, Kobayashi DK, Pierce RA, Shapiro SD. Matrix metalloproteinases generate angiostatin: effects on neovascularization. J Immunol. 1998;161:6845–6852. [PubMed] [Google Scholar]
- 9.Patterson BC, Sang QA. Angiostatin-converting enzyme activities of human matrilysin (MMP-7) and gelatinase B/type IV collagenase (MMP-9) J Biol Chem. 1997;272:28823–28825. doi: 10.1074/jbc.272.46.28823. [DOI] [PubMed] [Google Scholar]
- 10.Wang WS, Chen PM, Wang HS, Liang WY, Su Y. Matrix metalloproteinase-7 increases resistance to Fas-mediated apoptosis and is a poor prognostic factor of patients with colorectal carcinoma. Carcinogenesis. 2006;27:1113–1120. doi: 10.1093/carcin/bgi351. [DOI] [PubMed] [Google Scholar]
- 11.Martin MD, Matrisian LM. The other side of MMPs: protective roles in tumor progression. Cancer Metastasis Rev. 2007;26:717–724. doi: 10.1007/s10555-007-9089-4. [DOI] [PubMed] [Google Scholar]
- 12.Shimajiri S, Arima N, Tanimoto A, Murata Y, Hamada T, Wang KY, Sasaguri Y. Shortened microsatellite d(CA)21 sequence down-regulates promoter activity of matrix metalloproteinase 9 gene. FEBS Lett. 1999;455:70–74. doi: 10.1016/s0014-5793(99)00863-7. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Zhang L, Luo H, Zhu Z, Zhang C, Hou Y. Association of matrix metalloproteinases-9 gene polymorphisms with genetic susceptibility to esophageal squamous cell carcinoma. DNA Cell Biol. 2008;27:553–557. doi: 10.1089/dna.2008.0732. [DOI] [PubMed] [Google Scholar]
- 14.Kubben FJ, Sier CF, Meijer MJ, van den Berg M, van der Reijden JJ, Griffioen G, van de Velde CJ, Lamers CB, Verspaget HW. Clinical impact of MMP and TIMP gene polymorphisms in gastric cancer. Br J Cancer. 2006;95:744–751. doi: 10.1038/sj.bjc.6603307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu C, Zhou Y, Miao X, Xiong P, Tan W, Lin D. Functional haplotypes in the promoter of matrix metalloproteinase-2 predict risk of the occurrence and metastasis of esophageal cancer. Cancer Res. 2004;64:7622–7628. doi: 10.1158/0008-5472.CAN-04-1521. [DOI] [PubMed] [Google Scholar]
- 16.Ouyang G, Yao P, Hu W, Chen Q, Wang H, Wang L and Li J. A non-synonymous coding SNP lys45Glu of mmp3 associated with ESCC genetic susceptibility in population of Henan, China. Chinese-German J Clin Oncol. 2009;8:510–515. [Google Scholar]
- 17.Jormsjö S, Whatling C, Walter DH, Zeiher AM, Hamsten A, Eriksson P. Allele-specific regulation of matrix metalloproteinase-7 promoter activity is associated with coronary artery luminal dimensions among hypercholesterolemic patients. Arterioscler Thromb Vasc Biol. 2001;21:1834–1839. doi: 10.1161/hq1101.098229. [DOI] [PubMed] [Google Scholar]
- 18.Rutter JL, Mitchell TI, Butticè G, Meyers J, Gusella JF, Ozelius LJ, Brinckerhoff CE. A single nucleotide polymorphism in the matrix metalloproteinase-1 promoter creates an Ets binding site and augments transcription. Cancer Res. 1998;58:5321–5325. [PubMed] [Google Scholar]
- 19.Ye S, Eriksson P, Hamsten A, Kurkinen M, Humphries SE, Henney AM. Progression of coronary atherosclerosis is associated with a common genetic variant of the human stromelysin-1 promoter which results in reduced gene expression. J Biol Chem. 1996;271:13055–13060. doi: 10.1074/jbc.271.22.13055. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Ye S, Herrmann SM, Eriksson P, de Maat M, Evans A, Arveiler D, Luc G, Cambien F, Hamsten A, et al. Functional polymorphism in the regulatory region of gelatinase B gene in relation to severity of coronary atherosclerosis. Circulation. 1999;99:1788–1794. doi: 10.1161/01.cir.99.14.1788. [DOI] [PubMed] [Google Scholar]
- 21.Jormsjö S, Ye S, Moritz J, Walter DH, Dimmeler S, Zeiher AM, Henney A, Hamsten A, Eriksson P. Allele-specific regulation of matrix metalloproteinase-12 gene activity is associated with coronary artery luminal dimensions in diabetic patients with manifest coronary artery disease. Circ Res. 2000;86:998–1003. doi: 10.1161/01.res.86.9.998. [DOI] [PubMed] [Google Scholar]
- 22.Price SJ, Greaves DR, Watkins H. Identification of novel, functional genetic variants in the human matrix metalloproteinase-2 gene: role of Sp1 in allele-specific transcriptional regulation. J Biol Chem. 2001;276:7549–7558. doi: 10.1074/jbc.M010242200. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Parry S, Macones G, Sammel MD, Ferrand PE, Kuivaniemi H, Tromp G, Halder I, Shriver MD, Romero R, et al. Functionally significant SNP MMP8 promoter haplotypes and preterm premature rupture of membranes (PPROM) Hum Mol Genet. 2004;13:2659–2669. doi: 10.1093/hmg/ddh287. [DOI] [PubMed] [Google Scholar]
- 24.Yoon S, Kuivaniemi H, Gatalica Z, Olson JM, Butticè G, Ye S, Norris BA, Malcom GT, Strong JP, Tromp G. MMP13 promoter polymorphism is associated with atherosclerosis in the abdominal aorta of young black males. Matrix Biol. 2002;21:487–498. doi: 10.1016/s0945-053x(02)00053-7. [DOI] [PubMed] [Google Scholar]
- 25.Joos L, He JQ, Shepherdson MB, Connett JE, Anthonisen NR, Paré PD, Sandford AJ. The role of matrix metalloproteinase polymorphisms in the rate of decline in lung function. Hum Mol Genet. 2002;11:569–576. doi: 10.1093/hmg/11.5.569. [DOI] [PubMed] [Google Scholar]
- 26.Hinterseher I, Krex D, Kuhlisch E, Schmidt KG, Pilarsky C, Schneiders W, Saeger HD, Bergert H. Tissue inhibitor of metalloproteinase-1 (TIMP-1) polymorphisms in a Caucasian population with abdominal aortic aneurysm. World J Surg. 2007;31:2248–2254. doi: 10.1007/s00268-007-9209-x. [DOI] [PubMed] [Google Scholar]
- 27.Harendza S, Lovett DH, Panzer U, Lukacs Z, Kuhnl P, Stahl RA. Linked common polymorphisms in the gelatinase a promoter are associated with diminished transcriptional response to estrogen and genetic fitness. J Biol Chem. 2003;278:20490–20499. doi: 10.1074/jbc.M211536200. [DOI] [PubMed] [Google Scholar]
- 28.Vasků A, Goldbergová M, Izakovicová Hollá L, Sisková L, Groch L, Beránek M, Tschöplová S, Znojil V, Vácha J. A haplotype constituted of four MMP-2 promoter polymorphisms (-1575G/A, -1306C/T, -790T/G and -735C/T) is associated with coronary triple-vessel disease. Matrix Biol. 2004;22:585–591. doi: 10.1016/j.matbio.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Shagisultanova EI, Novikova IA, Sidorenko YS, Marchenko GN, Strongin AY, Malkhosyan SR. The matrix metalloproteinase-21 gene 572C/T polymorphism and the risk of breast cancer. Anticancer Res. 2004;24:199–201. [PubMed] [Google Scholar]
- 30.Hirano K, Sakamoto T, Uchida Y, Morishima Y, Masuyama K, Ishii Y, Nomura A, Ohtsuka M, Sekizawa K. Tissue inhibitor of metalloproteinases-2 gene polymorphisms in chronic obstructive pulmonary disease. Eur Respir J. 2001;18:748–752. doi: 10.1183/09031936.01.00102101. [DOI] [PubMed] [Google Scholar]
- 31.Sun D-L, Duan Y-N, Zhang X-J, Wang N, Zhou R-M, Chen Z-F and Li Y. Association of single nucleotide polymorphisms in the promoter region of MMP-2 gene with susceptibility to esophageal squamous cell carcinoma in high prevalence area. Tumor. 2009;29:354–357. [Google Scholar]
- 32.Früh M, Zhou W, Zhai R, Su L, Heist RS, Wain JC, Nishioka NS, Lynch TJ, Shepherd FA, Christiani DC, et al. Polymorphisms of inflammatory and metalloproteinase genes, Helicobacter pylori infection and the risk of oesophageal adenocarcinoma. Br J Cancer. 2008;98:689–692. doi: 10.1038/sj.bjc.6604234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradbury PA, Zhai R, Hopkins J, Kulke MH, Heist RS, Singh S, Zhou W, Ma C, Xu W, Asomaning K, et al. Matrix metalloproteinase 1, 3 and 12 polymorphisms and esophageal adenocarcinoma risk and prognosis. Carcinogenesis. 2009;30:793–798. doi: 10.1093/carcin/bgp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samantaray S, Sharma R, Chattopadhyaya TK, Gupta SD, Ralhan R. Increased expression of MMP-2 and MMP-9 in esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2004;130:37–44. doi: 10.1007/s00432-003-0500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi M, Li JS, Li Y. [Clinicopathological significance of expressions of RECK and MMP-2 in esophageal squamous carcinoma] Zhonghua Zhong Liu Za Zhi. 2010;32:283–285. [PubMed] [Google Scholar]
- 36.Li Y, Ma J, Guo Q, Duan F, Tang F, Zheng P, Zhao Z, Lu G. Overexpression of MMP-2 and MMP-9 in esophageal squamous cell carcinoma. Dis Esophagus. 2009;22:664–667. doi: 10.1111/j.1442-2050.2008.00928.x. [DOI] [PubMed] [Google Scholar]
- 37.Ishibashi Y, Matsumoto T, Niwa M, Suzuki Y, Omura N, Hanyu N, Nakada K, Yanaga K, Yamada K, Ohkawa K, et al. CD147 and matrix metalloproteinase-2 protein expression as significant prognostic factors in esophageal squamous cell carcinoma. Cancer. 2004;101:1994–2000. doi: 10.1002/cncr.20593. [DOI] [PubMed] [Google Scholar]
- 38.Peng B, Cao L, Ma X, Wang W, Wang D, Yu L. Meta-analysis of association between matrix metalloproteinases 2, 7 and 9 promoter polymorphisms and cancer risk. Mutagenesis. 2010;25:371–379. doi: 10.1093/mutage/geq015. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Sun DL, Duan YN, Zhang XJ, Wang N, Zhou RM, Chen ZF, Wang SJ. Association of functional polymorphisms in MMPs genes with gastric cardia adenocarcinoma and esophageal squamous cell carcinoma in high incidence region of North China. Mol Biol Rep. 2010;37:197–205. doi: 10.1007/s11033-009-9593-4. [DOI] [PubMed] [Google Scholar]
- 40.Chen XB, Chen GL, Liu JN, Yang JZ, Yu DK, Lin DX, Tan W. [Genetic polymorphisms in STK15 and MMP-2 associated susceptibility to esophageal cancer in Mongolian population] Zhonghua Yu Fang Yi Xue Za Zhi. 2009;43:559–564. [PubMed] [Google Scholar]
- 41.Zhang XJ, Guo W, Wang N, Zhou RM, Dong XJ, Li Y. [The association of MMP-13 polymorphism with susceptibility to esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma] Yi Chuan. 2006;28:1500–1504. doi: 10.1360/yc-006-1500. [DOI] [PubMed] [Google Scholar]
- 42.Jin X, Kuang G, Wei LZ, Li Y, Wang R, Guo W, Wang N, Fang SM, Wen DG, Chen ZF, et al. No association of the matrix metalloproteinase 1 promoter polymorphism with susceptibility to esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma in northern China. World J Gastroenterol. 2005;11:2385–2389. doi: 10.3748/wjg.v11.i16.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia P, Chang D-M, Dang C-X, Meng L, Xue H and Liu Y. Association between the -1562 C/T polymorphism in the MMP-9 promoter and phenotype of esophageal squamous cell carcinoma in northern Chinese population. Academic J Xi'an Jiaotong University. 2010;22:39–43. [Google Scholar]
- 44.Zhang J, Jin X, Fang S, Li Y, Wang R, Guo W, Wang N, Wang Y, Wen D, Wei L, et al. The functional SNP in the matrix metalloproteinase-3 promoter modifies susceptibility and lymphatic metastasis in esophageal squamous cell carcinoma but not in gastric cardiac adenocarcinoma. Carcinogenesis. 2004;25:2519–2524. doi: 10.1093/carcin/bgh269. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Jin X, Fang S, Wang R, Li Y, Wang N, Guo W, Wang Y, Wen D, Wei L, et al. The functional polymorphism in the matrix metalloproteinase-7 promoter increases susceptibility to esophageal squamous cell carcinoma, gastric cardiac adenocarcinoma and non-small cell lung carcinoma. Carcinogenesis. 2005;26:1748–1753. doi: 10.1093/carcin/bgi144. [DOI] [PubMed] [Google Scholar]
- 46.Ii M, Yamamoto H, Adachi Y, Maruyama Y, Shinomura Y. Role of matrix metalloproteinase-7 (matrilysin) in human cancer invasion, apoptosis, growth, and angiogenesis. Exp Biol Med (Maywood) 2006;231:20–27. doi: 10.1177/153537020623100103. [DOI] [PubMed] [Google Scholar]
- 47.Tanioka Y, Yoshida T, Yagawa T, Saiki Y, Takeo S, Harada T, Okazawa T, Yanai H, Okita K. Matrix metalloproteinase-7 and matrix metalloproteinase-9 are associated with unfavourable prognosis in superficial oesophageal cancer. Br J Cancer. 2003;89:2116–2121. doi: 10.1038/sj.bjc.6601372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohashi K, Nemoto T, Nakamura K, Nemori R. Increased expression of matrix metalloproteinase 7 and 9 and membrane type 1-matrix metalloproteinase in esophageal squamous cell carcinomas. Cancer. 2000;88:2201–2209. [PubMed] [Google Scholar]
- 49.Yamashita K, Mori M, Shiraishi T, Shibuta K, Sugimachi K. Clinical significance of matrix metalloproteinase-7 expression in esophageal carcinoma. Clin Cancer Res. 2000;6:1169–1174. [PubMed] [Google Scholar]
- 50.González CA, López-Carrillo L. Helicobacter pylori, nutrition and smoking interactions: their impact in gastric carcinogenesis. Scand J Gastroenterol. 2010;45:6–14. doi: 10.3109/00365520903401959. [DOI] [PubMed] [Google Scholar]
- 51.Sier CF, Kubben FJ, Ganesh S, Heerding MM, Griffioen G, Hanemaaijer R, van Krieken JH, Lamers CB, Verspaget HW. Tissue levels of matrix metalloproteinases MMP-2 and MMP-9 are related to the overall survival of patients with gastric carcinoma. Br J Cancer. 1996;74:413–417. doi: 10.1038/bjc.1996.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kubben FJ, Sier CF, van Duijn W, Griffioen G, Hanemaaijer R, van de Velde CJ, van Krieken JH, Lamers CB, Verspaget HW. Matrix metalloproteinase-2 is a consistent prognostic factor in gastric cancer. Br J Cancer. 2006;94:1035–1040. doi: 10.1038/sj.bjc.6603041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang XM, Miao XP, Xiong P, Yu CY, Tan W, Qu SN, Sun T, Zhou YF, Lin DX. [Association of functional polymorphisms in matrix metalloproteinase-2 (MMP-2) and MMP-9 genes with risk of gastric cancer in a Chinese population] Ai Zheng. 2004;23:1233–1237. [PubMed] [Google Scholar]
- 54.Miao X, Yu C, Tan W, Xiong P, Liang G, Lu W, Lin D. A functional polymorphism in the matrix metalloproteinase-2 gene promoter (-1306C/T) is associated with risk of development but not metastasis of gastric cardia adenocarcinoma. Cancer Res. 2003;63:3987–3990. [PubMed] [Google Scholar]
- 55.Wu CY, Wu MS, Chen YJ, Chen CJ, Chen HP, Shun CT, Chen GH, Huang SP, Lin JT. Clinicopathological significance of MMP-2 and TIMP-2 genotypes in gastric cancer. Eur J Cancer. 2007;43:799–808. doi: 10.1016/j.ejca.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 56.Alakus H, Afriani N, Warnecke-Eberz U, Bollschweiler E, Fetzner U, Drebber U, Metzger R, Hölscher AH, Mönig SP. Clinical impact of MMP and TIMP gene polymorphisms in gastric cancer. World J Surg. 2010;34:2853–2859. doi: 10.1007/s00268-010-0761-4. [DOI] [PubMed] [Google Scholar]
- 57.Matsumura S, Oue N, Nakayama H, Kitadai Y, Yoshida K, Yamaguchi Y, Imai K, Nakachi K, Matsusaki K, Chayama K, et al. A single nucleotide polymorphism in the MMP-9 promoter affects tumor progression and invasive phenotype of gastric cancer. J Cancer Res Clin Oncol. 2005;131:19–25. doi: 10.1007/s00432-004-0621-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu CY, Wu MS, Chen HP, Chen CJ, Chen GH and Lin JT. MMP-9-1562 C/T promoter polymorphism associated with gastric cancer development in female. Gastroenterology. 2007;132:A299. [Google Scholar]
- 59.Tang Y, Zhu J, Chen L, Chen L, Zhang S, Lin J. Associations of matrix metalloproteinase-9 protein polymorphisms with lymph node metastasis but not invasion of gastric cancer. Clin Cancer Res. 2008;14:2870–2877. doi: 10.1158/1078-0432.CCR-07-4042. [DOI] [PubMed] [Google Scholar]
- 60.Ajisaka H, Yonemura Y, Miwa K. Correlation of lymph node metastases and expression of matrix metalloproteinase-7 in patients with gastric cancer. Hepatogastroenterology. 2004;51:900–905. [PubMed] [Google Scholar]
- 61.Liu XP, Kawauchi S, Oga A, Tsushimi K, Tsushimi M, Furuya T, Sasaki K. Prognostic significance of matrix metalloproteinase-7 (MMP-7) expression at the invasive front in gastric carcinoma. Jpn J Cancer Res. 2002;93:291–295. doi: 10.1111/j.1349-7006.2002.tb02171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koskensalo S, Mrena J, Wiksten JP, Nordling S, Kokkola A, Hagström J, Haglund C. MMP-7 overexpression is an independent prognostic marker in gastric cancer. Tumour Biol. 2010;31:149–155. doi: 10.1007/s13277-010-0020-1. [DOI] [PubMed] [Google Scholar]
- 63.Sugimoto M, Furuta T, Kodaira C, Nishino M, Yamade M, Ikuma M, Sugimura H, Hishida A. Polymorphisms of matrix metalloproteinase-7 and chymase are associated with susceptibility to and progression of gastric cancer in Japan. J Gastroenterol. 2008;43:751–761. doi: 10.1007/s00535-008-2221-6. [DOI] [PubMed] [Google Scholar]
- 64.Li JY, Tian MM and Zhao AL. Polymorphism in the promoter region of the metalloproteinase-7 increases susceptibility and risk of metastasis of gastric adenocarcinoma. Gastroenterology. 2008;134:A603. [Google Scholar]
- 65.Matsumura S, Oue N, Kitadai Y, Chayama K, Yoshida K, Yamaguchi Y, Toge T, Imai K, Nakachi K, Yasui W. A single nucleotide polymorphism in the MMP-1 promoter is correlated with histological differentiation of gastric cancer. J Cancer Res Clin Oncol. 2004;130:259–265. doi: 10.1007/s00432-004-0543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang L, Gu HJ, Zhu HJ, Sun QM, Cong RH, Zhou B, Tang NP, Wang B. Tissue inhibitor of metalloproteinase-2 G-418C polymorphism is associated with an increased risk of gastric cancer in a Chinese population. Eur J Surg Oncol. 2008;34:636–641. doi: 10.1016/j.ejso.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 67.Voland P, Besig S, Rad R, Braun T, Baur DM, Perren A, Langer R, Höfler H, Prinz C. Correlation of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinase expression in ileal carcinoids, lymph nodes and liver metastasis with prognosis and survival. Neuroendocrinology. 2009;89:66–78. doi: 10.1159/000151482. [DOI] [PubMed] [Google Scholar]
- 68.Pryczynicz A, Guzińska-Ustymowicz K, Dymicka-Piekarska V, Czyzewska J, Kemona A. Expression of matrix metalloproteinase 9 in pancreatic ductal carcinoma is associated with tumor metastasis formation. Folia Histochem Cytobiol. 2007;45:37–40. [PubMed] [Google Scholar]
- 69.Harvey SR, Hurd TC, Markus G, Martinick MI, Penetrante RM, Tan D, Venkataraman P, DeSouza N, Sait SN, Driscoll DL, et al. Evaluation of urinary plasminogen activator, its receptor, matrix metalloproteinase-9, and von Willebrand factor in pancreatic cancer. Clin Cancer Res. 2003;9:4935–4943. [PubMed] [Google Scholar]
- 70.Nakamura H, Horita S, Senmaru N, Miyasaka Y, Gohda T, Inoue Y, Fujita M, Meguro T, Morita T, Nagashima K. Association of matrilysin expression with progression and poor prognosis in human pancreatic adenocarcinoma. Oncol Rep. 2002;9:751–755. [PubMed] [Google Scholar]
- 71.Yamamoto H, Itoh F, Iku S, Adachi Y, Fukushima H, Sasaki S, Mukaiya M, Hirata K, Imai K. Expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases in human pancreatic adenocarcinomas: clinicopathologic and prognostic significance of matrilysin expression. J Clin Oncol. 2001;19:1118–1127. doi: 10.1200/JCO.2001.19.4.1118. [DOI] [PubMed] [Google Scholar]
- 72.Ito T, Ito M, Shiozawa J, Naito S, Kanematsu T, Sekine I. Expression of the MMP-1 in human pancreatic carcinoma: relationship with prognostic factor. Mod Pathol. 1999;12:669–674. [PubMed] [Google Scholar]
- 73.Jones LE, Humphreys MJ, Campbell F, Neoptolemos JP, Boyd MT. Comprehensive analysis of matrix metalloproteinase and tissue inhibitor expression in pancreatic cancer: increased expression of matrix metalloproteinase-7 predicts poor survival. Clin Cancer Res. 2004;10:2832–2845. doi: 10.1158/1078-0432.ccr-1157-03. [DOI] [PubMed] [Google Scholar]
- 74.Gurevich LE. Role of matrix metalloproteinases 2 and 9 in determination of invasive potential of pancreatic tumors. Bull Exp Biol Med. 2003;136:494–498. doi: 10.1023/b:bebm.0000017103.79321.0b. [DOI] [PubMed] [Google Scholar]
- 75.Giannopoulos G, Pavlakis K, Parasi A, Kavatzas N, Tiniakos D, Karakosta A, Tzanakis N, Peros G. The expression of matrix metalloproteinases-2 and -9 and their tissue inhibitor 2 in pancreatic ductal and ampullary carcinoma and their relation to angiogenesis and clinicopathological parameters. Anticancer Res. 2005;28:1875–1881. [PubMed] [Google Scholar]
- 76.Wiencke K, Louka AS, Spurkland A, Vatn M, Schrumpf E, Boberg KM. Association of matrix metalloproteinase-1 and -3 promoter polymorphisms with clinical subsets of Norwegian primary sclerosing cholangitis patients. J Hepatol. 2004;41:209–214. doi: 10.1016/j.jhep.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 77.Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 78.Okamoto K, Ishida C, Ikebuchi Y, Mandai M, Mimura K, Murawaki Y, Yuasa I. The genotypes of IL-1 beta and MMP-3 are associated with the prognosis of HCV-related hepatocellular carcinoma. Intern Med. 2010;49:887–895. doi: 10.2169/internalmedicine.49.3268. [DOI] [PubMed] [Google Scholar]
- 79.Okamoto K, Mandai M, Mimura K, Murawaki Y, Yuasa I. The association of MMP-1, -3 and -9 genotypes with the prognosis of HCV-related hepatocellular carcinoma patients. Res Commun Mol Pathol Pharmacol. 2005;117-118:77–89. [PubMed] [Google Scholar]
- 80.Zhai Y, Qiu W, Dong XJ, Zhang XM, Xie WM, Zhang HX, Yuan XY, Zhou GQ, He FC. Functional polymorphisms in the promoters of MMP-1, MMP-2, MMP-3, MMP-9, MMP-12 and MMP-13 are not associated with hepatocellular carcinoma risk. Gut. 2007;56:445–447. doi: 10.1136/gut.2006.112706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qiu W, Zhou G, Zhai Y, Zhang X, Xie W, Zhang H, Yang H, Zhi L, Yuan X, Zhang X, et al. No association of MMP-7, MMP-8, and MMP-21 polymorphisms with the risk of hepatocellular carcinoma in a Chinese population. Cancer Epidemiol Biomarkers Prev. 2008;17:2514–2518. doi: 10.1158/1055-9965.EPI-08-0557. [DOI] [PubMed] [Google Scholar]
- 82.Wu LM, Zhang F, Xie HY, Xu X, Chen QX, Yin SY, Liu XC, Zhou L, Xu XB, Sun YL, et al. MMP2 promoter polymorphism (C-1306T) and risk of recurrence in patients with hepatocellular carcinoma after transplantation. Clin Genet. 2008;73:273–278. doi: 10.1111/j.1399-0004.2007.00955.x. [DOI] [PubMed] [Google Scholar]
- 83.Okamoto K, Ikebuchi Y, Ishida C, Murawaki Y. The association of IL-10 and TIMP-2 gene polymorphisms with HCC carcinogenesis and the prognosis in chronic hepatitis C patients. J Hepatol. 2010;52:S228. [Google Scholar]
- 84.Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59:366–378. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- 85.Decock J, Paridaens R, Ye S. Genetic polymorphisms of matrix metalloproteinases in lung, breast and colorectal cancer. Clin Genet. 2008;73:197–211. doi: 10.1111/j.1399-0004.2007.00946.x. [DOI] [PubMed] [Google Scholar]
- 86.Arakaki PA, Marques MR, Santos MC. MMP-1 polymorphism and its relationship to pathological processes. J Biosci. 2009;34:313–320. doi: 10.1007/s12038-009-0035-1. [DOI] [PubMed] [Google Scholar]
- 87.Peng B, Cao L, Wang W, Xian L, Jiang D, Zhao J, Zhang Z, Wang X, Yu L. Polymorphisms in the promoter regions of matrix metalloproteinases 1 and 3 and cancer risk: a meta-analysis of 50 case-control studies. Mutagenesis. 2010;25:41–48. doi: 10.1093/mutage/gep041. [DOI] [PubMed] [Google Scholar]
- 88.Kouhkan F, Motovali-Bashi M, Hojati Z. The influence of interstitial collagenase-1 genotype polymorphism on colorectal cancer risk in Iranian population. Cancer Invest. 2008;26:836–842. doi: 10.1080/07357900801953204. [DOI] [PubMed] [Google Scholar]
- 89.Woo M, Park K, Nam J, Kim JC. Clinical implications of matrix metalloproteinase-1, -3, -7, -9, -12, and plasminogen activator inhibitor-1 gene polymorphisms in colorectal cancer. J Gastroenterol Hepatol. 2007;22:1064–1070. doi: 10.1111/j.1440-1746.2006.04424.x. [DOI] [PubMed] [Google Scholar]
- 90.Elander N, Söderkvist P, Fransén K. Matrix metalloproteinase (MMP) -1, -2, -3 and -9 promoter polymorphisms in colorectal cancer. Anticancer Res. 2006;26:791–795. [PubMed] [Google Scholar]
- 91.Hinoda Y, Okayama N, Takano N, Fujimura K, Suehiro Y, Hamanaka Y, Hazama S, Kitamura Y, Kamatani N, Oka M. Association of functional polymorphisms of matrix metalloproteinase (MMP)-1 and MMP-3 genes with colorectal cancer. Int J Cancer. 2002;102:526–529. doi: 10.1002/ijc.10750. [DOI] [PubMed] [Google Scholar]
- 92.Ghilardi G, Biondi ML, Mangoni J, Leviti S, DeMonti M, Guagnellini E, Scorza R. Matrix metalloproteinase-1 promoter polymorphism 1G/2G is correlated with colorectal cancer invasiveness. Clin Cancer Res. 2001;7:2344–2346. [PubMed] [Google Scholar]
- 93.Biondi ML, Turri O, Leviti S, Seminati R, Cecchini F, Bernini M, Ghilardi G, Guagnellini E. MMP1 and MMP3 polymorphisms in promoter regions and cancer. Clin Chem. 2000;46:2023–2024. [PubMed] [Google Scholar]
- 94.Fang WL, Liang WB, He H, Zhu Y, Li SL, Gao LB, Zhang L. Association of matrix metalloproteinases 1, 7, and 9 gene polymorphisms with genetic susceptibility to colorectal carcinoma in a Han Chinese population. DNA Cell Biol. 2010;29:657–661. doi: 10.1089/dna.2010.1017. [DOI] [PubMed] [Google Scholar]
- 95.de Lima JM, de Souza LG, da Silva ID, Forones NM. E-cadherin and metalloproteinase-1 and -7 polymorphisms in colorectal cancer. Int J Biol Markers. 2009;24:99–106. doi: 10.1177/172460080902400206. [DOI] [PubMed] [Google Scholar]
- 96.Hettiaratchi A, Hawkins NJ, McKenzie G, Ward RL, Hunt JE, Wakefield D, Di Girolamo N. The collagenase-1 (MMP-1) gene promoter polymorphism - 1607/2G is associated with favourable prognosis in patients with colorectal cancer. Br J Cancer. 2007;96:783–792. doi: 10.1038/sj.bjc.6603630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu E, Lai M, Lŭ B, Xing X, Huang Q. No association between the polymorphisms in matrix metalloproteinase-1 and matrix metalloproteinase-3 promoter regions and colorectal cancer in Chinese. Dis Colon Rectum. 2006;49:1439–1444. doi: 10.1007/s10350-006-0652-9. [DOI] [PubMed] [Google Scholar]
- 98.Przybylowska K, Blasiak J. A single nucleotide polymorphism in the matrix metalloproteinase-1 gene promoter in colorectal cancer. Exp Oncol. 2002;24:25–27. [Google Scholar]
- 99.Zinzindohoué F, Lecomte T, Ferraz JM, Houllier AM, Cugnenc PH, Berger A, Blons H, Laurent-Puig P. Prognostic significance of MMP-1 and MMP-3 functional promoter polymorphisms in colorectal cancer. Clin Cancer Res. 2005;11:594–599. [PubMed] [Google Scholar]
- 100.McColgan P, Sharma P. Polymorphisms of matrix metalloproteinases 1, 2, 3 and 9 and susceptibility to lung, breast and colorectal cancer in over 30,000 subjects. Int J Cancer. 2009;125:1473–1478. doi: 10.1002/ijc.24441. [DOI] [PubMed] [Google Scholar]
- 101.Lièvre A, Milet J, Carayol J, Le Corre D, Milan C, Pariente A, Nalet B, Lafon J, Faivre J, Bonithon-Kopp C, et al. Genetic polymorphisms of MMP1, MMP3 and MMP7 gene promoter and risk of colorectal adenoma. BMC Cancer. 2006;6:270. doi: 10.1186/1471-2407-6-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu E, Lai M, Lv B, Xing X, Huang Q, Xia X. A single nucleotide polymorphism in the matrix metalloproteinase-2 promoter is associated with colorectal cancer. Biochem Biophys Res Commun. 2004;324:999–1003. doi: 10.1016/j.bbrc.2004.09.150. [DOI] [PubMed] [Google Scholar]
- 103.Ohtani H, Maeda N and Murawaki Y. Functional polymorphisms in the promoter regions of matrix metalloproteinase-2,-3,-7,-9 and TNF-alpha Genes, and the risk of colorectal neoplasm in Japanese. Yonago Acta Med. 2009;52:47–56. [Google Scholar]
- 104.Xu E, Xia X, Lü B, Xing X, Huang Q, Ma Y, Wang W, Lai M. Association of matrix metalloproteinase-2 and -9 promoter polymorphisms with colorectal cancer in Chinese. Mol Carcinog. 2007;46:924–929. doi: 10.1002/mc.20323. [DOI] [PubMed] [Google Scholar]
- 105.Langers AM, Sier CF, Hawinkels LJ, Kubben FJ, van Duijn W, van der Reijden JJ, Lamers CB, Hommes DW, Verspaget HW. MMP-2 geno-phenotype is prognostic for colorectal cancer survival, whereas MMP-9 is not. Br J Cancer. 2008;98:1820–1823. doi: 10.1038/sj.bjc.6604380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xing LL, Wang ZN, Jiang L, Zhang Y, Xu YY, Li J, Luo Y, Zhang X. Matrix metalloproteinase-9-1562C& gt; T polymorphism may increase the risk of lymphatic metastasis of colorectal cancer. World J Gastroenterol. 2007;13:4626–4629. doi: 10.3748/wjg.v13.i34.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu EP, Huang Q, Lu BJ, Xing XM, Lai MD. [The correlation between polymorphisms of matrix metalloproteinase-2 and -9 genes and colorectal cancer of Chinese patients] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23:78–81. [PubMed] [Google Scholar]
- 108.Sillanpää S, Anttila M, Voutilainen K, Ropponen K, Turpeenniemi-Hujanen T, Puistola U, Tammi R, Tammi M, Sironen R, Saarikoski S, et al. Prognostic significance of matrix metalloproteinase-9 (MMP-9) in epithelial ovarian cancer. Gynecol Oncol. 2007;104:296–303. doi: 10.1016/j.ygyno.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 109.Liu PY, Li YH, Chan SH, Lin LJ, Wu HL, Shi GY, Chen JH. Genotype-phenotype association of matrix metalloproteinase-3 polymorphism and its synergistic effect with smoking on the occurrence of acute coronary syndrome. Am J Cardiol. 2006;98:1012–1017. doi: 10.1016/j.amjcard.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 110.Langers A, Verspaget HW, Hawinkels L, Hommes DW, Lamers CB and Sier CF. Protein level and gene promoter polymorphism of Matrilysin (Mmp-7) are independent prognostic factors for survival of colorectal cancer patients. Gastroenterology. 2008;134:A66. [Google Scholar]
- 111.Ghilardi G, Biondi ML, Erario M, Guagnellini E, Scorza R. Colorectal carcinoma susceptibility and metastases are associated with matrix metalloproteinase-7 promoter polymorphisms. Clin Chem. 2003;49:1940–1942. doi: 10.1373/clinchem.2003.018911. [DOI] [PubMed] [Google Scholar]
- 112.Ye S. Polymorphism in matrix metalloproteinase gene promoters: implication in regulation of gene expression and susceptibility of various diseases. Matrix Biol. 2000;19:623–629. doi: 10.1016/s0945-053x(00)00102-5. [DOI] [PubMed] [Google Scholar]
- 113.Koch O, Kwiatkowski DP, Udalova IA. Context-specific functional effects of IFNGR1 promoter polymorphism. Hum Mol Genet. 2006;15:1475–1481. doi: 10.1093/hmg/ddl071. [DOI] [PubMed] [Google Scholar]
- 114.Wang B, Ngoi S, Wang J, Chong SS, Lee CG. The promoter region of the MDR1 gene is largely invariant, but different single nucleotide polymorphism haplotypes affect MDR1 promoter activity differently in different cell lines. Mol Pharmacol. 2006;70:267–276. doi: 10.1124/mol.105.019810. [DOI] [PubMed] [Google Scholar]
- 115.Lacchini R, Metzger IF, Luizon M, Ishizawa M, Tanus-Santos JE. Interethnic differences in the distribution of matrix metalloproteinases genetic polymorphisms are consistent with interethnic differences in disease prevalence. DNA Cell Biol. 2010;29:649–655. doi: 10.1089/dna.2010.1056. [DOI] [PubMed] [Google Scholar]
- 116.Zhou P, Du LF, Lv GQ, Yu XM, Gu YL, Li JP, Zhang C. Current evidence on the relationship between four polymorphisms in the matrix metalloproteinases (MMP) gene and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2011;127:813–818. doi: 10.1007/s10549-010-1294-0. [DOI] [PubMed] [Google Scholar]
- 117.Houlston RS, Cheadle J, Dobbins SE, Tenesa A, Jones AM, Howarth K, Spain SL, Broderick P, Domingo E, Farrington S, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]