Abstract
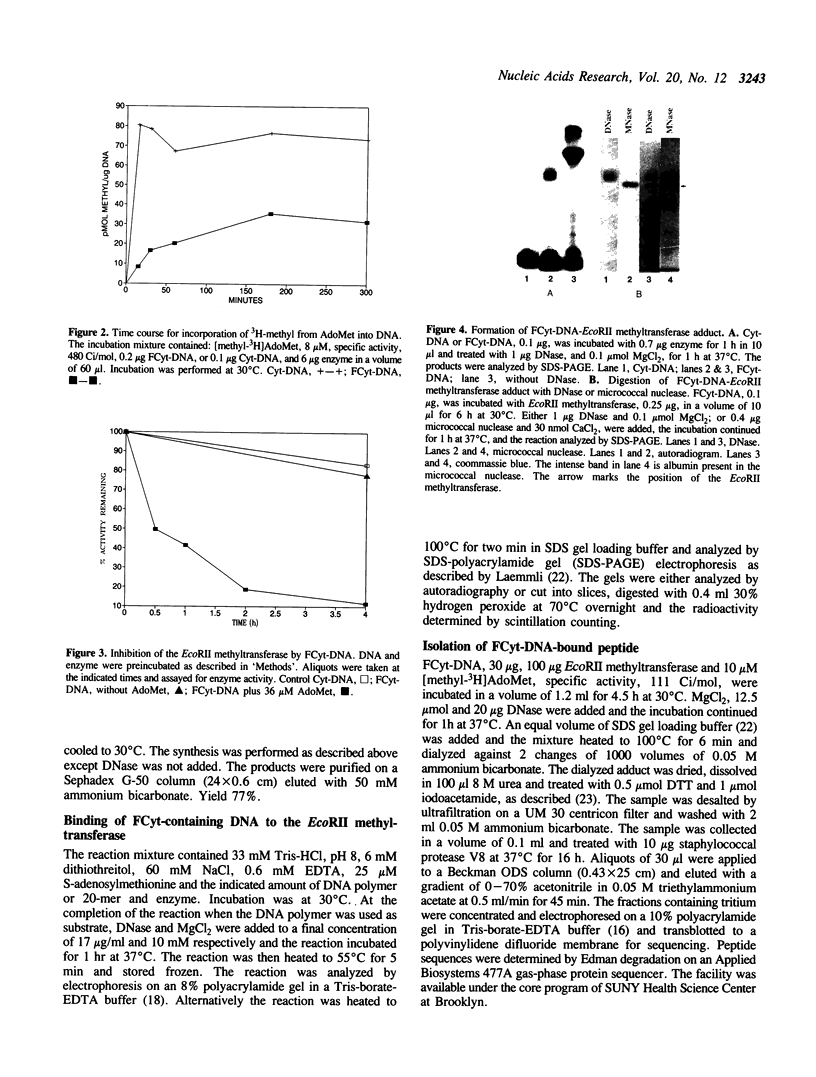
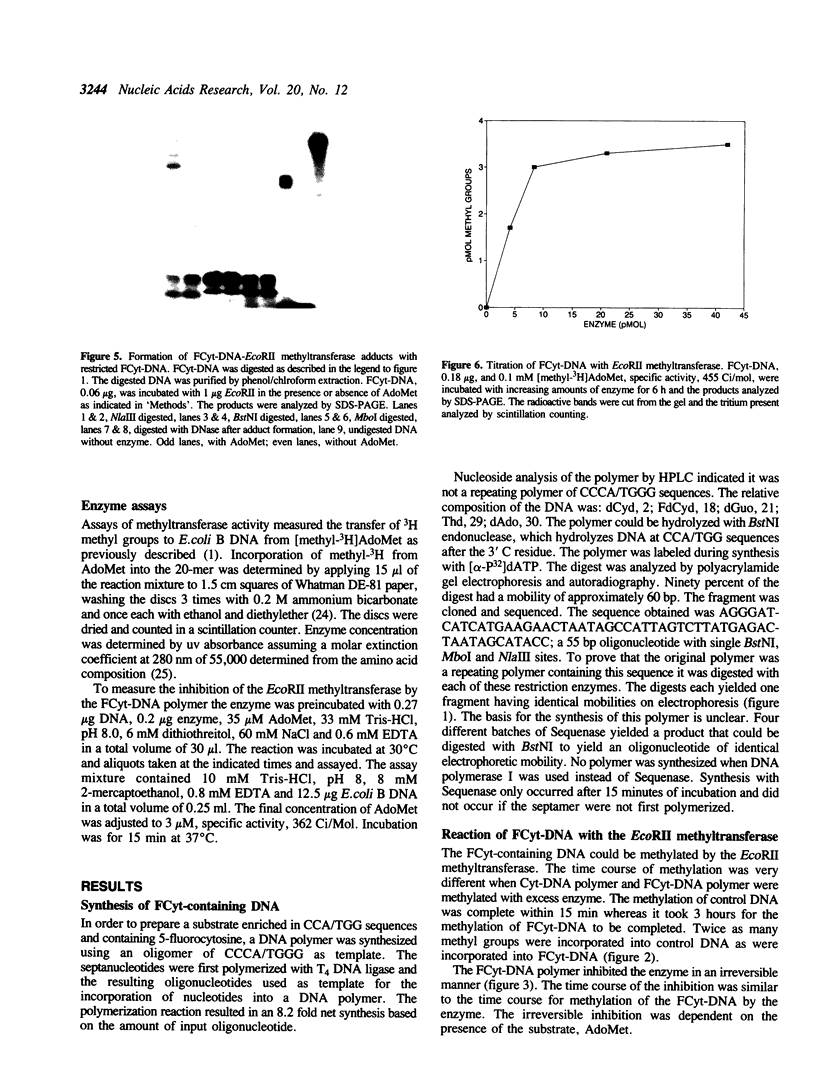
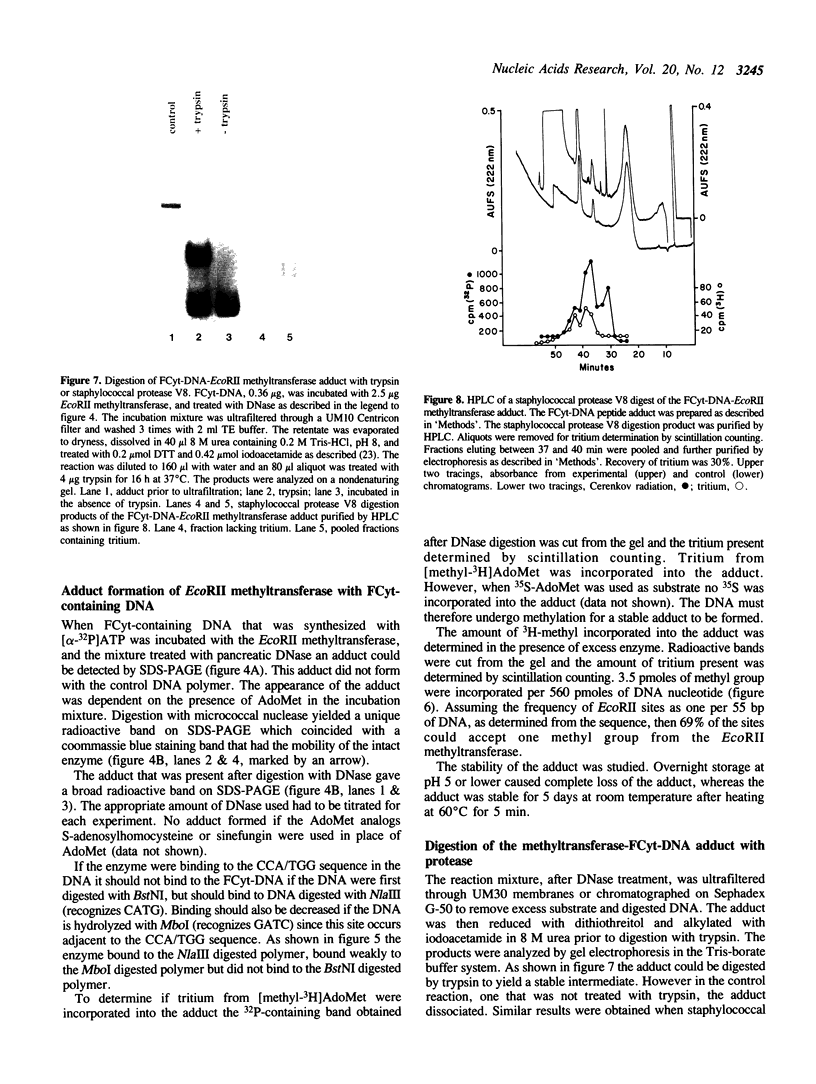
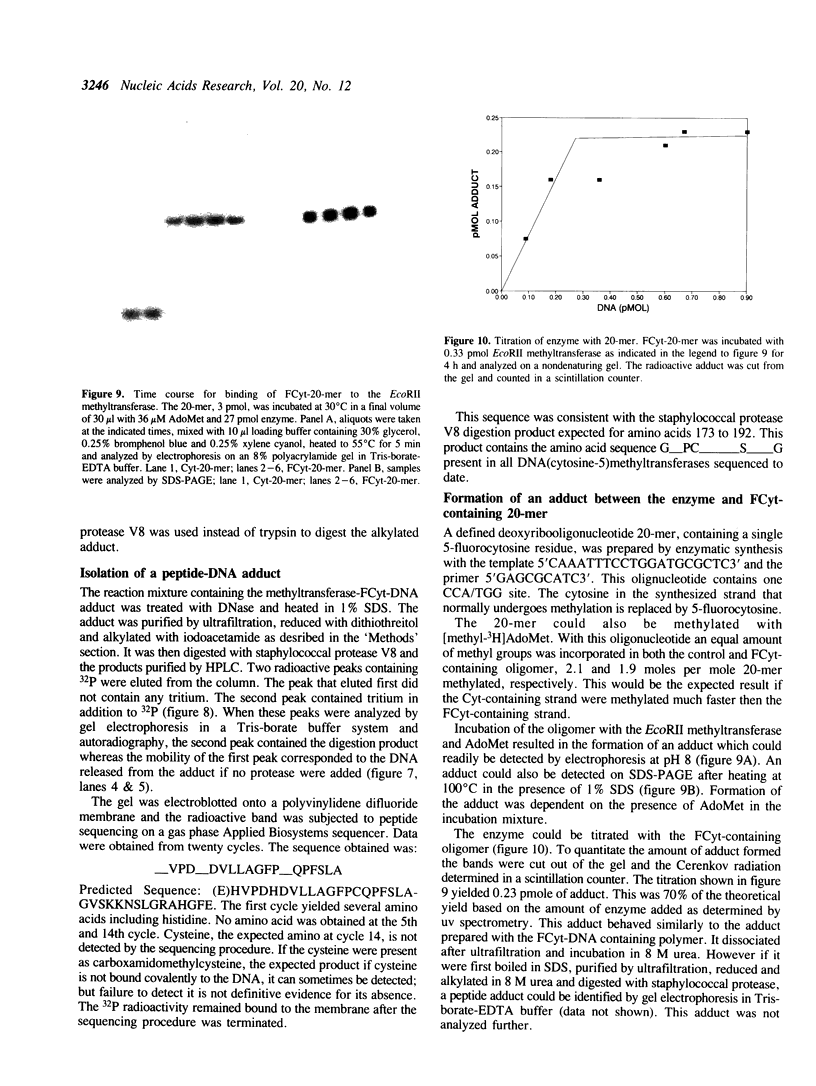
The properties of the interaction of 5-fluorocytosine-containing DNA with the EcoRII methyltransferase were studied. The DNA used was either a polymer synthesized in vitro, or a 20-mer containing one CCA/TGG sequence. The DNA could be methylated by the enzyme. In the process the enzyme formed a tight binding adduct with the DNA that could be identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Enzyme activity was inhibited by this interaction. The 20-mer could be used to titrate the active site of the enzyme. The DNA polymer formed a tight binding complex that could be identified following digestion of the DNA with pancreatic deoxyribonuclease or micrococcal nuclease. A peptide-DNA adduct could be isolated after digestion of the EcoRII-DNA adduct with staphylococcal protease V8 by high pressure liquid chromatography and polyacrylamide gel electrophoresis. Sequencing of the peptide indicated the DNA bound to a region of the protein that is conserved in all procaryotic DNA(cytosine-5)-methyltransferases. We have previously shown that this region contains a cysteine that can be photomethylated with adenosylmethionine. This region, in addition to forming part of, or being adjacent to, the AdoMet binding site, also forms part of the DNA binding site.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bellisario R. L., Maley G. F., Galivan J. H., Maley F. Amino acid sequence at the FdUMP binding site of thymidylate synthetase. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1848–1852. doi: 10.1073/pnas.73.6.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard J., Momparler R. L. Incorporation of 5-Aza-2'-deoxycytidine-5'-triphosphate into DNA. Interactions with mammalian DNA polymerase alpha and DNA methylase. Mol Pharmacol. 1983 Jul;24(1):109–114. [PubMed] [Google Scholar]
- Friedman S. Binding of the EcoRII methylase to azacytosine-containing DNA. Nucleic Acids Res. 1986 Jun 11;14(11):4543–4556. doi: 10.1093/nar/14.11.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S. The inhibition of DNA(cytosine-5)methylases by 5-azacytidine. The effect of azacytosine-containing DNA. Mol Pharmacol. 1981 Mar;19(2):314–320. [PubMed] [Google Scholar]
- Friedman S. The irreversible binding of azacytosine-containing DNA fragments to bacterial DNA(cytosine-5)methyltransferases. J Biol Chem. 1985 May 10;260(9):5698–5705. [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980 May;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Kunitani M. G., Santi D. V. On the mechanism of 2'-deoxyuridylate hydroxymethylase. Biochemistry. 1980 Apr 1;19(7):1271–1275. doi: 10.1021/bi00548a001. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matthews D. A., Appelt K., Oatley S. J., Xuong N. H. Crystal structure of Escherichia coli thymidylate synthase containing bound 5-fluoro-2'-deoxyuridylate and 10-propargyl-5,8-dideazafolate. J Mol Biol. 1990 Aug 20;214(4):923–936. doi: 10.1016/0022-2836(90)90346-N. [DOI] [PubMed] [Google Scholar]
- Negishi K., Tamanoi K., Ishii M., Kawakami M., Yamashita Y., Hayatsu H. Mutagenic nucleoside analog N4-aminocytidine: metabolism, incorporation into DNA, and mutagenesis in Escherichia coli. J Bacteriol. 1988 Nov;170(11):5257–5262. doi: 10.1128/jb.170.11.5257-5262.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterman D. G., DePillis G. D., Wu J. C., Matsuda A., Santi D. V. 5-Fluorocytosine in DNA is a mechanism-based inhibitor of HhaI methylase. Biochemistry. 1988 Jul 12;27(14):5204–5210. doi: 10.1021/bi00414a039. [DOI] [PubMed] [Google Scholar]
- Rubin R. A., Modrich P. EcoRI methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1977 Oct 25;252(20):7265–7272. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi D. V., Garrett C. E., Barr P. J. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983 May;33(1):9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- Santi D. V., Hardy L. W. Catalytic mechanism and inhibition of tRNA (uracil-5-)methyltransferase: evidence for covalent catalysis. Biochemistry. 1987 Dec 29;26(26):8599–8606. doi: 10.1021/bi00400a016. [DOI] [PubMed] [Google Scholar]
- Santi D. V., Norment A., Garrett C. E. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som S., Bhagwat A. S., Friedman S. Nucleotide sequence and expression of the gene encoding the EcoRII modification enzyme. Nucleic Acids Res. 1987 Jan 12;15(1):313–332. doi: 10.1093/nar/15.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som S., Friedman S. Direct photolabeling of the EcoRII methyltransferase with S-adenosyl-L-methionine. J Biol Chem. 1990 Mar 15;265(8):4278–4283. [PubMed] [Google Scholar]
- Som S., Friedman S. Identification of a highly conserved domain in the EcoRII methyltransferase which can be photolabeled with S-adenosyl-L-[methyl-3H]methionine. Evidence for UV-induced transmethylation of cysteine 186. J Biol Chem. 1991 Feb 15;266(5):2937–2945. [PubMed] [Google Scholar]
- Subramaniam R., Wang Y., Mathews C. K., Santi D. V. On the inhibition of deoxycytidylate hydroxymethylase by 5-fluoro-2'-deoxycytidine 5'-monophosphate. Arch Biochem Biophys. 1989 Nov 15;275(1):11–15. doi: 10.1016/0003-9861(89)90343-3. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Yoshida S., Saneyoshi M., Yamaguchi T. Utilization of 5-fluoro-2'-deoxyuridine triphosphate and 5-fluoro-2'-deoxycytidine triphosphate in DNA synthesis by DNA polymerases alpha and beta from calf thymus. Cancer Res. 1981 Oct;41(10):4132–4135. [PubMed] [Google Scholar]
- Wells R. D., Jacob T. M., Narang S. A., Khorana H. G. Studies on polynucleotides. LXIX. Synthetic deoxyribopolynucleotides as templates for the DNA polymerase of Escherichia coli: DNA-like polymers containing repeating trinucleotide sequences. J Mol Biol. 1967 Jul 28;27(2):237–263. doi: 10.1016/0022-2836(67)90018-6. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]
- Wyszynski M. W., Gabbara S., Bhagwat A. S. Substitutions of a cysteine conserved among DNA cytosine methylases result in a variety of phenotypes. Nucleic Acids Res. 1992 Jan 25;20(2):319–326. doi: 10.1093/nar/20.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]