Abstract
It is well known that scrapie is a fatal, neurodegenerative disease in sheep and goat, which belongs to the group of transmissible spongiform encephalopathies (TSEs) or prion diseases. It has been confirmed that the polymorphisms of prion protein gene (PRNP) at codons 136, 154, and 171 have strong relationship with scrapie in sheep. In the present study, nine polymorphisms of PRNP at codons 136, 154, and 171 and other six loci (at codons 101, 112, 127, 137, 138, and 152) were detected in 180 Chinese Hu sheep. All the alleles at codons 136, 154, and 171 have been identified and resulted in three new genotypes. The frequencies of predominant alleles were 85% (A136), 99.40% (R154), and 37.78% (Q171), respectively. The predominant haplotype ARQ has a relatively high frequency of 57.77%. The frequencies of dominant genotypes of ARR/ARQ and ARQ/ARQ were 30 and 26.67%, respectively. Three new found genotypes named ARQ/TRK, ARQ/TRR, and TRR/TRQ had the same lower frequencies (0.56%). The relationship of PRNP genotype with scrapie risk and litter size showed that the predominant genotypes are corresponded to the risk score of R1 (1.67%), R2 (32.22%), and R3 (42.22%). Just at the first parity, the individuals with ARH/ARH genotype had significantly larger litter size than the mean value and those with ARQ/ARQ and ARR/ARQ genotypes. In short, this study provided preliminary information about alleles and genotypes of PRNP in Chinese Hu sheep. It could be concluded that Hu sheep has a low susceptibility to natural scrapie, and the predominant PRNP genotype at least has no significant effect on litter size.
Keywords: Hu sheep, PRNP gene, Polymorphisms, Litter size, Scrapie susceptibility
Introduction
Scrapie is a fatal neurodegenerative disease occurring in sheep and goat belonging to the group of transmissible spongiform encephalopathies (TSEs) or prion diseases [37]. The common feature of all TSEs is the aberrant metabolism of the prion protein (PrP). It has been reported that the polymorphisms of sheep prion protein gene (PRNP) at codons 136A/V/T, 154 R/H, and 171Q/R/H/K were strongly associated with susceptibility or resistance to classical scrapie in sheep [15, 21]. The VRQ/VRQ genotype was regarded as the most susceptible to scrapie [32, 14, 4], while ARR/ARR genotype was associated with the most resistant to scrapie [14, 3, 18]. In addition, some studies have reported that other SNPs in PRNP gene also affected incubation period and clinical symptom of scrapie, such as codons 102, 112, 137, and so on [5, 41].
Increasing ARR/ARR genotype frequency is considered as an effective way to eradicate scrapie in sheep breeding and selection program, but the efficiency depends on several factors such as population size, ARR allele frequency, and their relationship with economic performance [13]. Litter size is one of the key economic traits represented the productivity of commercial ewe flocks. It has reported that there was no significant relationship between PRNP genotypes and litter size in sheep except for a few breeds [7, 9, 12, 31]. Breeding selection program was carried out and focused on resistance PRNP genotypes in many European states such as Great Britain and Netherlands. However, how the PRNP genotypes affect scrapie susceptibility and litter size still remains unknown in many sheep breeds, especially for prolific breed of Chinese Hu sheep.
In China, no scrapie case has been reported since the worldwide incidence. So there are a few studies about the PRNP genotypes and risk evaluation in Chinese sheep breeds. Recently, sheep scrapie or BSE have been detected in many states around China, which is a great risk to Chinese animal husbandry. Up to now, little information about PRNP polymorphisms had been reported in three Chinese Mongolia native breeds (Ujumqin, Sunite, and Mongolian sheep), small Tailed Han, Wadi, Tong sheep, and even Hu sheep. It had been demonstrated that these breeds have low diversity and medium-high and/or high level susceptibility to scrapie. However, the sample sizes in those studies were less than 50 [20, 33, 39, 41]. Neither the relationship between PRNP genotypes and reproduction traits nor detailed scrapie evaluation is studied in Chinese sheep breeds.
The objectives of the present study are to identify the PRNP polymorphisms and evaluate their effects on risk of classical scrapie and litter size in Chinese Hu sheep. The aim is to provide informative protocol for breeding and selection programs in this breed.
Materials and methods
Animals and DNA extraction
Hu sheep is one of the famous Chinese native prolific breeds in the world because of its high fertility up to about 300% [19]. Now this breed has been widely cultivated throughout the South of China such as Jiangsu province, Zhejiang province, and Shanghai city, where they are well adapted to the hot humid climate. Hu sheep mostly carried the prolific FecB gene and so was widely selected for female parent in crossbreed [19].
A total of 180 female Hu sheep (3–5 years old) were collected from Jiangsu Province (100) and Shanghai city (80). The number of live lambs at the first and second, the third and even fourth were recorded. Genomic DNA was extracted from blood using a standard phenol–chloroform extraction method, and then stored at −20°C until used.
Polymorphisms analysis
Polymorphisms of PRNP at codons 136, 154, and 171 were genotyped by polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) method using the restriction enzymes BspH1 and BspD1 (New England Biolabs) according to the method described by Luhken et al. [27]. Two different length PCR fragments (197 bp/196 bp) were obtained and then digested by BspH1 and/or BspD1 according to the manufacture. PCR-RFLP products were separated by 8% polyacrylamide gel and then stained by silver.
A 557 bp PCR fragment of PRNP including the entire nine polymorphisms at codons 101, 112, 127, 136, 137, 138, 152, 154, and 171 was amplified using the following primers (forward: 5′-ACCTGGCGGAGGATGGAACACTG-3′ and reverse: 5′-GCTCCACCACTCGCTCCATTATCTT-3′). PCR reactions were performed in a final 20 μl volume containing 50 ng genomic DNA, 2.0 μl 10× buffer, 1.2 mM MgCL2, 0.2 mM each of dNTPs, 1.0 U of Taq polymerase, 20 pM each primer. Amplification conditions: 94°C denaturation for 5 min; 30 cycles at 94°C for 40 s, annealing at 60°C for 30 s, and extension at 72°C for 45 s followed by a final extension at 72°C for 5 min. The PCR products were cloned into the pGEM-T vector and sequenced directly. The DNA sequencing was performed by Shanghai Sangon Biotechnology Co. Ltd.
Statistical analysis
The sequencing results were analyzed using the Chromas Lite (2.0) software. Allelic and genotypic frequencies were calculated by the simple gene counting method. The average litter size in different genotypes were presented as mean ± SE and compared by T-test.
Risk evaluation for scrapie
According to an assessment of clinical scrapie risk scores (R) to the PRNP genotype classes described by the Scrapie Information Group in England [11], the common 15 PRNP genotypes were divided into five scores (R1–R5), with the risk of susceptibility to scrapie increasing with an increase in the score. Each risk score is associated with certain peculiar PRNP genotypes.
Results
Allele and genotype frequency
The results showed that all the alleles at codons A136V/T, R154H, and Q171R/H/K previously detected in other sheep breeds were identified in Hu sheep in the present study. The allele frequencies are summarized in Table 1. The predominant alleles were A, R, and Q, respectively. The frequencies of PRNP polymorphisms at codons 101, 112, 127, 137, 138, and 152 are summarized in Table 2. Nine haplotypes at codons 136, 154, and 171 were detected in this study and were summarized in Table 3. Two new haplotypes of TRR and TRK were detected. The frequencies of different haplotypes varied a lot. The haplotypes ARQ had the highest frequency (57.77%) followed with ARR (18.33%), ARH (9.72%), and TRQ (6.67%).
Table 1.
The frequencies of PRNP alleles at codons 136, 154, and 171 in Hu sheep
| Condon | PrP Allele | Number | Frequency (%) |
|---|---|---|---|
| 136 | A | 331 | 91.94 |
| V | 1 | 0.28 | |
| T | 28 | 7.78 | |
| 154 | R | 358 | 99.44 |
| H | 2 | 0.56 | |
| 171 | Q | 232 | 64.45 |
| R | 71 | 19.72 | |
| H | 35 | 9.72 | |
| K | 22 | 6.11 |
Table 2.
Allelic frequencies of additional PRNP polymorphisms in Hu sheep
| Codon | PrP allele | Number | Frequency (%) |
|---|---|---|---|
| 101 | 177 | 98.33 | |
| QR | 3 | 1.67 | |
| 112 | MM | 179 | 99.44 |
| MT | 1 | 0.56 | |
| 127 | GG | 156 | 86.67 |
| GS | 24 | 13.33 | |
| 137 | MM | 171 | 95 |
| MT | 9 | 5 | |
| 138 | SS | 179 | 99.44 |
| SR | 1 | 0.56 | |
| 152 | YY | 177 | 98.33 |
| YF | 3 | 1.67 |
Table 3.
Haplotypic frequencies of PNPR at codons 136, 154, and 171
| Genotype | Number | Frequency (%) |
|---|---|---|
| ARQ | 208 | 57.77 |
| ARR | 66 | 18.33 |
| ARH | 35 | 9.72 |
| TRQ | 24 | 6.67 |
| ARK | 21 | 5.83 |
| AHQ | 2 | 0.56 |
| TRR | 2 | 0.56 |
| TRK | 1 | 0.28 |
| VRQ | 1 | 0.28 |
In this study, a total of 19 PRNP genotypes were detected (Table 4). And the corresponding DNA sequences were submitted to the GenBank database with Access Numbers from HM639748 to HM639766. The most frequent genotype at codons 136, 154, and 171 was ARR/ARQ (30%), followed by ARQ/ARQ (26.67%) and ARQ/ARH (12.22%). Three new genotypes of ARQ/TRK (HM639758), ARQ/TRR (HM639760), and TRR/TRQ (HM639765) were identified with the same low frequency of 0.56%.
Table 4.
PRNP genotypes and frequencies in Hu sheep
| Genotype | Number | Frequency (%) |
|---|---|---|
| ARR/ARQ | 54 | 30.00 |
| ARQ/ARQ | 48 | 26.67 |
| ARH/ARQ | 22 | 12.22 |
| ARQ/TRQ | 17 | 9.44 |
| ARQ/ARK | 14 | 7.78 |
| ARH/ARH | 4 | 2.22 |
| ARH/ARR | 4 | 2.22 |
| ARR/ARR | 3 | 1.67 |
| ARK/ARK | 2 | 1.11 |
| ARQ/AHQ | 2 | 1.11 |
| TRQ/TRQ | 2 | 1.11 |
| ARH/ARK | 1 | 0.56 |
| ARK/TRQ | 1 | 0.56 |
| ARQ/TRK | 1 | 0.56 |
| ARQ/TRR | 1 | 0.56 |
| ARR/ARK | 1 | 0.56 |
| ARR/TRQ | 1 | 0.56 |
| TRR/TRQ | 1 | 0.56 |
| VRQ/ARQ | 1 | 0.56 |
| Total | 180 | 100 |
Risk level estimation
Chinese Hu sheep had the high frequency of predominant genotypes of ARR/ARQ (30.00%) and ARQ/ARQ (26.67%) which corresponded to risk scores R2 and risk R3, respectively. The rare genotype ARR/ARR (1.67%) corresponded to risk score R1. Eight of nineteen PRNP genotypes in Hu sheep corresponded to risk score R1 (1.67%), R2 (32.22%), R3 (42.22%), and R5 (0.56%), and other 11 genotypes (23.36%) could not be classified to date, because their susceptibility to scrapie has not yet been established. The risk scores and genotype frequencies were shown in Fig. 1.
Fig. 1.
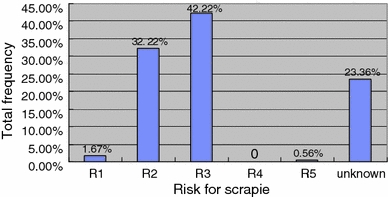
Risk level for scrapie and frequencies within Hu sheep. Associated scrapie risk for R1–5: R1 very low, R2 low, R3 moderate, especially in ARQ/ARQ, R4 moderate, R5 high, especially in ARQ/VRQ and VRQ/VRQ
Association between polymorphisms at codons 136, 154, and 171 and litter size
The comparison of the mean litter size in parities 1 and 2 and average among different PRNP genotype flocks is showed in Table 5. The individuals without informative protocol and a genotype with individual numbers less than three were excluded from the association analysis. The results showed that the litter size in ARH/ARH genotype was significantly higher than those in ARQ/ARQ and ARR/ARQ genotypes in the 1st parity. There was no significant difference in the second parity and the average live lambs among different genotypes.
Table 5.
Comparison of litter size among flocks with different PRNP genotypes
| Genotype | Number | Mean litter size | ||
|---|---|---|---|---|
| 1st parity | 2nd parity | Average | ||
| ARR/ARQ | 33 | 1.67 ± 0.14b | 2.34 ± 0.14 | 2.17 ± 0.04 |
| ARQ/ARQ | 32 | 1.59 ± 0.11Ab | 2.00 ± 0.12 | 2.05 ± 0.07 |
| ARH/ARQ | 16 | 1.83 ± 0.12 | 2.22 ± 0.21 | 2.19 ± 0.10 |
| ARQ/TRQ | 9 | 1.78 ± 0.22 | 2.22 ± 0.22 | 2.10 ± 0.13 |
| ARQ/ARK | 5 | 1.80 ± 0.37 | 1.80 ± 0.20 | 2.07 ± 0.18 |
| ARH/ARH | 3 | 2.67 ± 0.33Ba | 2.33 ± 0.33 | 2.50 ± 0.22 |
| ARR/ARR | 3 | 1.67 ± 0.67 | 2.23 ± 0.33 | 2.22 ± 0.28 |
| Total | 101 | 1.72 ± 0.07b | 2.17 ± 0.07 | 2.17 ± 0.04 |
Note: The number of a group with one genotype lower than 3 was not analyzed. Values in the same columns with small letters a and b means P < 0.05, with capital letters A and B mean P < 0.01
Discussion
It has been reported that PRNP polymorphisms at codons A136V/T, R154H, and Q171H/R/K have a strong correlation with scrapie susceptibility/resistance in sheep [15]. The allele V136 was associated with the susceptibility to scrapie, while A136 was associated with the resistance to scrapie (but not absolutely) [16, 23]. The rare allele T136 was previously only reported in Greece sheep, but its effect on sheep susceptibility has not been defined yet [5]. In this study, the allele A at codon 136 had the absolutely high frequency in Hu sheep (91.94%). This result was consistent with the previous reports in other Chinese sheep breeds [39].
The second allele (R154) has been reported due to its high association with delayed onset of symptoms for classical scrapie strains [21]. The effect of H154 on scrapie remains controversial, because it presented protection against scrapie in some breeds [21], but it was susceptible in other breeds [1, 2] and even presented a positive risk to Nor98 scrapie [10]. The allele R was absolutely predominant at codon 154 (99.44%) in Hu sheep and only two heterozygotes with H154 were detected. It seems that Hu sheep have a low risk to scrapie according to this allele distribution, which seems to delay the progression of scrapie [34].
The third allele, R171 presented resistant to scrapie and was detected mostly with heterozygotic genotype [18, 22]. The homozygote QQ171 was associated with the susceptibility to scrapie, but in some breeds it was not related with scrapie cases [3, 5, 36]. Previous studies showed that RR171 or QR171 conferred resistance when Suffolk sheep were at risk of exposure to the scrapie agent. In Hu sheep, the frequencies of allele Q171 and the predominant genotype QQ were 64.45 and 37.78%, respectively. Other two alleles named H and K at this code previously reported by Goldmann [15] were also detected in Hu sheep. Furthermore, the codon K171 in Hu sheep had a higher frequency than other Chinese sheep breeds as suggested in previous reports [25, 39].
Fifteen common PRNP genotypes identified had been divided into five risk levels, R1–5 [11]. In National Scrapie Plan (NSP), the rams with levels of R1, 2, and 3 are selected, with which levels of R4 and 5 are culled. Among 19 genotypes in Hu sheep detected in this study, two and four genotypes belonged to risk level R2 (32.22%) and R3 (42.22%), respectively. In addition, ARR/ARR and VRQ/ARQ genotypes belonged to risk level R1 (1.67%) and R5 (0.56%), respectively. And other 11 genotypes (total 23.36%) were unknown because they carried the uncertain alleles for scrapie risk. The genotypes of VRQ/VRQ and ARR/ARR were, respectively, regarded as most susceptible [4] and most resistant to scrapie [3, 14, 18], while the latter had low frequency (1.67%) and no VRQ/VRQ was detected in Hu sheep. The ARR/ARQ genotype with the highest frequency belonged to risk level R2. The wild genotype ARQ/ARQ with the second higher frequency in Hu sheep, which was regarded as middle susceptibility to scrapie [4, 5], and belonged to risk level R3. The distribution of PRNP genotypes in Hu sheep was consistent with many other sheep breeds [24]. The ARQ/TRQ genotype had high frequency (9.44%) in Hu sheep, which was considered as susceptible genotype similar to ARQ/ARQ and ARQ/AHQ [5]. The study on the scrapie risks estimation of ARK-encoding genotype is still in progress to be explored. Although the frequency of ARR/ARR genotype in Hu sheep is low, this result suggests that Hu sheep has a low or middle risk level to scrapie in a certain extent,
In addition, other six loci in PRNP including Q/R101, M/T112, G/S127, M/T137, S/R138, and Y/F152 were detected (Table 3). These polymorphisms were called as rare alleles and affected susceptibility/resistance to scrapie in sheep. It is remarkable that all the variants detected are heterozygotes in Hu sheep. T112 was found in both scrapie-free and scrapie-affected flocks with the lower frequency [17]. T137 has been reported to offer protective effects on sheep which were experimentally challenged with both scrapie and BSE [17, 29, 35]. The frequency of these two mutations is low in Hu sheep. And the detailed effects of these rare alleles on susceptibility/resistance to scrapie are required further study.
To date, studies on the association of PRNP polymorphisms with litter size formed different conclusions [6–9, 12, 26, 28, 31]. Most studies revealed that no significant associations existed [9, 12, 31, 38]. In contrast, it was reported that a small positive effect of ARR/ARR genotype on litter size in Texel [6]. Likewise, the ewes with ARR/ARR genotype had a higher litter size at birth than those with ARR/ARQ and ARQ/ARQ genotypes [28]. The allele ARH conferred a significantly larger litter size than that of with ARQ or ARR allele in an autochthonous Spain breed [8]. In this study, the litter size at first parity for ARH/ARH genotype was significantly higher than those with ARQ/ARQ and ARR/ARQ genotypes. Other reproduction performances had no significant difference among different genotype flocks.
Selection of individuals with the resistant genotype ARR/ARR is an effective way to eradicate scrapie in sheep. However, breeding programs could encounter three main problems in procedure. Firstly, a large number of breeds have a low frequency of the ARR allele. Secondly, selection for the PRNP genotype will yield selection pressure on other economical traits and thus may reduce the whole genetic improvement. Thirdly, there may be a negative association between the ARR allele and other animal performance traits [30]. Although the frequency of ARR/ARR in Hu sheep is low, the second highest frequency of ARR/ARQ genotype could be helpful for increasing the number of ARR/ARR genotype and reducing the risk of genetic defects within population [40].
Although there have been no scrapie and BSE reports in China, it is still necessary to select individuals with resistant genotypes for securing animal breeding program and food safety in future. However, it is difficult to select a higher resistance flock with low ARR/ARR frequency, which would influence the selection of resistant sheep breeds. Fortunately, the results show that at least there is no significantly negative effect between the resistant PRNP genotypes and litter size in Hu sheep.
Conclusions
Polymorphisms at codons 136, 154, 171, and other six loci in sheep PRNP were detected in Chinese Hu sheep, the risk level for scrapie was evaluated, too. Moreover, the association of different genotypes on litter size were analyzed. The results showed that all the alleles at codons 136A/V/T, 154 R/H, and 171Q/R/H/K were detected. Nine haplotypes and nineteen genotypes were detected at codons 136, 154, and 171. Most of Hu sheep individuals belonged to the risk levels of R2 (32.22%) and R3 (42.22%). Just the litter size at the first parity was significantly affected by the variance genotypes of ARH/ARH, ARQ/ARQ, and ARR/ARQ. It could be concluded that Hu sheep has a lower susceptibility to natural scrapie, and the predominant PRNP genotype has no significant effects on litter size.
Acknowledgments
We appreciated the farmers for samples collection. This work was financially supported by grants from the National Technological System of wool and cashmere Industry (No. nycytx-40-06), the National Transgenic fine wool sheep Breeding Program (No. 2009ZX08008-001B), and the National Natural Science Foundation of China (No. C120103).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Feng Guan and Lei Pan contributed equally to this work.
The GenBank access numbers of nineteen different Hu PRNP genotypes DNA sequences were HM639748 to HM639766.
Contributor Information
Feng Guan, Email: jlguanfeng@yahoo.com.cn.
Guoqing Shi, Email: nkkxyxms@163.com.
References
- 1.Acutis PL, Bossers A, Priem J, Riina MV, Peletto S, Mazza M, Casalone C, Forloni G, Ru G, Caramelli M. J. Gen. Virol. 2006;87(4):1029–1033. doi: 10.1099/vir.0.81440-0. [DOI] [PubMed] [Google Scholar]
- 2.Acutis PL, Sbaiz L, Verburg F, Riina MV, Ru G, Moda G, Caramelli M, Bossers A. J. Gen. Virol. 2004;85:3165–3172. doi: 10.1099/vir.0.80053-0. [DOI] [PubMed] [Google Scholar]
- 3.Agrimi U, Conte M, Morelli L, Di Bari MA, Di Guardo G, Ligios C, Antonucci G, Aufiero GM, Pozzato N, Mutinelli F, Nonno R, Vaccari G. Vet. Res. Commun. 2003;27(Suppl 1):31–38. doi: 10.1023/B:VERC.0000014115.18327.26. [DOI] [PubMed] [Google Scholar]
- 4.Baylis M, Chihota C, Stevenson E, Goldmann W, Smith A, Sivam K, Tongue S, Gravenor MB. J. Gen. Virol. 2004;85:2735–2740. doi: 10.1099/vir.0.79876-0. [DOI] [PubMed] [Google Scholar]
- 5.Billinis C, Psychas V, Leontides L, Spyrou V, Argyroudis S, Vlemmas I, Leontides S, Sklaviadis T, Papadopoulos O. J. Gen. Virol. 2004;85:547–554. doi: 10.1099/vir.0.19520-0. [DOI] [PubMed] [Google Scholar]
- 6.Brandsma JH, Janss LLG, Visscher AH. Livest. Prod. Sci. 2003;85:59–64. doi: 10.1016/S0301-6226(03)00116-7. [DOI] [Google Scholar]
- 7.Brandsma JH, Janss LLG, Visscher AH. Livest. Prod. Sci. 2005;95(1–2):89–94. doi: 10.1016/j.livprodsci.2004.12.011. [DOI] [Google Scholar]
- 8.Casellas J, Caja G, Bach R, Francino O, Piedrafita J. J. Anim. Sci. 2007;85(3):592–597. doi: 10.2527/jas.2006-308. [DOI] [PubMed] [Google Scholar]
- 9.Chase-Topping ME, Kruuk LE, Lajous D, Touzeau S, Matthews L, Simm G, Foster JD, Rupp R, Eychenne F, Hunter N, Elsen JM, Woolhouse ME. J. Gen. Virol. 2005;86:1229–1238. doi: 10.1099/vir.0.80277-0. [DOI] [PubMed] [Google Scholar]
- 10.Colussi S, Vaccari G, Maurella C, Bona C, Lorenzetti R, Troiano P, Casalinuovo F, Di Sarno A, Maniaci MG, Zuccon F, Nonno R, Casalone C, Mazza M, Ru G, Caramelli M, Agrimi U, Acutis PL. J. Gen. Virol. 2008;89:3173–3176. doi: 10.1099/vir.0.2008/004150-0. [DOI] [PubMed] [Google Scholar]
- 11.Dawson M, Moore RC, Bishop SC. Vet. Res. 2008;39(4):25. doi: 10.1051/vetres:2007064. [DOI] [PubMed] [Google Scholar]
- 12.De Vries F, Hamann H, Drögemüller C, Ganter M, Distl O. J. Dairy Sci. 2005;88(1):392–398. doi: 10.3168/jds.S0022-0302(05)72699-0. [DOI] [PubMed] [Google Scholar]
- 13.Drogemuller C, Leeb T, Distl O. Vet. Rec. 2001;149(12):349–352. doi: 10.1136/vr.149.12.349. [DOI] [PubMed] [Google Scholar]
- 14.Elsen JM, Amigues Y, Schelcher F, Ducrocq V, Andreoletti O, Eychenne F, Khang JV, Poivey JP, Lantier F, Laplanche JL. Arch. Virol. 1999;144(3):431–445. doi: 10.1007/s007050050516. [DOI] [PubMed] [Google Scholar]
- 15.Goldmann W. Vet. Res. 2008;39(3):1–14. doi: 10.1051/vetres:2008010. [DOI] [PubMed] [Google Scholar]
- 16.Goldmann W, Hunter N, Benson G, Foster JD, Hope J. J. Gen. Virol. 1991;72(Pt 10):2411–2417. doi: 10.1099/0022-1317-72-10-2411. [DOI] [PubMed] [Google Scholar]
- 17.Goldmann W, Baylis M, Chihota C, Stevenson E, Hunter N. J. Appl. Microbiol. 2005;98(6):1294–1302. doi: 10.1111/j.1365-2672.2005.02568.x. [DOI] [PubMed] [Google Scholar]
- 18.Goldmann W, Hunter N, Smith G, Foster J, Hope J. J. Gen. Virol. 1994;75:989–995. doi: 10.1099/0022-1317-75-5-989. [DOI] [PubMed] [Google Scholar]
- 19.Guan F, Liu SR, Shi GQ, Yang LG. Anim. Reprod. Sci. 2007;99(1–2):44–52. doi: 10.1016/j.anireprosci.2006.04.048. [DOI] [PubMed] [Google Scholar]
- 20.C.X. Han, H.X. Liu, Y.X. Lu, M.X. Song, D.M. Zhao, X.M. Zhou, L.F. Yang, X.Y. Li, Virus Genes 42(1), 153–155 (2011) [DOI] [PubMed]
- 21.Hunter N. Trends Microbiol. 1997;5(8):331–334. doi: 10.1016/S0966-842X(97)01081-0. [DOI] [PubMed] [Google Scholar]
- 22.Ibeagha-Awemu EM, Kgwatalala P, Ibeagha AE, Zhao X. Mamm. Genome. 2008;19(4):226–245. doi: 10.1007/s00335-008-9101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeda T, Horiuchi M, Ishiguro N, Muramatsu Y, Kai-Uwe GD, Shinagawa M. J. Gen. Virol. 1995;76:2577–2581. doi: 10.1099/0022-1317-76-10-2577. [DOI] [PubMed] [Google Scholar]
- 24.Kutzer T, Pfeiffer I, Brenig B. J. Anim. Breed. Genet. 2002;119(4):201–208. doi: 10.1046/j.1439-0388.2002.00335.x. [DOI] [Google Scholar]
- 25.Lan Z, Wang ZL, Liu Y, Zhang X. Arch. Virol. 2006;151:2095–2101. doi: 10.1007/s00705-006-0758-3. [DOI] [PubMed] [Google Scholar]
- 26.Lipsky S, Brandt H, Lühken G, Erhardt G. Anim. Reprod. Sci. 2008;103(1–2):69–77. doi: 10.1016/j.anireprosci.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Luhken G, Buschmann A, Groschup MH, Erhardt G. Arch. Virol. 2004;149(8):1571–1580. doi: 10.1007/s00705-004-0303-1. [DOI] [PubMed] [Google Scholar]
- 28.Man WY, Nicholls N, Woolhouse ME, Lewis RM, Villanueva B. Prev. Vet. Med. 2009;91(2–4):161–171. doi: 10.1016/j.prevetmed.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 29.Saunders GC, Cawthraw S, Mountjoy SJ, Hope J, Windl O. J. Gen. Virol. 2006;87:3141–3149. doi: 10.1099/vir.0.81779-0. [DOI] [PubMed] [Google Scholar]
- 30.Sweeney T, Hanrahan JP. Vet. Res. 2008;39(4):28. doi: 10.1051/vetres:2008004. [DOI] [PubMed] [Google Scholar]
- 31.Sweeney T, Hanrahan JP, O’Doherty E. Anim. Reprod. Sci. 2007;101(1–2):153–157. doi: 10.1016/j.anireprosci.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Thorgeirsdottir S, Sigurdarson S, Thorisson HM, Georgsson G, Palsdottir A. J. Gen. Virol. 1999;80:2527–2534. doi: 10.1099/0022-1317-80-9-2527. [DOI] [PubMed] [Google Scholar]
- 33.Tsunoda K, Namikawa T, Sato K, Hasnath MA, Nyunt MM, Rajbandary HB, Loc CB, Zanchiv Ts, Chang H, Sun W, Dorji T. Biochem. Genet. 2010;48(1–2):13–25. doi: 10.1007/s10528-009-9287-6. [DOI] [PubMed] [Google Scholar]
- 34.Vaccari G, Di Bari MA, Morelli L, Nonno R, Chiappini B, Antonucci G, Marcon S, Esposito E, Fazzi P, Palazzini N, Troiano P, Petrella A, Di Guardo G, Agrimi U. J. Gen. Virol. 2006;87:1395–1402. doi: 10.1099/vir.0.81485-0. [DOI] [PubMed] [Google Scholar]
- 35.Vaccari G, D’Agostino C, Nonno R, Rosone F, Conte M, Di Bari MA, Chiappini B, Esposito E, De Grossi L, Giordani F, Marcon S, Morelli L, Borroni R, Agrimi U. J. Virol. 2007;81(13):7306–7309. doi: 10.1128/JVI.02880-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaccari G, Petraroli R, Agrimi U, Eleni C, Perfetti MG, Di Bari MA, Morelli L, Ligios C, Busani L, Nonno R, Di Guardo G. Arch. Virol. 2001;146:2029–2037. doi: 10.1007/s007050170050. [DOI] [PubMed] [Google Scholar]
- 37.Vaccari G, Panagiotidis CH, Acin C, Peletto S, Barillet F, Acutis P, Bossers A, Langeveld J, van Keulen L, Sklaviadis T, Badiola JJ, Andreéoletti O, Groschup MH, Agrimi U, Foster J, Goldmann W. Vet. Res. 2009;40(5):48. doi: 10.1051/vetres/2009031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitezica ZG, Moreno CR, Bodin L, François D, Barillet F, Brunel JC, Elsen JM. J. Anim. Sci. 2006;84(6):1317–1322. doi: 10.2527/2006.8461317x. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Qin Z, Qiao J, Zhao D. Virus Genes. 2008;37(1):128–130. doi: 10.1007/s11262-008-0244-y. [DOI] [PubMed] [Google Scholar]
- 40.Windig JJ, Meuleman H, Kaal L. Prev. Vet. Med. 2007;78(2):161–171. doi: 10.1016/j.prevetmed.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Li N, Fan B, Fang M, Xu W. Anim. Genet. 2004;35(6):457–461. doi: 10.1111/j.1365-2052.2004.01204.x. [DOI] [PubMed] [Google Scholar]


