Abstract
Although the concept of Reverse Vaccinology was first pioneered for sepsis and meningococcal meningitidis causing bacterium, Neisseria meningitides, no broadly effective vaccine against serogroup B meningococcal disease is yet available. In the present investigation, HLA distribution analysis was undertaken to select three most promiscuous T-cell epitopes out of ten computationally validated epitopes of Iron acquisition proteins from Neisseria MC58 by using the population coverage tool of Immune Epitope Database (IEDB). These epitopes have been determined on the basis of their binding ability with maximum number of HLA alleles along with highest population coverage rate values for all the geographical areas studied. The comparative population coverage analysis of moderately immunogenic and high immunogenic peptides suggests that the former may activate T-cell response in a fairly large proportion of people in most geographical areas, thus indicating their potential for development of epitope-based vaccine.
Keywords: Neisseria meningitidis, Population Coverage, T cell epitopes, Immune Epitope Database
Background
The availability of a large and ever increasing volume of microbial genomic information have brought forth Reverse Vaccinology as a faster and cheaper approach to design vaccines in vivo compared to traditional empirical methods. Serogroup B meningococcus represents the first example of the successful application of Reverse Vaccinology [1]. Invasive meningococcal disease is now endemic worldwide and most cases are caused by five of the thirteen meningococcal subgroups A, B, C, Y and W135 [2]. Serogroup B is the main cause of invasive meningococcal disease in most temperate countries and accounts for 32% of all meningococcal disease cases in the United States, 45- 80% of the cases in Europe and more than 50% of the cases in the rest of the world [3, 4]. Applications of new approaches for vaccine design including genome mining [5–9] and proteomics [10, 11] have identified several vaccine targets against Men B as listed in (see Table 1) [12–37]. However, the vaccine potential of nearly all of these candidates has been limited either by antigenic variables, phase variability, presence of auto antigens or low constitutive expression of the antigen by some strains [38]. Hence, the development of a universally safe and effective serogroup B Neisseria meningitidis vaccine still remains a challenge for researchers. T-cell recognises a complex between a specific major histocompatibility complex (MHC) molecules and a particular pathogen derived epitope [39]. MHC molecules are highly polymorphic. Till date, in human more than 225 HLA Class I and 986 HLA Class II allelic sequences have been identified [40]. This polymorphism is concentrated in the region encoding the peptide-binding groove of HLA molecules and as a result, MHC molecules exhibit a widely varying binding specificity. Moreover, in the design of epitope-based vaccines, the issue of population coverage in relation to MHC polymorphism is further complicated by the fact that frequency of expression of different HLA types varies in different ethnicities [39]. Thus the main desirable characteristics for meningococcal vaccine candidate is that the selected epitopes should elicit Tcell immune response and that they should bind to several alleles of HLA super type for maximal population coverage. In the present study an effort has been made to perform population coverage analysis of total ten promiscuous T-cell epitopes (Class I and Class II) of Iron acquisition proteins (FrpB; Acc no NMB 1988 and FbpA; Acc no NMB0634) that has been computationally validated using different immunoinformatics tools to elicit immune response against the pathogen Neisseria meningitidis MC58 [41]. The resultant epitopes of the present study would be a relevant representative of a large proportion of human population.
Methodology
HLA Distribution Analysis
In order to determine the population coverage rate of the computationally validated ten promiscuous high or moderate immunogenic T- cell epitopes, the predicted putative epitopic core sequences with the corresponding HLA alleles (Class I and Class II) were submitted to the population coverage analysis tool of the Immune Epitope Database (IEDB) [39] by keeping the default parameters on (Population/Area= 78 populations grouped into 11 different geographical areas). Population coverage analysis tool calculates the fraction of individuals predicted to respond to a given set of epitopes with known MHC restrictions. For individual population coverage, the tool computes the following: (1) projected population coverage, (2) average number of epitope hits/ HLA combinations recognised by the populations and (3) minimum number of epitope hits/ HLA combinations recognised by 90% of the population (PC90). These calculations are made on the basis of HLA genotypic frequencies assuming non-linkage disequilibrium between HLA loci.
Blast Screening
In order to avoid generation of auto immune disease the final epitopic core sequences showing affinity to maximum number of HLA molecules with high population rates were searched against the human proteome using HLAPred tool [42].
Results and Discussion
Population coverage analysis plays an important role for the design of peptide based vaccine. The frequency of expression of different HLA types varies in different ethnicities as the MHC molecule is highly polymorphic [43, 44]. Extreme polymorphism restricts the proportion of the human population that may respond to a particular antigen [45, 46]. Thus a peptide which functions as T-cell epitope in a population with certain HLA make up may not be effective in another population with a different HLA allelic distribution. The aim of the study was to select promiscuous T-cell epitopes that bind to several alleles of HLA super types for maximal population coverage.
The population coverage rate of the predicted epitopes of Neisseria meningitidis Iron acquisition proteins (FrpB and FbpA) were analyzed by submitting the promiscuous epitopic core sequences (Class I and class II) with their corresponding HLA alleles to IEDB population coverage analysis tool. A total of 27 alleles were shared by all the epitopes and approximately 90 alleles bind to maximum number of peptides (see Table 4 & 5). The IEDB results reveal a strong positive correlation between HLA binding affinity and population coverage rate among the peptides. For example the peptides 148KTVDAQDLLK157 and 60ATDMRELLK68 with maximum (83.13%) and minimum (59.55%) population coverage rate were found to be correlated with highest (121 HLA alleles) and lowest (62 HLA alleles) HLA binding epitopes, respectively. However, similar type of correlation does not exist among the moderately predicted epitopes (44RATGIKVKL52 and 488HGKRGSII496). Interestingly, these epitopes, although exhibited to bind with lower number of HLA alleles in comparison to high immunogenic peptide, elicit wider population coverage rate (Figure 1, see Table 2 & 3). In order to further pin-point the most potential epitopes comparative HLA distribution analysis was preformed and the values of two moderate and five high immunogenic epitopes indicates that the former are equally potential candidates for vaccine design (Figure 2). Finally three epitopes have been conclusively selected one each from moderate immunogenic (44RATGIKVKL52) and high immunogenic (148KTVDAQDLLK157) Class I epitopes and one from Class II epitopes (629VQKAVGSILVAG643), on the basis of high population coverage rate and maximum affinity to HLA alleles. These epitopes exhibited coverage rates exceeding 99% in five ethnic groups and between 96 to 98% for six other ethnic groups (Figure 3). Maximum population coverage rate (99.79%) was observed in Europe indicating that a future vaccine based on these putative epitopes might work most efficiently for majority of population in Europe where the incidence of Meningococcal disease is highest (> 90%) 47].
Figure 1.
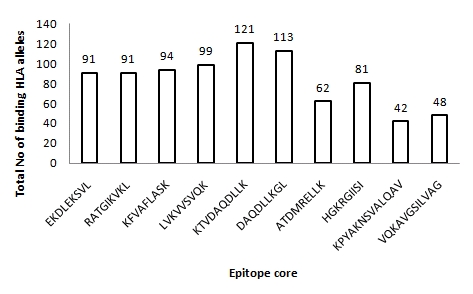
HLA binding affinity for the putative promiscuous T-cell epitopes of FrpB and FbpA proteins.
Figure 2.
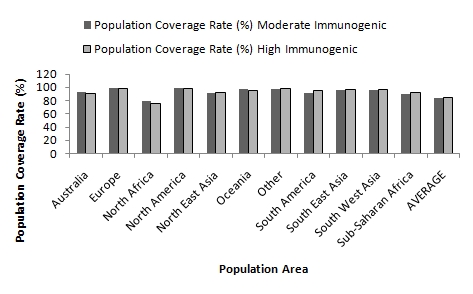
Comparative Population coverage rate analysis between Moderate (RATGIKVKL and HGKRGSII) and High (EKDLEKSVL, DAQDLLKGL, ATDMRELLK, KFVAFLASK, KTVDAQDLLK and LVKVVSVQK) immunogenic peptides.
Figure 3.
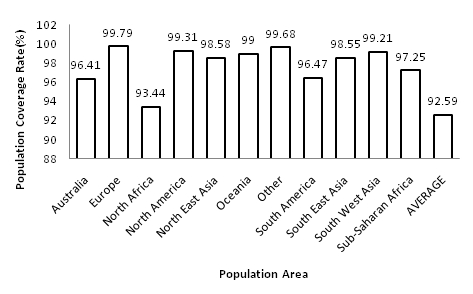
Predicted population coverage rate (%) of final selected three promiscuous T-cell epitopes viz RATGIKVKL, KTVDAQDLLK (Class I) and VQKAVGSILVAG (Class II) of FrpB and FbpA MenB proteins
Conclusion
Vaccination programs are the most versatile means of prophylactic infectious diseases control majors for vulnerable human population. Immunoinformatic epitope-based vaccine development approach using computational tools, databases and web servers is time and cost effective as compared to traditional laboratory experiments. In the present investigation three promiscuous putative epitopes viz., 148KTVDAQDLLK157, 44RATGIKVKL52 and 629VQKAVGSILVAG640 from iron acquisition proteins (FrpB and FbpA) have been computationally validated as potential MenB vaccine candidates across diverse ethnicities. This study may stimulate further in vitro investigations to ascertain the immunogenicity of these putative epitopes for designing effective vaccines against Neisseria meningitidis serogroup B.
Supplementary material
Acknowledgments
Thanks are due to Director, IMMT for providing necessary facilities to carry out the work.
Footnotes
Citation:Misra et al, Bioinformation 6(7): 255-261 (2011)
References
- 1.R Rappuoli. Curr Opin Microbiol. 2000;3:445. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 2.M Sabrangini, AJ Pollard. Lancet Infect Dis. 2010;10:112. [Google Scholar]
- 3.CE Frasch, et al. Clin Microbiol Rev. 1989;2:S134. doi: 10.1128/cmr.2.suppl.s134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.AJ Kvalsvig, DJ Unsworth. J Clin Pathol. 2003;56:417. doi: 10.1136/jcp.56.6.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.M Pizza, et al. Science. 2000;287:1816. [Google Scholar]
- 6.R Rappuoli, A Covacci. Science. 2003;302:602. doi: 10.1126/science.1092329. [DOI] [PubMed] [Google Scholar]
- 7.AS DeGroot, R Rappuoli. Expert Rev Vaccines. 2004;3:59. doi: 10.1586/14760584.3.1.59. [DOI] [PubMed] [Google Scholar]
- 8.B Capecchi, et al. Curr Issues Mol Biol. 2004;6:17. [PubMed] [Google Scholar]
- 9.R Grifantini, et al. Proc Natl Acad Sci U S A. 2003;100:9542. [Google Scholar]
- 10.G Bernardini, et al. Expert Rev Proteomics. 2009;6:135. doi: 10.1586/epr.09.3. [DOI] [PubMed] [Google Scholar]
- 11.G Bernardini, et al. Expert Rev Proteomics. 2007;4:667. doi: 10.1586/14789450.4.5.667. [DOI] [PubMed] [Google Scholar]
- 12.FA Wyle, et al. J Infect Dis. 1972;126:514. doi: 10.1093/infdis/126.5.514. [DOI] [PubMed] [Google Scholar]
- 13.J Bruge, et al. Vaccine. 2004;22:1087. [Google Scholar]
- 14.CE Frasch, et al. Methods Mol Med. 2001;66:81. doi: 10.1385/1-59259-148-5:81. [DOI] [PubMed] [Google Scholar]
- 15.GV Sierra, et al. NIPH Ann. 1991;14:195. [PubMed] [Google Scholar]
- 16.JC deMoraes, et al. Lancet. 1992;340:1074. [Google Scholar]
- 17.CP Noronha, et al. Int J Epidemiol. 1995;24:1050. doi: 10.1093/ije/24.5.1050. [DOI] [PubMed] [Google Scholar]
- 18.J Holst, et al. Vaccine. 2003;21:734. [Google Scholar]
- 19.G Bjune, et al. Lancet. 1991;338:1093. [Google Scholar]
- 20.B Feiring, et al. Clin Vaccine Immunol. 2006;13:790. doi: 10.1128/CVI.00047-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.P Oster, et al. Vaccine. 2005;23:2191. [Google Scholar]
- 22.P Oster, et al. Vaccine. 2007;25:3075. [Google Scholar]
- 23.AR Gorringe, et al. Clin Vaccine Immunol. 2009;16:1113. doi: 10.1128/CVI.00118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.J O'Hallahan, et al. Vaccine. 2005;23:2197. [Google Scholar]
- 25.J Boslego, et al. Vaccine. 1995;13:821. [Google Scholar]
- 26.ED deKleijn, et al. Vaccine. 2000;18:1456. [Google Scholar]
- 27.K Cartwright, et al. Vaccine. 1999;17:2612. [Google Scholar]
- 28.CC Peeters, et al. Vaccine. 1996;14:1009. [Google Scholar]
- 29.ED deKleijn, et al. J Infect Dis. 2001;184:98. doi: 10.1086/320993. [DOI] [PubMed] [Google Scholar]
- 30.ER van der Voort, et al. Infect Immun. 1996;64:2745. doi: 10.1128/iai.64.7.2745-2751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.S Sandbu, et al. Clin Vaccine Immunol. 2007;14:1062. doi: 10.1128/CVI.00094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D Boutriau, et al. Clin Vaccine Immunol. 2007;14:65. doi: 10.1128/CVI.00230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.B Haneberg, et al. Infect Immun. 1998;66:1334. doi: 10.1128/iai.66.4.1334-1341.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.JJ Drabick, et al. Vaccine. 1999;18:160. [Google Scholar]
- 35.RK Katial, et al. Infect Immun. 2002;70:702. doi: 10.1128/iai.70.2.702-707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.SA Halperin, et al. Vaccine. 2007;25:450. [Google Scholar]
- 37.JS Plested, et al. Clin Vaccine Immunol. 2009;16:785. doi: 10.1128/CVI.00007-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.SK Gupta, et al. Vaccine. 2010;28:7092. doi: 10.1016/j.vaccine.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 39.HH Bui, et al. BMC Bioinformatics. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.J Robinson, et al. Nucleic Acids Res. 2009;37:D1013. doi: 10.1093/nar/gkn662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.N Mishra, et al. Journal of Proteins and Proteomics. 2010;1:53. [Google Scholar]
- 42.HP Adams, JA Koziol. J Immunol Methods. 1995;185:181. doi: 10.1016/0022-1759(95)00111-m. [DOI] [PubMed] [Google Scholar]
- 43.PA Reche, EL Reinherz. J Mol Biol. 2003;331:623. doi: 10.1016/s0022-2836(03)00750-2. [DOI] [PubMed] [Google Scholar]
- 44.K Maenaka, EY Jones. Curr Opin Struct Biol. 1999;9:745. doi: 10.1016/s0959-440x(99)00039-1. [DOI] [PubMed] [Google Scholar]
- 45.V Brusic, JT August. Pharmacogenomics. 2004;5:597. [Google Scholar]
- 46.IG Ovsyannikova, et al. Pharmacogenomics. 2004;5:417. [Google Scholar]
- 47.DM Granoff. Clin Infect Dis. 2010;50:S54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


