Abstract
The emergence of new strains of Influenza virus have caused several pandemics over the last hundred years with the latest being the H1N1 Swine flu pandemic of 2009. The Hemagglutinin (HA) protein of the Influenza virus is the primary target of human immune system and is responsible for generation of protective antibodies in humans. Mutations in this protein results in change in antigenic regions (antigenic drift) which consequently leads to loss of immunity in hosts even in vaccinated population (herd immunity). This necessitates periodic changes in the Influenza vaccine composition. In this paper, we investigate the molecular basis of the reported loss of herd immunity in vaccinated population (vaccine component: Influenza A/X-31/1968 (H3N2)) which resulted in the outbreak due to strain Influenza A/Port Chalmers/1/1973 (H3N2). Also, the effects of antigenic drift in HA protein (H3N2 vaccine strains 1968-2007) on the 3D structures as well as interactions with BH151, a 1968 antibody, has been studied. Rigid body molecular docking protocol has been used to study the antigen-antibody interactions. We believe that the present study will help in better understanding of host-pathogen interactions at the molecular level.
Keywords: Influenza virus, H3N2, hemagglutinin, antibody, molecular docking, antigenic drift, host-pathogen interactions, BH151
Background
Influenza continues to be a major cause for concern worldwide due to the considerable human suffering and economic burden afflicted by the periodic epidemics. The occurrence of the repeated epidemics is due to effective evasion of herd immunity in host populations by the Influenza A viruses through antigenic variation in the surface proteins. Influenza A viruses have two surface glycoproteins– hemaglutinin (HA) and neuraminidase (NA), both of which undergo extensive antigenic variation. Thus, Influenza A viruses escape neutralization by host antibodies through accumulated mutations (antigenic drift) in these proteins or by introducing new variants of these by genetic reassortments (antigenic shift) as in case of the recent H1N1 Swine flu pandemic of 2009 [1]. The antigenic variations in HA and NA proteins necessitates new vaccine strains to be updated frequently and hence constitutes a major challenge to effective vaccine design [2]. The HA protein is a trimer of identical subunits each of which consists of two chains HA1 and HA2 with 329 and 175 amino acids respectively. Of these, HA1 has been found to mutate more frequently [3–6]. Though NA also acquires mutations, it is considered less relevant because of its relatively less prevalence on the virion surface and limited interactions with antibodies [6]. Hence, studies on antigenic variability have mostly concentrated on HA proteins [4, 5, 7, 8].
Periodic outbreaks of influenza epidemics due to H3N2 subtype strains have also mostly occurred due to antigenic drifts or shifts, which did effectively overcome the existing herd immunity in the population. However, a reassortant virus possessing an avian H3 hemaglutinin and all other genes of the previously circulating human strain were responsible for the 1968 H3N2 pandemic [9]. Further, the 2003-2004 epidemic of H3N2 was primarily due to the proliferation of the new H3N2 subtype strain A/Fujian/411/2002 (an antigenic drift mutant of the A/Panama/2007/99 strain), with the existing vaccine failing to offer any protection [2, 10]. Though evolution of H3N2 in terms of antigenic variability has been studied, to the best of our knowledge, reports about antigen-antibody interactions involving HA proteins of H3N2 viruses are sparse. In order to understand the antigenic drift and host-pathogen interactions, we investigated the antigenic variability of HA proteins in strains of influenza A/H3N2 in the light of antigen-antibody interactions using bioinformatics tools for molecular docking. The motivation for the present study originated from the reported ineffectiveness of a vaccine based on Hong Kong strain of the 1968 pandemic (equivalent of A/Aichi/2/1968 (X31)) to provide protection against the infection due to strain A/Port Chalmers/1/1973 [11].
Methodology
The dataset
The HA amino acid sequences for the following WHO recommended vaccine strains of influenza A/H3N2 virus were downloaded from the SwissProt database: A/Duck/Ukraine/1/1963 (UKR63), A/Aichi/2/1968 (X31), A/Albany/1/1970 (ALB70), A/Port Chalmers/1/1973 (PC73), A/Philippines/2/1982 (PH82), A/Sydney/5/1997 (SYD97), A/Moscow/10/1999 (MOS99), A/Wisconsin/67/2005 (WIS05), A/Brisbane/10/2007 (BR07) and A/Perth/16/2009 (PR09). The names in parenthesis will be used henceforth in the present paper. The structural information for the strains X-31 and UKR63 were retrieved from the Protein Databank (PDB IDs: 1EO8 and 1MQL respectively). The known structure of X-31 HA protein complexed with antibody BH151 (derived from a survivor of the 1968 influenza A/H3N2 pandemic) has been considered as a reference for the study.
Phylogenetic analyses
Sequence-based phylogenetic analysis was carried out using the Molecular Evolutionary Genetic Analyses (MEGA 4.0) package. Multiple sequence alignment was carried out with ClustalW implementation in MEGA4 [12] with default parameters. The phylogenetic tree was constructed using the neighborjoining algorithm and bootstrap (10,000 replications) was used as a test of phylogeny.
Antigenic variability
In silico prediction of antigenic determinants was performed for each sequence of the dataset using the Kolaskar method [13] as implemented in the B-cell epitope prediction server at Immune Epitope Database (IEDB; www.immuneepitope.org/). The known antigenic regions [13] were also compared with the predictions. The antigenic distance between any two strains (newer vs older) can be measured in terms of the fraction of amino acids that differ between them in the epitope regions. Such a measure is defined by pepitope [2]. Calculation is given in Supplementary material.
3D structure prediction and analyses
The 3D structures of the HA proteins from strains ALB70, PC73, PH82, MOS99, WIS05 and BR07 have been predicted using the SwissModel online workstation. The template chosen (based on the automatic template selection mode) was the HA protein of X-31 (PDB ID: 1EO8). Energy minimization of the modeled structures and structural comparisons were performed using the GROMOS96 force field application in Swiss PDB-Viewer (SPDBV) [14]. Predicted structures were evaluated using PROCHECK analyses [15]. Visualization of all the molecules and rendering of images was carried out in Discovery Studio v.2.0 (Accelyrs Inc., USA). The surface electrostatics of the proteins was studied using NOC software [16].
Molecular docking
Computational docking, a method which predicts the preferred stable orientation of one molecule when bound to a second one, was carried out to determine the binding site and the residues interacting between the antigens and antibody through hydrogen bonds (H-bonds) and salt bridges. In silico docking of antibody BH151against the HA proteins of ALB70, PC73, PH82, MOS99 and BR07 has been carried out using the ZDOCK server with default parameters [17, 18]. As a precondition for docking in each case, the following amino acids on the variable heavy chain of antibody BH151 were desired to form the binding interface between antigen and antibody: T31, Y32, R94, R98 and W100. However, no such constrains were imposed on any amino acid on the antigens. From the ten solutions returned by ZDOCK in each case, the best solution was selected based on the following conditions: i) the complimentarity determining regions (CDRs) of the antibody interacting with the antigen at the antigen-antibody interface and ii) the value of minimized energy of the complex being the least (energetically favourable). The details of contacts between amino acids of antigen and antibody for each complex were obtained by Contacts of Structural Units (CSU) analyses [19].
Discussion
The HA sequences from various strains were subjected to pairwise comparison (all possible pairs within the data set) using the ALIGN tool (EMBL server). Identity (%) and similarity (%) were calculated from these alignments ( see Table 1). Maximum identity (98.6%) and similarity (99.1%) in amino acid composition has been observed between the strains X-31 and ALB70. Phylogenetic analyses further indicated the clustering of the sequences and placed both X-31 and ALB70 in one clade (see Figure S1). The multiple sequence alignment indicated the occurrence of mutations in the various strains with respect to X-31. Compared to the X-31 there were 21, 7, 18, 41, 66, 64, 78, 76 & 76 mutations in the sequences UKR63, ALB70, PC73, PH82, SYD97, MOS99, WIS05, BR07 and PR09 respectively. The number of mutations between X-31 and ALB70 were found to be 7 with only 3 mutations in the epitope regions (see Figure S2). The stretches that record maximum variations in amino acid composition are: 151-162, 170-180 and 200-210 (as per numbering of complete amino acid sequences as in Figure S2).
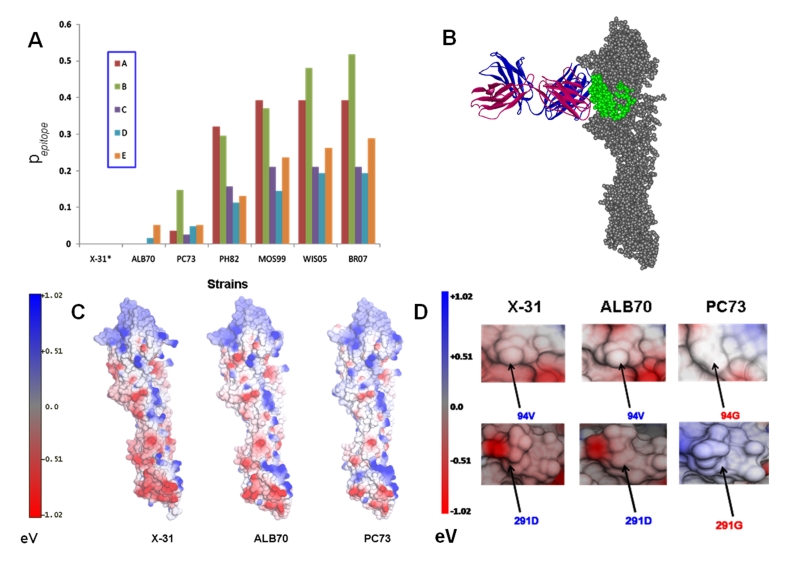
The antigenic evolution has been determined in terms of mutations in the epitope regions. Figure 1A describes the antigenic distances of various strains with respect to X-31, for the known epitopes A, B, C, D and E, evaluated in terms of pepitope. It can be seen from the graph that antigenic distance is least between X-31 and ALB70 with no mutations in the epitopes A, B and C. Comparatively, the antigenic distance is higher between X-31 and PC73 with many mutations in the epitope regions. The antigenic distance of PH82, MOS99, WIS05 and BR07 with respect to X-31 is high for all the epitopes. Maximum antigenic variability is observed in the epitope B within the data set considered (Figure 1A). The number of mutations with respect to X-31 in the known epitopes of HA proteins from various strains has been enlisted in (see Table 2). Other studies have also revealed the high mutation rates in epitopes A and B [2] with the emergence of fresh outbreaks coinciding with drastic changes in one or more epitopes simultaneously [20]. For structure-based analyses, following representative strains were considered: ALB70, PC73, PH82, MOS99, WIS05 and BR07. The 3D structures of these proteins were predicted by homology modelling with HA of X-31 (1E08) as the default template. The target–template identity of sequence composition along with the results of PROCHECK analyses (amino acid occupancy in Ramachandran Plot) and the minimized energy values of structures have been summarized in (see Table 3). For the predicted structures of HA, the occupancy of amino acids in Ramachandran plot was above 96% indicating that the models were good and reliable.
Figure 1.
(A) Antigenic distances of all the epitopes (A, B, C, D and E) on HA proteins from selected vaccine strains of Influenza A/H3N2 computed with X- 31 as a standard. (B) The docking of BH151 Mab into the E epitope of ALB70 HA protein. The VH and VL of the Bh151 are indicated in magenta and indigo colors respectively in Ribbon mode display, whereas, the HA protein of ALB70 is displayed in spacefill mode in Grey color with the epitope E highlighted in Green. (C) The comparison of Surface electrostatics (outputs from NOC program) reveals similarities between the X-31 and ALB70 whereas considerable differences existed between X-31 and PC73. (D) Figure highlighting the altered surface electrostatics in HA protein from PC73 due to mutations V94G and D291G w.r.t ALB70 (and X-31). N.B. The amino acid numbering followed in the text is in accordance with the sequence nomenclature of Influenza A/X-31/1968 HA protein (SwissProt id: P03438).
Docking of BH151 antibody onto the 3D structures of HA proteins was carried out to determine the binding site and the residues interacting within the antigen-antibody complex through hydrogen bonds (H-bonds) and salt bridges. The BH151 antibody docked into the antigenic site E of ALB70 HA and the complex was stabilized by the formation of H-bonds. The minimized total energy of the X-31 HA-BH151 co-crystal was -43891.53 KJ/mol while that of the ALB70 HA-BH151 complex was -40219.512 KJ/mol. However, in comparison with X-31 HA-BH151 co-crystal, some changes were observed in the orientation of the antibody (Figure 1B). The root mean squared deviation (RMSD) between the two complexes was found to be ˜1.8 Ǻ (considering 664 backbone atoms of both antigen and antibody). The Contacts of Structural Units analyses (CSU Analyses) (Sobolev et al., 1999) of the ALB70 HABH151 docked complex revealed that only four residues of the HCDRs are interacting with the antigen through hydrogen bonding. Formation of Hbonds was observed between the residues Y32 and T31 of (HCDR1) of BH151 with the residues K156 and N112 of the site E of HA respectively. Also, the residues R98 and G97 of HCDR3 of BH151 respectively formed Hbonds with the residues D93 and P90 of site E of HA (see Table 4) (Please refer to Figure S2 for the annotations of epitopes). The mutations D79N and N97D in the epitope E of ALB70 did not drastically affect the 3D conformation or the surface electrostatics, though the first one resulted in a loss of Hbond formation with antibody. The D79N mutation created a potential glycosylation site at position 79, whereas the N97D resulted in the loss of glycosylation site at position in ALB70 compared to X-31.
On the other hand, when the docking of BH151onto the HA of PC73, UKR63, PH82, MOS99, WIS05 and BR07 was carried out, the antibody could not recognize antigenic site E or other known antigenic sites. In each case, the ZDOCK output returned docking orientations that are physically unrealistic and non-feasible under natural physiological conditions. This is because some of the amino acids on the HA proteins of these strains interacting with the docked antibody actually remain inaccessible for antibodies in the HA prefusion trimer under natural physiological conditions. The failure of BH151 to recognize and bind to the HA of PC73 is (probably) due to the mutations V94G (w.r.t both ALB70 and X-31) at site E and D291G in the vicinity, which not only altered the surface contour without drastically changing the fold of the backbone but also changed the surface electrostatics from negative to positive potential (Figure 1C & D). These mutations were retained in the subsequent strains (PH82, MOS99, WIS05 and BR07). However, the comparison of the surface electrostatics (Figure 1C) revealed that the HAs of X31 and ALB70 are almost identical whereas those from PC73 and others are very dissimilar in spite of the antigenic distance being less (pepitope =0.0526). The varied surface electrostatics along with the conformational deviation in the E site may be responsible for BH151 not recognizing the HA proteins from PC73 and other strains. The mutation D79N in PC73 (w.r.t X-31) also resulted in the creation of a potential glycosylation site at position 79. Thus, the E epitope of PC73 HA possesses two glycosylation sites. Also, the antibody failed to recognize and bind to the HA of UKR63 because of dissimilar surface electrostatics and differences in amino acid compositions at antigenic site E as well as overall. The antigenic distance in terms of pepitope value between UKR63 and X-31 for all the antigenic regions ranged from 0.078 to 0.107 (Figure 1A). This outcome, however, is not far from expectation since the X-31 is not a direct descendant from the UKR63 strain and possessed an avian H3 HA [9].
Overall, antigenic variability between the various strains of H3N2 viruses have been studied. Though the epitopes of ALB70 were very similar to that on X-31, considerable differences existed between those on X-31 and other strains evolving from 1973 onwards. The docking of BH151 onto the ALB70 HA revealed that the antibody recognizes the similar antigenic determinant on HA of strain ALB70. Compared to the co-crystal of X-31 HA − BH151, we noted a loss of few contacts and a gain of a few in the ALB70 HA − BH151 docked complex. The ALB70 HA − BH151complex was stable and comparable to the original X31 HA − BH151 co-crystal. However, the antibody failed to recognize and bind to HA from PC73 and subsequent strains. It was noted that even two amino acid changes in the epitope E (of PC73 w.r.t X-31) resulted in altered surface electrostatics sufficient to affect the nature of interactions with antibody BH151. The HA proteins of strains with greater antigenic distances from the X-31 strain could not be recognized by BH151.
Conclusion
The antigenic evolution of HA proteins from vaccine strains of influenza A/H3N2 has been studied over the period 1968-2007 and variability in terms of antigenic distances have been observed for all the epitopes. The structural basis for the antibody BH151 not recognizing the HAs of 1973 and subsequently evolved strains could be explained through molecular docking studies. The results revealed the molecular basis for reported failure of the vaccine based on the Hong Kong strain of the 1968 pandemic to provide protection against strain A/Port Chalmers/1/1973. Further, even two amino acid changes were found to be sufficient to alter the antigenicity and surface properties of the epitopes in HA proteins. Overall, our study reflects the highly specific nature of antigenantibody interactions and gives insight into the molecular basis of host-immune evasion by influenza viruses.
Supplementary material
Footnotes
Citation:Shil et al, Bioinformation 6(7): 266-270 (2011)
References
- 1.VA Potdar, et al. PloS ONE. 2010;5(3):e9693. doi: 10.1371/journal.pone.0009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ET Munoz, MW Deem. Vaccine. 2005;23:1144. [Google Scholar]
- 3.YC Liao, et al. Bioinformatics. 2008;24(4):505. doi: 10.1093/bioinformatics/btm638. [DOI] [PubMed] [Google Scholar]
- 4.RM Bush, et al. Science. 1999;286:1921. [Google Scholar]
- 5.RM Bush, et al. Mol Biol Evol. 1999;16:1457. doi: 10.1093/oxfordjournals.molbev.a026057. [DOI] [PubMed] [Google Scholar]
- 6.JB Plotkin, J Dushoff. Proc Natl Acad Sci USA. 2003;100:7152. [Google Scholar]
- 7.MS Lee, JS Chen. Emerg Infect Dis. 2004;10:1385. doi: 10.3201/eid1008.040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DJ Smith, et al. Science. 2004;305:371. [Google Scholar]
- 9.M Matrosovich, et al. J Virol. 2000;74:8502. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S Dolan, et al. Center Dis Control Prevent, MMWR Morbid Mortal Wkly Rep. 2004;53(1):8. [Google Scholar]
- 11.Wkly Epidem Rec. 1974;49:41. WHO Selection Document. [Google Scholar]
- 12.K Tamura, et al. Mol Biol Evol. 2007;24:1596. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 13.Kolaskar AS, PC Tongaonkar. FEBS Lett. 1990;276:172. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 14. http://spdbv.vital-it.ch/
- 15. http://www.ebi.ac.uk/thornton-srv/software/PROCHECK.
- 16. http://noch.sourceforge.net/
- 17. http://zdock.bu.edu/
- 18.K Wiehe, et al. Proteins. 2007;69:719. doi: 10.1002/prot.21747. [DOI] [PubMed] [Google Scholar]
- 19. http://bip.weizmann.ac.il/oca-bin/lpccsu.
- 20.JB Plotkin, et al. Proc Natl Acad Sci USA. 22;99:6263. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.