Abstract
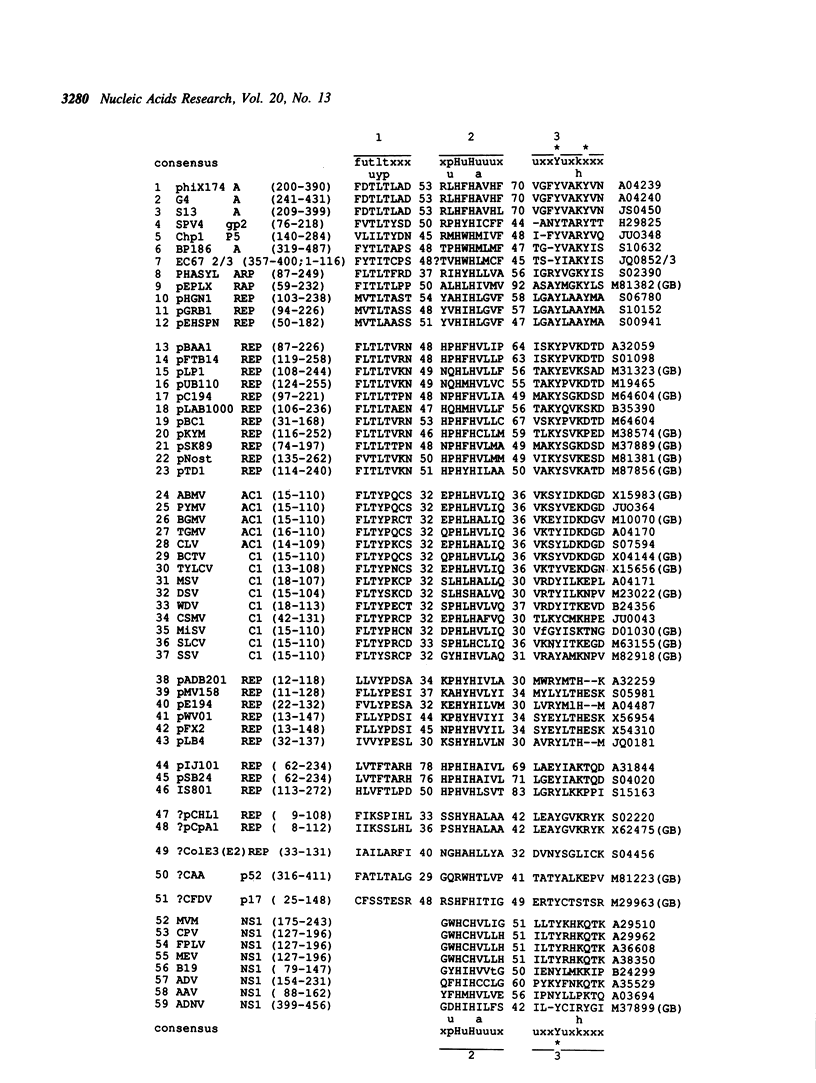
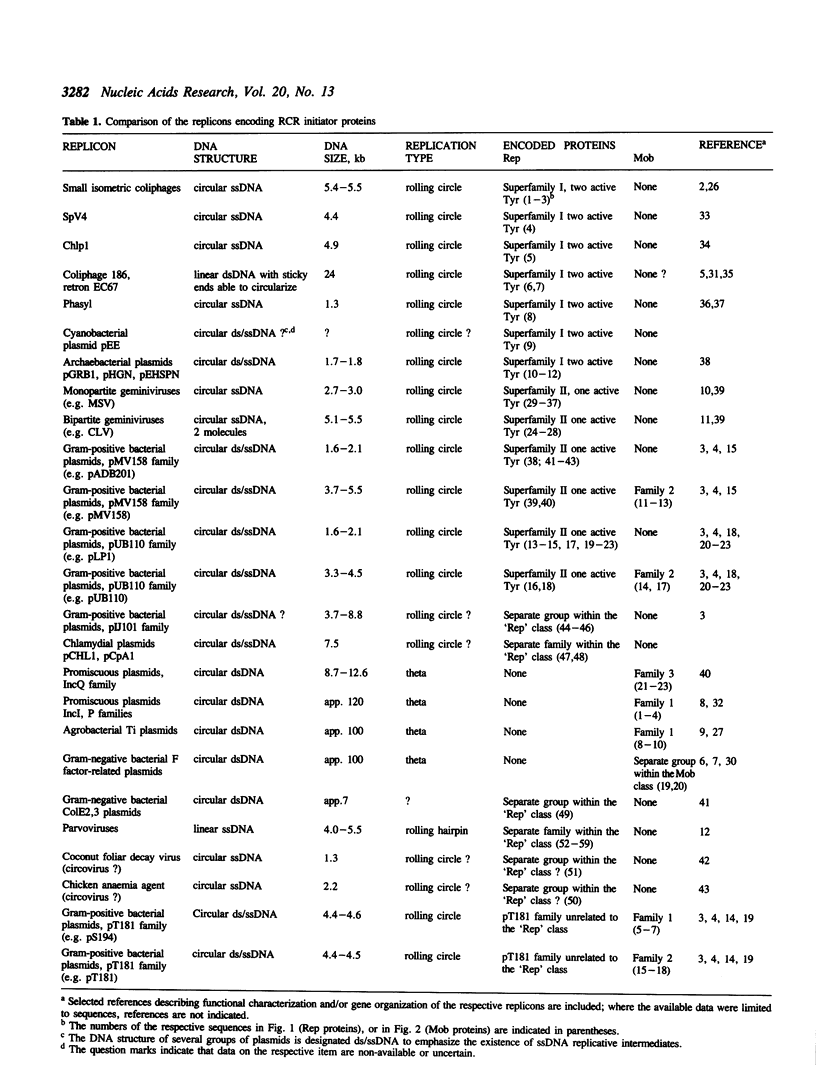
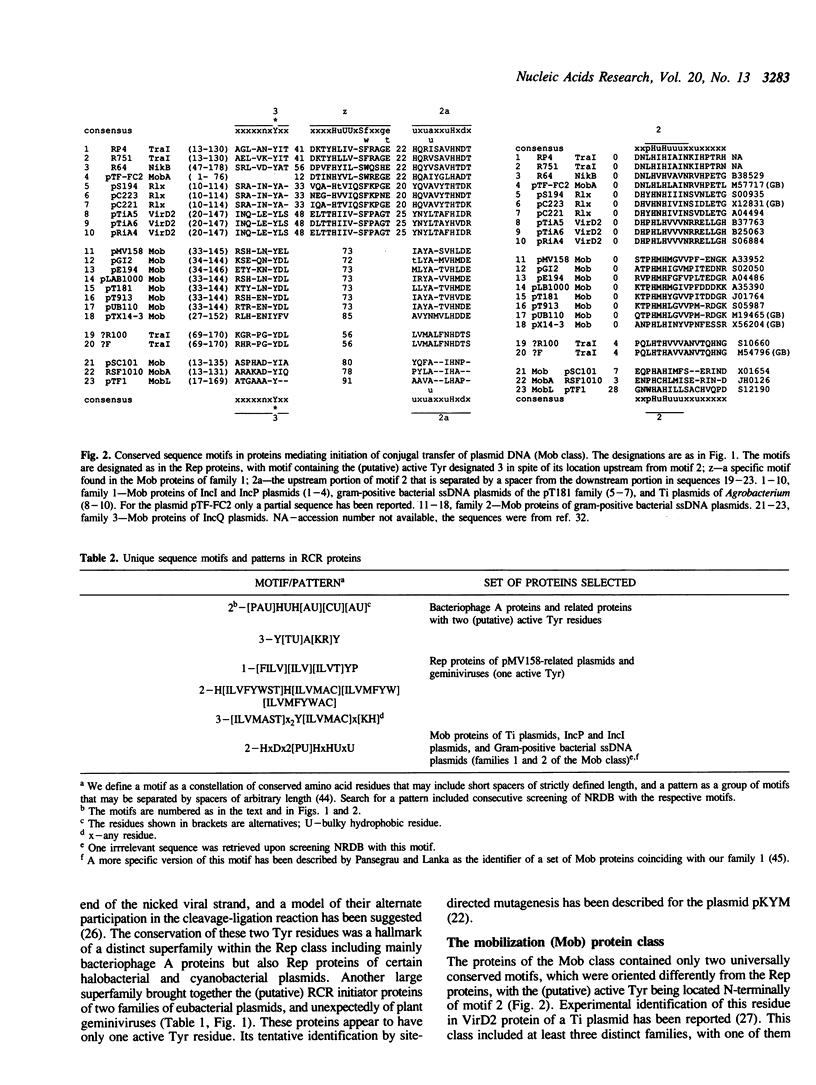
An amino acid motif was identified that consists of the sequence HisHydrHisHydrHydrHydr (Hydr--bulky hydrophobic residue) and is conserved in two vast classes of proteins, one of which is involved in initiation and termination of rolling circle DNA replication, or RCR (Rep proteins), and the other in mobilization (conjugal transfer) of plasmid DNA (Mob proteins). Based on analogies with metalloenzymes, it is hypothesized that the two conserved His residues in this motif may be involved in metal ion coordination required for the activity of the Rep and Mob proteins. Rep proteins contained two additional conserved motifs, one of which was located upstream, and the other downstream from the 'two His' motif. The C-terminal motif encompassed the Tyr residue(s) forming the covalent link with nicked DNA. Mob proteins were characterized by the opposite orientation of the conserved motifs, with the (putative) DNA-linking Tyr being located near their N-termini. Both Rep and Mob protein classes further split into several distinct families. Although it was not possible to find a motif or pattern that would be unique for the entire Rep or Mob class, unique patterns were derived for large subsets of the proteins of each class. These observations allowed the prediction of the amino acid residues involved in DNA nicking, which is required for the initiation of RCR or conjugal transfer of single-stranded (ss) DNA, in Rep and Mob proteins encoded by a number of replicons of highly diverse size, structure and origin. It is conjectured that recombination has played a major part in the dissemination of genes encoding related Rep or Mob proteins among the replicons exploiting RCR. It is speculated that the eucaryotic small ssDNA replicons encoding proteins with the conserved RCR motifs and replicating via RCR-related mechanisms, such as geminiviruses and parvoviruses, may have evolved from eubacterial replicons.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baas P. D., Jansz H. S. Single-stranded DNA phage origins. Curr Top Microbiol Immunol. 1988;136:31–70. doi: 10.1007/978-3-642-73115-0_3. [DOI] [PubMed] [Google Scholar]
- Berns K. I. Parvovirus replication. Microbiol Rev. 1990 Sep;54(3):316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A. P., Sorokin A. V., Aleksandrov N. N., Danilenko V. N., Kozlov Iu I. Nukleotidnaia posledovatel'nost' DNK aktinomitsetnoi plazmidy pSB24.2. Dokl Akad Nauk SSSR. 1985;283(4):1014–1017. [PubMed] [Google Scholar]
- Bouia A., Bringel F., Frey L., Kammerer B., Belarbi A., Guyonvarch A., Hubert J. C. Structural organization of pLP1, a cryptic plasmid from Lactobacillus plantarum CCM 1904. Plasmid. 1989 Nov;22(3):185–192. doi: 10.1016/0147-619x(89)90001-2. [DOI] [PubMed] [Google Scholar]
- Chakrabarti P. Geometry of interaction of metal ions with histidine residues in protein structures. Protein Eng. 1990 Oct;4(1):57–63. doi: 10.1093/protein/4.1.57. [DOI] [PubMed] [Google Scholar]
- Claessens J. A., Schrier C. C., Mockett A. P., Jagt E. H., Sondermeijer P. J. Molecular cloning and sequence analysis of the genome of chicken anaemia agent. J Gen Virol. 1991 Aug;72(Pt 8):2003–2006. doi: 10.1099/0022-1317-72-8-2003. [DOI] [PubMed] [Google Scholar]
- Davies J. W., Stanley J. Geminivirus genes and vectors. Trends Genet. 1989 Mar;5(3):77–81. doi: 10.1016/0168-9525(89)90030-9. [DOI] [PubMed] [Google Scholar]
- De Rossi E., Milano A., Brigidi P., Bini F., Riccardi G. Structural organization of pBC1, a cryptic plasmid from Bacillus coagulans. J Bacteriol. 1992 Jan;174(2):638–642. doi: 10.1128/jb.174.2.638-642.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet M., Zanga P., Lau P. C. The mobilization and origin of transfer regions of a Thiobacillus ferrooxidans plasmid: relatedness to plasmids RSF1010 and pSC101. Mol Microbiol. 1990 Aug;4(8):1381–1391. doi: 10.1111/j.1365-2958.1990.tb00717.x. [DOI] [PubMed] [Google Scholar]
- Gielow A., Diederich L., Messer W. Characterization of a phage-plasmid hybrid (phasyl) with two independent origins of replication isolated from Escherichia coli. J Bacteriol. 1991 Jan;173(1):73–79. doi: 10.1128/jb.173.1.73-79.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Dressler D. DNA replication: the rolling circle model. Cold Spring Harb Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Blinov V. M., Donchenko A. P., Koonin E. V. An NTP-binding motif is the most conserved sequence in a highly diverged monophyletic group of proteins involved in positive strand RNA viral replication. J Mol Evol. 1989 Mar;28(3):256–268. doi: 10.1007/BF02102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987 Dec 1;6(12):3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgman T. C. The elucidation of protein function by sequence motif analysis. Comput Appl Biosci. 1989 Feb;5(1):1–13. doi: 10.1093/bioinformatics/5.1.1. [DOI] [PubMed] [Google Scholar]
- Hsu M. Y., Inouye M., Inouye S. Retron for the 67-base multicopy single-stranded DNA from Escherichia coli: a potential transposable element encoding both reverse transcriptase and Dam methylase functions. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9454–9458. doi: 10.1073/pnas.87.23.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamoto S., Yoshioka Y., Ohtsubo E. Site- and strand-specific nicking in vitro at oriT by the traY-traI endonuclease of plasmid R100. J Biol Chem. 1991 Jun 5;266(16):10086–10092. [PubMed] [Google Scholar]
- Jones C., Holland I. B. Role of the SulB (FtsZ) protein in division inhibition during the SOS response in Escherichia coli: FtsZ stabilizes the inhibitor SulA in maxicells. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6045–6049. doi: 10.1073/pnas.82.18.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josson K., Soetaert P., Michiels F., Joos H., Mahillon J. Lactobacillus hilgardii plasmid pLAB1000 consists of two functional cassettes commonly found in other gram-positive organisms. J Bacteriol. 1990 Jun;172(6):3089–3099. doi: 10.1128/jb.172.6.3089-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido M., Yasueda H., Itoh T. Identification of a plasmid-coded protein required for initiation of ColE2 DNA replication. Nucleic Acids Res. 1991 Jun 11;19(11):2875–2880. doi: 10.1093/nar/19.11.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- Pansegrau W., Lanka E. Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 1991 Jun 25;19(12):3455–3455. doi: 10.1093/nar/19.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projan S. J., Novick R. Comparative analysis of five related Staphylococcal plasmids. Plasmid. 1988 May;19(3):203–221. doi: 10.1016/0147-619x(88)90039-x. [DOI] [PubMed] [Google Scholar]
- Renaudin J., Pascarel M. C., Bové J. M. Spiroplasma virus 4: nucleotide sequence of the viral DNA, regulatory signals, and proposed genome organization. J Bacteriol. 1987 Nov;169(11):4950–4961. doi: 10.1128/jb.169.11.4950-4961.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde W., Randles J. W., Langridge P., Hanold D. Nucleotide sequence of a circular single-stranded DNA associated with coconut foliar decay virus. Virology. 1990 Jun;176(2):648–651. doi: 10.1016/0042-6822(90)90038-s. [DOI] [PubMed] [Google Scholar]
- Romantschuk M., Richter G. Y., Mukhopadhyay P., Mills D. IS801, an insertion sequence element isolated from Pseudomonas syringae pathovar phaseolicola. Mol Microbiol. 1991 Mar;5(3):617–622. doi: 10.1111/j.1365-2958.1991.tb00732.x. [DOI] [PubMed] [Google Scholar]
- Saraste M. Structural features of cytochrome oxidase. Q Rev Biophys. 1990 Nov;23(4):331–366. doi: 10.1017/s0033583500005588. [DOI] [PubMed] [Google Scholar]
- Saunders K., Lucy A., Stanley J. DNA forms of the geminivirus African cassava mosaic virus consistent with a rolling circle mechanism of replication. Nucleic Acids Res. 1991 May 11;19(9):2325–2330. doi: 10.1093/nar/19.9.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W., Lurz R., Messer W. A novel replicon occurring naturally in Escherichia coli is a phage-plasmid hybrid. EMBO J. 1988 Dec 1;7(12):4005–4010. doi: 10.1002/j.1460-2075.1988.tb03289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M., Baldacci G., Forterre P., de Recondo A. M. Novobiocin induces accumulation of a single strand of plasmid pGRB-1 in the archaebacterium Halobacterium GRB. Nucleic Acids Res. 1988 Aug 25;16(16):7833–7842. doi: 10.1093/nar/16.16.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaprasad A. V., Jarvinen R., Puspurs A., Egan J. B. DNA replication studies with coliphage 186. III. A single phage gene is required for phage 186 replication. J Mol Biol. 1990 Jun 5;213(3):449–463. doi: 10.1016/S0022-2836(05)80207-4. [DOI] [PubMed] [Google Scholar]
- Stenger D. C., Revington G. N., Stevenson M. C., Bisaro D. M. Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling-circle replication of a plant viral DNA. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8029–8033. doi: 10.1073/pnas.88.18.8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey C. C., Lusher M., Richmond S. J. Analysis of the complete nucleotide sequence of Chp1, a phage which infects avian Chlamydia psittaci. J Gen Virol. 1989 Dec;70(Pt 12):3381–3390. doi: 10.1099/0022-1317-70-12-3381. [DOI] [PubMed] [Google Scholar]
- Symons R. H. Self-cleavage of RNA in the replication of small pathogens of plants and animals. Trends Biochem Sci. 1989 Nov;14(11):445–450. doi: 10.1016/0968-0004(89)90103-5. [DOI] [PubMed] [Google Scholar]
- Vogel A. M., Das A. Mutational analysis of Agrobacterium tumefaciens virD2: tyrosine 29 is essential for endonuclease activity. J Bacteriol. 1992 Jan;174(1):303–308. doi: 10.1128/jb.174.1.303-308.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N., Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiol Rev. 1984 Mar;48(1):24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa H., Hase T., Sakai A., Masamune Y. Rolling-circle replication of the plasmid pKYM isolated from a gram-negative bacterium. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10282–10286. doi: 10.1073/pnas.88.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski P. Basic processes underlying Agrobacterium-mediated DNA transfer to plant cells. Annu Rev Genet. 1988;22:1–30. doi: 10.1146/annurev.ge.22.120188.000245. [DOI] [PubMed] [Google Scholar]
- Ziegelin G., Pansegrau W., Strack B., Balzer D., Kröger M., Kruft V., Lanka E. Nucleotide sequence and organization of genes flanking the transfer origin of promiscuous plasmid RP4. DNA Seq. 1991;1(5):303–327. doi: 10.3109/10425179109020786. [DOI] [PubMed] [Google Scholar]
- de la Campa A. G., del Solar G. H., Espinosa M. Initiation of replication of plasmid pLS1. The initiator protein RepB acts on two distant DNA regions. J Mol Biol. 1990 May 20;213(2):247–262. doi: 10.1016/S0022-2836(05)80188-3. [DOI] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Are single-stranded circles intermediates in plasmid DNA replication? EMBO J. 1986 Mar;5(3):631–637. doi: 10.1002/j.1460-2075.1986.tb04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mansfeld A. D., van Teeffelen H. A., Baas P. D., Jansz H. S. Two juxtaposed tyrosyl-OH groups participate in phi X174 gene A protein catalysed cleavage and ligation of DNA. Nucleic Acids Res. 1986 May 27;14(10):4229–4238. doi: 10.1093/nar/14.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]



