SUMMARY
The ESCRT complexes are required for multivesicular body biogenesis, macroautophagy, cytokinesis, and the budding of HIV-1. The final step in the ESCRT cycle is the disassembly of the ESCRT-III lattice by the AAA ATPase Vps4. Vps4 assembles on its membrane-bound ESCRT-IIII substrate with its cofactor, Vta1. The crystal structure of the dimeric VSL domain of yeast Vta1 with the small ATPase and the β domains of Vps4 was determined. Residues involved in structural interactions are conserved and are required for binding in vitro and for Cps1 sorting in vivo. Modeling of the Vta1 complex in complex with the lower hexameric ring of Vps4 indicates that the 2-fold axis of the Vta1 VSL domain is parallel to within ~20 degrees of the 6-fold axis of the hexamer. This suggests that Vta1 might not crosslink the two hexameric rings of Vps4, but rather stabilizes an array of Vps4-Vta1 complexes for ESCRT-III disassembly.
INTRODUCTION
The ESCRT-III complex (Babst et al., 2002) and the Vps4 AAA ATPase (Babst et al., 1997; Babst et al., 1998) comprise an ancient membrane scission machinery, present throughout eukaryotes and in the Crenarchaea (Hanson et al., 2009; Samson and Bell, 2009). The ESCRT-III complex is responsible for cleaving narrow membrane necks from the side contiguous with the interior of the neck (Wollert et al., 2009). The neck is cleaved following the assembly of soluble ESCRT-III monomers into a tightly packed helical lattice on the membrane, which uses its affinity for the membrane to pull the sides of the membrane close enough for a hemiscission intermediate to form (Fabrikant et al., 2009). Following scission, ESCRT-III monomers do not disassemble spontaneously, but require the ATP-dependent action of Vps4 for release (Babst et al., 1998; Ghazi-Tabatabai et al., 2008; Lata et al., 2008; Saksena et al., 2009; Wollert et al., 2009). The hydrolysis of ATP by Vps4 is the only direct energy input into the cycle and is an absolute requirement for its progression. Therefore, intense effort has been devoted to understanding this reaction.
Vps4 belongs to the AAA ATPase family, whose members use energy from ATP hydrolysis to remodel the conformation of its macromolecular substrates and are involved in many biological processes, such as microtubule severing, membrane fusion, protein disaggregation, etc (Erzberger and Berger, 2006). The Vps4 protein is composed of an N-terminal ESCRT-III-binding MIT domain (Scott et al., 2005b), followed by a flexible linker, a large ATPase mixed-α/β domain, a small ATPase helical domain, a β-domain, and a C-terminal helix (Gonciarz et al., 2008; Scott et al., 2005a; Xiao et al., 2007). Vps4 functions as an oligomer that is by most accounts a dodecamer (Inoue et al., 2008; Landsberg et al., 2009; Yu et al., 2008), although there has also been a report of a tetradecameric form (Hartmann et al., 2008). The dodecamer consists of two conformationally distinct hexameric rings (Yu et al., 2008). The lower ring has a constricted pore and has been modeled based on the structure of the p97 D1 domain hexamer. The upper ring has a wider pore and its detailed conformation is unknown. The central pore is required for function (Gonciarz et al., 2008; Scott et al., 2005a), and it is thought that ESCRT-III subunits physically enter the pore during disassembly, and perhaps pass all the way through it.
Vps4 does not function alone, but co-assembles onto membrane-bound ESCRT-III with another protein, Vta1(Lottridge et al., 2006; Shestakova et al., 2010; Shiflett et al., 2004; Ward et al., 2005; Yeo et al., 2003). The deletion of Vta1 results in the deficiency of cargo sorting in yeast cells but does not cause a severe defect as demonstrated by the appearance of class E compartment in Vps4 deficiency cells (Lottridge et al., 2006; Shiflett et al., 2004; Yeo et al., 2003). Vta1 enhances Vps4 ATPase activity (Azmi et al., 2006; Lottridge et al., 2006) and oligomerization in vitro, and cooperates in ESCRT-III disassembly in vitro (Azmi et al., 2008). Vta1 binds to Vps4 via the β domain of the latter protein (Scott et al., 2005a). Vta1 is composed of two MIT domains at its N-terminus (Xiao et al., 2008), a dimeric Vps4-binding VSL (Vps4, SBP1, LIP5) domain at its C-terminus (Azmi et al., 2006) (Xiao et al., 2008), and a long flexible linker connecting these two. The Vta1 MIT domains interact with various ESCRT-III proteins, but especially strongly with the late-acting ESCRT-III protein Vps60 (Bowers et al., 2004) (Azmi et al., 2008; Shiflett et al., 2004; Shim et al., 2008), and indeed Vps60 seems to function primarily as an adaptor for Vta1 to interact with the ESCRT-III assembly (Nickerson et al., 2010). Thus Vta1 has multiple roles in the disassembly process, from promoting the assembly and activity of the Vps4 dodecamer, to augmenting interactions with the ESCRT-III substrate.
In this paper, we probe the structural interface that joins Vps4 and Vta1. The minimal binary complex structure reported here confirms that the β domain in Vta1 makes direct contact with the VSL domain in Vps4. Disruptive mutations in this region leads to weakening or elimination of the Vps4-Vta1 interaction in vitro. Eliminating this interaction in yeast cells results in a phenotype similar vta1Δ, consistent with expectation. More surprisingly, we find that the 2-fold axis of the VSL domain dimer is nearly parallel to the 6-fold axis of the Vps4 hexamer. This implies that Vta1 promotes inter-dodecamer, not intra-dodecamer, dimerization, with implications for the role of Vta1 in promoting ESCRT-III disassembly by Vps4.
RESULTS
Structure of the Vps4-Vta1 interface
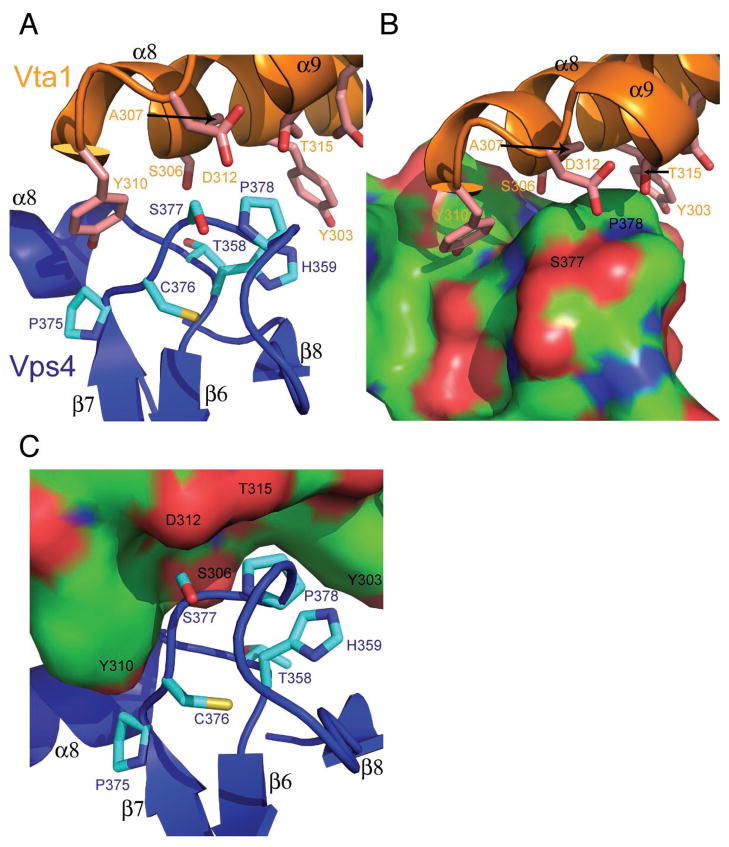
Binding of Vta1 and Vps4 fragments was measured (Fig. 1A, Table 1). A S. cerevisiae Vps4 construct consisting of the small ATPase and β domains, which we will refer to as the “SAB” fragment, bound to the VSL domain of S. cerevisiae Vta1 with an affinity only five-fold lower than that of the complete Vps4 catalytic domain. The Vps4 SAB construct was therefore co-crystallized with the Vta1 VSL domain and the structure was determined at 3.1 Å resolution by molecular replacement (Fig. 2A-C, Table 2). The asymmetric unit contains one copy each of Vta1-VSL and Vps4-SAB. Vta1-VSL is a crystallographic dimer that correspond precisely to the non-crystallographic dimer previously observed for Vta1-VSL in isolation (Xiao et al., 2008). The two copies of Vps4-SAB that bind to the same Vta1-VSL dimer have no direct contact with one another (Fig. 3). The Vps4-Vta1 minimal binary complex is formed by the interaction between helices α8 and α9 (numbered on the basis of full-length Vta1 (Xiao et al., 2008)) of the Vta1 C terminal domain (Vta1CTD) and the α8-β6 and β7- β8 loops in the Vps4 β domain (numbered on the basis of the yeast Vps4 catalytic domain, excluding the MIT domain (Gonciarz et al., 2008); Fig. 2B). The interaction buries 760 Å2 of solvent accessible surface area (Fig. 2C).
Fig. 1. Binding of Vps4 and Vta1 constructs and their mutants.
A. Binding curves from best fits to equilibrium binding model. B-I. SPR sensorgrams for the indicated constructs.
Table 1.
Binding constants determined by surface plasmon resonance
| Vta1 construct | Kd (μM ) | |
|---|---|---|
| WT SAB | WT VSL | 85 ± 13 |
| WT SAB | Y303A VSL | No binding |
| WT SAB | S306R VSL | >1000 |
| WT SAB | A307D VSL | No binding |
| WT SAB | D312A VSL | >1000 |
| WT SAB | T315R VSL | 440 ± 61 |
| H359D SAB | WT VSL | 280 ± 12 |
| WT full length | WT VSL | 17 ± 0.3 |
Fig. 2. Structure of the Vps4-Vta1 molecular interface.
A. Electron density calculated from a 2Fo-Fc synthesis in which the residues shown in orange were omitted. Density is contoured at 1 σ. B. Ribbon model for the 1:1 complex in the asymmetric unit. C. Interaction of Vta1 with the molecular surface of Vps4, with the latter colored by atom type (carbon, green; oxygen, red; nitrogen, blue).
Table 2.
Statistics of crystallographic data collection and refinement
| Data Collection | |
|---|---|
| Space Group | P2221 |
| Cell Dimensions | 37.98 Å, 70.32 Å, 88.34 Å 90º, 90º, 90º |
| Wavelength (Å) | 0.9719 |
| Resolution (Å) | 3.1 |
| Rsym | 0.085 (0.355) |
| I/σI | 19.2 (2.6) |
| Completeness (%) | 80.0 ( 62.1 ) |
| Redundancy | 3.7 (3.5) |
| Refinement | |
| Resolution (Å) | 3.1 |
| No. reflections | 3761 |
| Rwork/Rfree (%) | 30.0/34.3 |
| No. atoms | |
| Protein | 1138 |
| B-factor | 88.2 |
| R. m. s deviations | |
| Bond lenths (Å) | 0.011 |
| Bond angles (º) | 1.82 |
Fig. 3. The Vps4-Vta1 interacting complex is a dimer.

Vta1 is shown in orange and magenta, Vps4 in cyan and blue. The view is looking down the two-fold crystallographic axis related the two Vta1-Vps4 complexes.
A pair of tyrosine residues of Vta1, Tyr303 and Tyr310, delimit the major interactions of Vta1 helix α8 with the Vps4 β-domain (Fig. 2A, B, Fig. 4A). These two residues are essential for the binding of Vta1 of Vps4 (Xiao et al., 2008). Nearby residues of helix α8 Ser306, Ala307, and Asn309 also contact Vps4 (Fig. 5A-C). Outside of helix α8, Asp312 of the α8-α9 linker and Thr315 of helix α9 directly contact Vps4. Vps4 binds to Vta1 primarily through two loops, along with Lys353 of helix α8. The first loop is composed of conserved residues 356SATH359 (Fig. 4B) and corresponds to the α8-β6 linker and the first residue of β6. The second loop is composed of conserved residues 375PCSPGDDGAIE385, part of the long β7- β8 connector. The side-chains Vps4 Thr358, His359, Pro378, and Glu385 form the binding site for the side-chain of the key Vta1 Tyr303. Vps4 Lys353, Ala357, Pro375, Ser377, and the main chain of Cys376 form a pocket for the side-chain of Vta1 Tyr310. The involvement of this region in Vps4-Vta1 interaction is consistent with the earlier report that the S377A/D380A mutation in Vps4 abolishes the interaction (Scott et al., 2005a). As compared to the structures of Vta1-VSL and Vps4 in isolation, the structure of Vta1 is virtually unchanged, and the largest shift in Vps4 is a 2 Å movement of Pro378 to accommodate Vta1-Tyr303.
Fig. 4. Structure based alignment of interface residues.
Residues in Vta1 (A) and Vps4 (B) are marked by asterisks where they are involved in direct interactions.
Fig. 5. Details of the interface.
A. Ribbon model of the interface, with Vta1 in orange and Vps4 in blue. B. Surface representation of Vps4 colored by atom type, with Vta1 shown in a ribbon (orange) and ball-and-stick representation. C. Surface representation of Vta1 colored by atom type, with Vps4 shown in a ribbon (blue) and ball-and-stick representation.
Structural interface residues are important for binding in vitro
In order to expand upon and quantify previous mutational analyses of the Vps4-Vta1 interface (Azmi et al., 2006; Scott et al., 2005a; Xiao et al., 2008), interactions between wild-type and mutant Vps4-SAB and Vta1-VSL fragments were probed by SPR (Fig. 1A, Table 1). The wild-type Vps4-SAB and Vta1-VSL fragments interact with a Kd of 85 ± 13 μM. This relatively weak interaction is consistent with the modest amount of buried surface area in the interface and its partially polar character. Mutations in the interface result in a substantial increase in Kd. Vta1-A307D abolishes the interaction, while Vta1-S306R and D312A have Kd values above 1 mM. The Vta1-S306R mutation was designed to introduce the bulky Arg side-chain into a restricted space, and thus is more disruptive than the previously published benign mutation S306A (Azmi et al., 2006). Vta1-Y303A completely abolishes the Vps4-Vta1 interaction, consistent with earlier findings (Xiao et al., 2008). Vta1-T315R and Vps4- H359D have smaller effects on Kd, with values of 440 ± 61 μM and 280 ± 12 μM, respectively, consistent with their positions at the periphery of the interface.
Structural interface residues are required for Cps1 sorting
The loss of interaction in the Vta1 S306R and A307D mutants, which introduce charged residues into the sterically confined interface, led us to expect that these would be loss-of-function mutants in cargo so rting in yeast. Localization of the vacuolar protease Cps1 fused to GFP was used to assess function (Odorizzi et al., 1998). As observed previously (Shiflett et al., 2004), vta1Δ cells showed partial mislocalization of the cargo protein Cps1 (Fig. 6, column 1), and did not show the prominent class E puncta that are associated with the most severe defects in the ESCRT pathway. Cps1 mislocalization was completely rescued by a plasmid vector bearing wild type VTA1 (Fig. 6, column 2). The VTA1S306R and VTA1A307D failed to rescue (Fig. 6 columns 3–4), consistent with past results for other interface residues, Tyr303 and Tyr310 (Xiao et al., 2008). These findings validate that the structural interface can be used not only to rationalize previous biological observations, but is also predictive of the phenotypes of new mutations
Fig. 6. Structural interface residues are required for function.
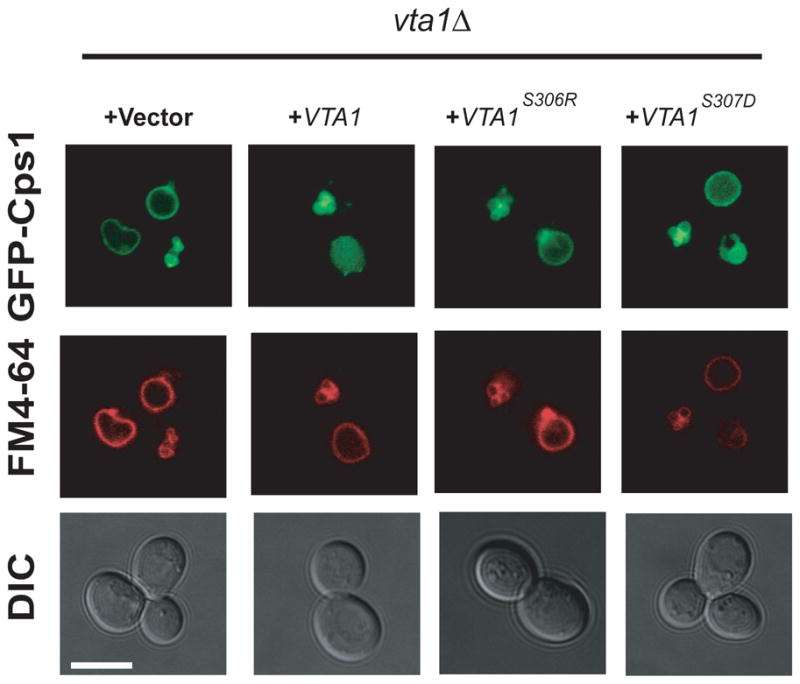
The indicated plasmids were transformed into vta1Δ yeast cells and imaged for GFP-Cps1 (cargo) and FM4-64 (membrane) fluorescence as shown. The scale bar is 5 μm.
Modeling the VSL dimer-Vps4 hexamer complex
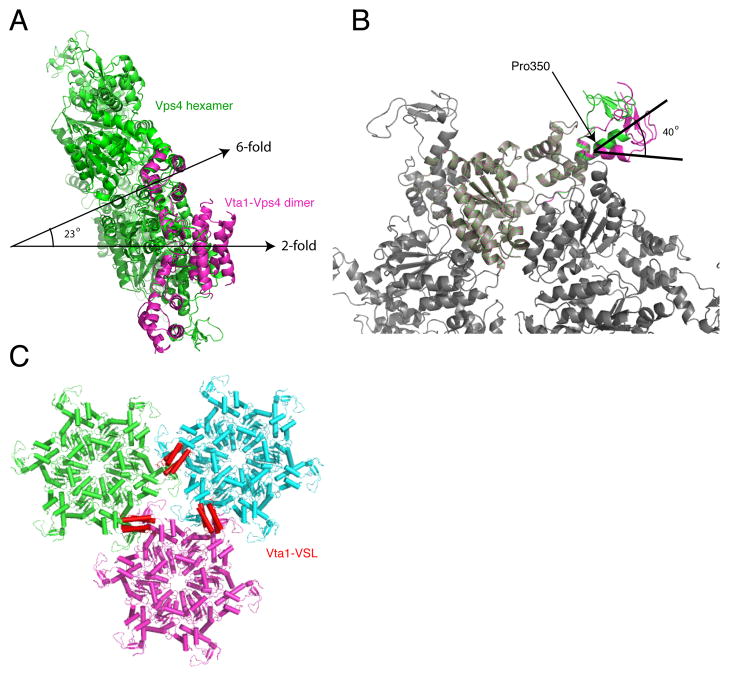
Vta1 functionally interacts with the oligomeric form of Vps4 (Azmi et al., 2006; Lottridge et al., 2006; Scott et al., 2005a), and we sought to model the complex. Yeast Vps4 has been visualized by EM as a dodecamer in the presence or absence of Vta1. The dodecamer consists of two non-equivalent hexameric rings. A pseudoatomic model of the lower ring of Vps4 consistent with the EM envelope has been constructed (Yu et al., 2008) by fitting to the atomic structure of the Vps4 catalytic domain monomer (Gonciarz et al., 2008) to that of the hexameric D1 catalytic domain of the p97 ATPase (Zhang et al., 2000). Cryo EM analysis indicates that Vta1 binding has effects on both the upper and lower rings, however there is only enough information on the lower ring to make molecular modeling of the Vta1-Vps4 complex feasible at present. The fragmentary Vps4-Vta1 complex was docked onto the lower hexameric ring model by superposition of the Vps4 SAB fragment. After docking, the Vta1-VSL two-fold axis of symmetry was 23 degrees from the hexamer six-fold (Fig. 7A). This was incompatible with what we had anticipated might be a role in crosslinking the upper and lower rings of a single dodecamer, which would require a 90 degree angle between the axes. This led us to assess by modeling whether Vta1 could cross link two separate lower rings in the same plane. The β domain is connected to the small ATPase domain by a helix α6, which is kinked at Pro350, and by a highly flexible loop from residues 391–398 (disordered in the present structure). The orientation of the β domain varies in different Vps4 catalytic domain structures, with the position differing by bending about a hinge axis roughly parallel to the hexamer six-fold axis. The β8- α9 loop is so flexible that helix α8 imposes the only meaningful constraint on the orientation of the β domain. The kink in helix α8 is essentially in the plane of the hexamer (Fig. 7B). A 40 degree rotation about the hinge axis within the plane of the hexamer can be achieved by removing the kink in α8. The energetic cost of straightening a Pro-induced kink in an α-helix is small (Yun et al., 1991). Thus at minimal energetic cost, the β-domain can reorient within the plane of the hexamer in a manner compatible with a p6 lattice crosslinked by the Vta1 dimer (Fig. 7C). In contrast, a reorientation such that the Vta1 could crosslink two Vps4 rings within the same dodecamer would require a 90 degree rotation of the β domain, which would require a complete break in helix α8.
Fig. 7.
A model for cross-linking of Vps4 hexamers by the Vta1-VSL domain. A. Following superposition of the Vps4 SAB domain on one subunit of the Vps4 hexamer (green), the two-fold axis of the Vta1-VLS:Vps4-SAB complex (magenta) is within 23° of the six-fold axis of the hexamer. Part of the second Vps4-SAB overlaps with another subunit in the hexamer. B. Model for a putative unkinked conformation of α8. This model rotates the Vps4 β-domain and Vta1 by 40°, eliminates the steric overlap between the second SAB fragment and the hexamer, and is compatible with continuous hexagonal lattice packing. C. Hypothetical p6 lattice arrangement of Vta1-VSL (red) cross-linked Vps4 lower ring hexamers. For illustration purposes, a model of the human VPS4B hexamer is used instead of yeast, as the longer β6- β7 loop in human makes the orientation of the β domain easier to visualize.
DISCUSSION
Here we have characterized the major molecular interface between Vps4 and its partner, Vta1. While the structure is at low resolution, having been determined from the edge of a single plate-like crystal within a large cluster, the identity of the interfaces and the symmetry and stoichiometry of the complex are not subject to doubt. The SAB-VSL fragment model for the interaction is sufficient for us to rationalize published mutational data (Azmi et al., 2006; Scott et al., 2005a; Xiao et al., 2008) and to predict the effect of new mutations on binding in vitro and sorting function in vivo.
The major finding from the modeling analysis, and the main insight obtained from this study, was that the two-fold axis of the Vta1-VSL dimer is nearly parallel to the sixfold axis of the Vps4 lower ring hexamer. One concept in the field had been that Vta1 could crosslink the two Vps4 monomers across the upper and lower rings. Such crosslinking is hard to reconcile with the striking asymmetry seen in the EM envelope (Yu et al., 2008), and cannot be reconciled with a parallel orientation of the two fold and six fold axes. A significant cautionary note is that the conformation of the upper ring is unknown, and adjustments in the conformation might be large enough to facilitate interring crosslinking in a way that is hard to anticipate based on current information.
The Vta1-VSL dimer interface observed in the present structure is essentially identical to that seen previously for the isolated VSL domain (Xiao et al., 2008), despite that the previous dimer interface used non-crystallographic symmetry, and this one is an exact crystallographic dimer. Dimer interface residues in Vta1 are essential for its biological function (Xiao et al., 2008). Thus there seems to be no question that Vta1 functions as a dimer, and that the present and previous structures accurately represent the physiological dimer. Taking together observations of symmetry and functional relevance of the Vta1-VSL dimer interface residues, the most reasonable interpretation- albeit a surprising one- is that the Vta1 dimer functionally links equivalent hexamers “in trans” in two different dodecameric assemblies. An attractive feature of the model is that lower ring hexamers can dock onto one another via knobs-into-holes packing of the β domains (Fig. 7C), suggesting that Vps4 hexamers might have some ability to form edge-on lattice interactions even in the absence of Vta1.
Direct biochemical and biological tests of this model will not necessarily be straightforward. Enzymological assessments of Vta1-Vps4 cooperativity do not readily discriminate between cooperative cross-linking in “cis” (within one dodecamer) vs. “trans”. Likewise, it is difficult to differentiate between physiological and non-physiological aggregation in vitro. In vitro biochemical and biophysical analyses of the Vta1-Vps4 complex to date have, of necessity, used conditions designed to inhibit aggregation. For example, EM analysis of Vps4 showed that most of the dodecamers were isolated particles (Yu et al., 2008). However, this same report noted a strong propensity for Vps4 to aggregate, and therefore all of the samples used for EM structural analysis had to be pretreated with glutaraldehyde to inhibit aggregation. While the concept that the Vps4-Vta1 complex might function in vivo as a planar p6 lattice has strong support on structural grounds, it must be considered speculative until means can devised to directly test this idea.
A cross-linked lattice would be structurally static, suggesting a low stoichiometry of Vps4 to ESCRT-III in vivo. Indeed, the estimated number of Vps4 molecules per cell is ~5000, exceeding the estimated ~3000 copies of its most abundant substrate, Snf7 (Huh et al., 2003). Roughly three Vps4 crosslinked hexamers would have a footprint whose width (26 nm) equals the diameter of a yeast ILV (24 nm) (Nickerson et al., 2006). The flexibility of the β domain linkage to the Vps4 catalytic core is such that the Vps4 lattice need not be strictly planar. It could bend to accommodate itself to the dome-like shape postulated for the ESCRT-III lattice responsible for membrane scission (Fabrikant et al., 2009). The lattice arrangement places the Vta1 MIT domains on the “top” side of the lower ring, distal to the Vps4 MIT domain. Most Vta1 molecules would, however, line the edge of the lattice, leaving their MIT domains with unobstructed access to the ESCRT-III substrate. The lattice model implies that ESCRT-III disassembly might occur dynamically, but in the context of a stable, laterally static lattice. This model thus potentially puts the action of Vps4-Vta1 in ESCRT-III disassembly into a new perspective.
Experimental procedures
Protein expression and purification
S. cerevisiae Vta1 (VSL domain fragment spanning residues 280 to 330) and Vps4 (full length and the small ATPase and β domain “SAB” fragment spanning residues 299 to 415) were cloned into pGST-Parallel2 (Sheffield et al., 1999). Proteins were expressed in BL21 cells (for the full length Vps4 E233Q mutant) and Rossetta 2 cells (all other proteins). In brief, cells were grown in LB media to log phase and the expression was induced by 1 mM IPTG at 37°C for 6 hours (for Vps4 proteins) or at 18°C for 20 hours (for Vta1 proteins). Cells were then harvested, resuspended in 1X PBS buffer with 7 mM β-mercaptoethanol and lysed by sonication. Lysates were applied to glutathione sepharose resin (GE healthcare) and eluted with on-column cleavage by TEV protease. To make a Vps4-Vta1 complex for crystallization, the Vps4 SAB fragment was incubated with the Vta1 VSL fragment at 4 °C overnight and then applied to a Superdex 75 size exclusion column. The complex was eluted with 20 mM Tris, 100 mM NaCl, 7 mM β-mercaptoethanol at pH 7.5. Point mutations were introduced using QuickChange II site-directed mutagenesis system (Stratagene), mutations were confirmed by sequencing the complete coding region, and mutant proteins were purified as above.
Crystallization and data collection
Crystals of Vps4-SAB with Vta1-VSL were produced by hanging drop vapor diffusion at 15 °C. Protein complexes at 27 mg/ml were mixed at a 1:1 ratio with a reservoir solution containing 16%- 24% PEG3350 and 0.1 -0.3 M sodium formate in a final volume of 2 μl, and the drop was equilibrated with the reservoir solution. Crystals grew to full size in several days and were in the form of clusters of plates. For data collection, crystals were transferred into a cryoprotection solution by mixing 7.5 μl reservoir solution with 2.5 μl glycerol or ethylene glycol. The data were collected at the Advanced Photon Source beamline 23-ID using a micro-beam in order to focus the X-ray beam selectively onto a singlet plate within the stack. The need to avoid interference from the rest of the plate cluster led to a missing wedge comprising some 20 % of the data. Data were integrated and scaled in HKL2000.
Structure solution
The structure was determined by molecular replacement using the previously solved crystal structures of yeast Vps4 (residues 299–415 of pdb entry 3EIE )(Gonciarz et al., 2008)) and the yeast Vta1 VSL domain (pdb entry 2RKL ) (Xiao et al., 2008) as the search model in programs MolRep (Vagin and Teplyakov, 1997) and Phaser (McCoy et al., 2007). There is one Vps4-Vta1 heterodimer in an asymmetric unit. The structural model was rebuilt in Coot (Emsley and Cowtan, 2004). Because the search models used for molecular replacement were determined at higher resolution and from more complete data than the present structure (1.5 and 2.7 Å for Vta1-VSL and Vps4 catalytic domain, respectively), and because there was little evidence for structural changes, minimal rebuilding was carried out. Essentially, Vps4 residues 377–381 were shifted by up to 2 Å to accommodate Vta1, and only minimal adjustments were made elsewhere. Again, due to the higher quality of the two parent structures, only a single pass of refinement was carried out in CNS (Brunger et al., 1998) using 5% randomly selected reflection for cross-validation. The residues of 280 to 288 in Vta1 are missing in the final structure due to presumed disorder. Residues 388–398 of Vps4 were omitted due to the fragmentary to nonexistent electron density for this flexible linker region.
SPR binding assay
Binding of Vta1 to Vps4 was analyzed using a Biacore T100 instrument operated at 25 ºC. Anti-GST (GE healthcare) was immobilized on a CM5 chip by amine coupling. GST and GST-Vps4-SAB proteins were captured on this surface to a density of 1000 units. The Vta1 VSL domain was injected at a flow rate of 20μl/min in 10mM HEPES pH 7.0, 150mM NaCl, 0.005% P20 at 25ºC. 10mM glycine-HCl pH 2.0 was used for surface regeneration. The data were processed using BiaEvaluation software (Biacore) and the dissociation constant was calculated by fitting the following equation:
where [MIT] is the protein concentration of the flowing analyte, Kd is the dissociation constant, Rmax is the maximal response, and ‘offset’ is the background signal. The data were fit using BiaEvaluation (Biacore) and SigmaPlot softwares.
Cps1 cargo sorting assay
The vps4Δ and vta1Δ strains were obtained from the Open Biosystems collection. The complete expression cassettes for VPS4 and VTA1 were amplified from yeast genomic DNA and cloned into YCplac111. Mutations in these genes were introduced as mentioned earlier. YCplac111 plasmids encoding VPS4 and VTA1 were co-transformed with the pRS426 GFP-Cps1 vector (Odorizzi et al., 1998) into vps4Δ and vta1Δ strains, respectively. Yeast transformants were selected on Ura− Leu− medium and single colonies were purified by restreaking. Cells were then grown to log phase, harvested, and labeled with FM4-64 for vacuolar membrane staining (Vida and Emr, 1995). Visualization was performed on an LSM510 fluorescence microscope (Carl Zeiss MicroImaging) with fluorescein isothiocyanate (FITC) and rhodamine filters.
HIGHLIGHTS.
Structure of the interacting fragments of Vps4 and Vta1 determined.
Interface residues are required for binding in vitro and Cps1 sorting in yeast.
Vta1 probably does not crosslink upper and lower Vps4 hexameric rings.
Modeling suggests Vta1 could crosslink different Vps4 dodecamers.
Acknowledgments
We thank B. Baibokov, X. Ren, and Y. Im for assistance with microscopy, J. Dean for microscope access, and D. Katzmann for helpful discussions. GM/CA CAT has been funded in whole or in part with Federal funds from the NCI (Y1-CO-1020) and the NIGMS (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. This work was supported by the NIDDK and IATAP programs of the NIH intramural research program. Crystallographic coordinates have been deposited in the Protein Data Bank with the accession code 3MHV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azmi I, Davies B, Dimaano C, Payne J, Eckert D, Babst M, Katzmann DJ. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta 1. J Cell Biol. 2006;172:705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi IF, Davies BA, Xiao J, Babst M, Xu Z, Katzmann DJ. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev Cell. 2008;14:50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: An endosome-associated heterooligomeric protein complex required for MVB sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr Sect D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA plus proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JAG, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLOS Comp Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, Crowther RA, Emr SD, Egelman EH, Williams RL. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Gonciarz MD, Whitby FG, Eckert DM, Kieffer C, Heroux A, Sundquist WI, Hill CP. Biochemical and Structural Studies of Yeast Vps4 Oligomerization. J Mol Biol. 2008;384:878–895. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Shim S, Merrill SA. Cell biology of the ESCRT machinery. Curr Opin Cell Biol. 2009;21:568–574. doi: 10.1016/j.ceb.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Chami M, Zachariae U, de Groot BL, Engel A, Grutter MG. Vacuolar protein sorting: Two different functional states of the AAA-ATPase Vps4p. J Mol Biol. 2008;377:352–363. doi: 10.1016/j.jmb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Inoue M, Kamikubo H, Kataoka M, Kato R, Yoshimori T, Wakatsuki S, Kawasaki M. Nucleotide-Dependent Conformational Changes and Assembly of the AAA ATPase SKD1/VPS4B. Traffic. 2008;9:2180–2189. doi: 10.1111/j.1600-0854.2008.00831.x. [DOI] [PubMed] [Google Scholar]
- Landsberg MJ, Vajjhala PR, Rothnagel R, Munn AL, Hankamer B. Three-Dimensional Structure of AAA ATPase Vps4: Advancing Structural Insights into the Mechanisms of Endosomal Sorting and Enveloped Virus Budding. Structure. 2009;17:427–437. doi: 10.1016/j.str.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger H, Weissenhorn W. Helical Structures of ESCRT-III are Disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottridge JM, Flannery AR, Vincelli JL, Stevens TH. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc Natl Acad Sci USA. 2006;103:6202–6207. doi: 10.1073/pnas.0601712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DP, West M, Henry R, Odorizzi G. Regulators of Vps4 activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol Biol Cell. 2010;21:1023–1032. doi: 10.1091/mbc.E09-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DP, West M, Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional Reconstitution of ESCRT-III Assembly and Disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Bell SD. Ancient ESCRTs and the evolution of binary fission. Trends Microbiol. 2009;17:507–513. doi: 10.1016/j.tim.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, Gaspar J, Holton JM, Viswanathan R, Ghaffarian S, Hill CP, Sundquist WI. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005a;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Gaspar J, Stuchell-Brereton MD, Alam SL, Skalicky JJ, Sundquist WI. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc Natl Acad Sci USA. 2005b;102:13813–13818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of "parallel" expression vectors. Prot Express Purific. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Shestakova A, Hanono A, Drosner S, Curtiss M, Davies BA, Katzmann DJ, Babst M. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol Biol Cell. 2010;21:1059–1071. doi: 10.1091/mbc.E09-07-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett SL, Ward DM, Huynh D, Vaughn MB, Simmons JC, Kaplan J. Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J Biol Chem. 2004;279:10982–10990. doi: 10.1074/jbc.M312669200. [DOI] [PubMed] [Google Scholar]
- Shim S, Merrill SA, Hanson PI. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol Biol Cell. 2008;19:2661–2672. doi: 10.1091/mbc.E07-12-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin AA, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- Vida TA, Emr SD. A New Vital Stain for Visualizing Vacuolar Membrane Dynamics and Endocytosis in Yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DMV, Vaughn MB, Shiflett SL, White PL, Pollock AL, Hill J, Schnegelberger R, Sundquist WI, Kaplan J. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J Biol Chem. 2005;280:10548–10555. doi: 10.1074/jbc.M413734200. [DOI] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Xia H, Zhou J, Azmi I, Davies BA, Katzmann DJ, Xu Z. Structural basis of Vta1 function in the multi-vesicular body sorting pathway. Dev Cell. 2008;14:37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao JY, Xia HC, Yoshino-Koh K, Zhou JH, Xu ZH. Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4. J Mol Biol. 2007;374:655–670. doi: 10.1016/j.jmb.2007.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo SCL, Xu LH, Ren JH, Boulton VJ, Wagle MD, Liu C, Ren G, Wong P, Zahn R, Sasajala P, et al. Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J Cell Sci. 2003;116:3957–3970. doi: 10.1242/jcs.00751. [DOI] [PubMed] [Google Scholar]
- Yu ZH, Gonciarz MD, Sundquist WI, Hill CP, Jensen GJ. Cryo-EM structure of dodecameric Vps4p and its 2: 1 complex with Vta1p. J Mol Biol. 2008;377:364–377. doi: 10.1016/j.jmb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun RH, Anderson A, Hermans J. Proline in Alpha-Helix - Stability and Conformation Studied by Dynamics Simulation. Proteins. 1991;10:219–228. doi: 10.1002/prot.340100306. [DOI] [PubMed] [Google Scholar]
- Zhang XD, Shaw A, Bates PA, Newman RH, Gowen B, Orlova E, Gorman MA, Kondo H, Dokurno P, Lally J, et al. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]